Transcription Factors in the Development and Function of Group 2 Innate Lymphoid Cells
Abstract
1. Introduction
2. Transcription Factors Involved in the Development of Group 2 Innate Lymphoid Cells (ILC2s)
2.1. GATA-3
2.2. Bcl11b
2.3. Gfi-1
2.4. RORα
2.5. Ets1
3. Runx Proteins and Immune Cells
3.1. Global Effects of Runx Proteins in Immune Cells
3.2. Runx3 Is Required for the Differentiation of ILC1s and ILC3s
4. The Function of Runx Proteins in ILC2s
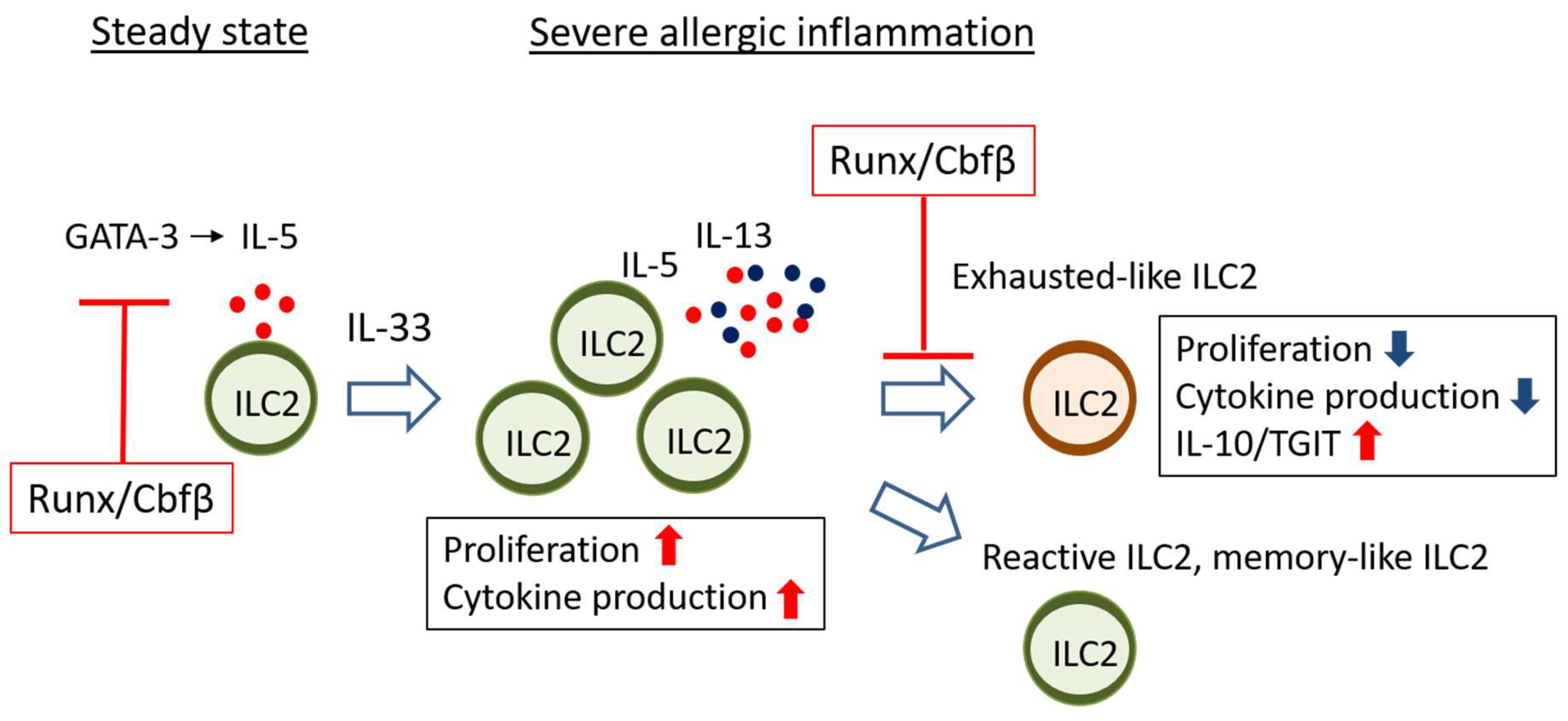
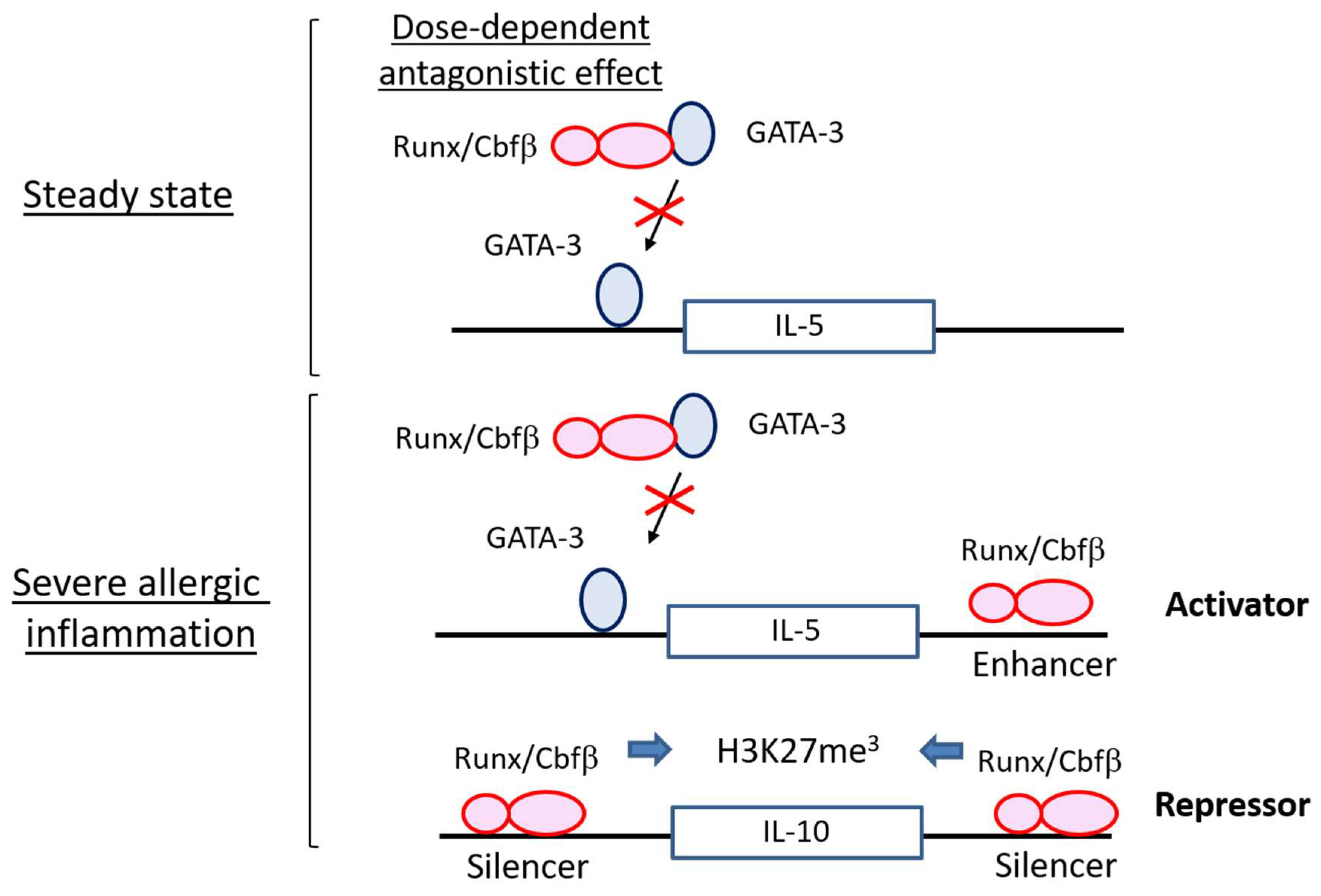
4.1. Runx Proteins Prevent Steady-State ILC2s from Overactivation
4.2. Runx Proteins Inhibit the Emergence of Exhausted-Like ILC2s during Allergic Inflammation
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| GATA-3 | GATA binding protein 3 |
| Bcl11b | B-Cell lymphoma/leukaemia 11B |
| Gfi1 | Growth factor independent 1 transcription repressor |
| RORα | RAR related orphan receptor A |
| Ets-1 | v-ets erythroblastosis virus E26 oncogene homolog 1 |
| Cbfb | Subunit b of core binding factor |
| Runx | Runt-related transcription factor |
| Tox | Thymocyte selection associated high mobility group box |
| NFIL3 | Nuclear factor, interleukin 3 regulated |
| PLZF | Promyelocytic leukemia zinc finger |
| T-bet | T-box–containing protein expressed in T cells |
| Eomes | Eomesodermin |
| RORγt | RAR-related orphan receptor gamma t |
| AhR | Aryl hydrocarbon receptor |
| TCF-1 | T cell factor 1 |
| ILC | Innate lymphoid cell |
| Sox4 | Sex determining region Y-box 4 |
| TH | Helper T cell |
| CLP | Common lymphoid progenitor |
| EILP | Early innate lymphoid progenitor |
| CHILP | Common helper innate lymphoid progenitors |
| ILCP | Innate lymphoid cell precursors |
| Lti | Lymphoid tissue inducer |
| IFNγ | Interferon gamma |
| IL | Interleukin |
| GM-CSF | Granulocyte-macrophage colony-stimulating factor |
| TSLP | Thymic stromal lymphopoietin |
| TL1A | TNF superfamily ligand TL1A |
| Vip | Vasoactive intestinal peptide |
| PD-1 | Programmed cell death 1 |
| Tigit | T cell immunoreceptor with Ig and ITIM domains |
| ICOS | Inducible costimulator |
| Lag3 | Lymphocyte activation gene-3 |
| KLRG1 | Killer cell lectin-like receptor G1 |
| GITR | Glucocorticoid-induced tumor necrosis factor receptor |
| ICOSL | Inducible costimulatory ligand |
| CCL17 | CC chemokine ligand 17 |
References
- Ebbo, M.; Crinier, A.; Vely, F.; Vivier, E. Innate lymphoid cells: major players in inflammatory diseases. Nat. Rev. Immunol. 2017, 17, 665–678. [Google Scholar] [CrossRef] [PubMed]
- Sonnenberg, G.F.; Artis, D. Innate lymphoid cells in the initiation, regulation and resolution of inflammation. Nat. Med. 2015, 21, 698–708. [Google Scholar] [CrossRef]
- Vivier, E.; Artis, D.; Colonna, M.; Diefenbach, A.; di Santo, J.P.; Eberl, G.; Koyasu, S.; Locksley, R.M.; McKenzie, A.N.J.; Mebius, R.E.; et al. Innate Lymphoid Cells: 10 Years On. Cell 2018, 174, 1054–1066. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, G.; Fan, X.; Dikiy, S.; Lee, S.Y.; Rudensky, A.Y. Tissue residency of innate lymphoid cells in lymphoid and nonlymphoid organs. Science 2015, 350, 981–985. [Google Scholar] [CrossRef] [PubMed]
- Klose, C.S.; Artis, D. Innate lymphoid cells as regulators of immunity, inflammation and tissue homeostasis. Nat. Immunol. 2016, 17, 765–774. [Google Scholar] [CrossRef] [PubMed]
- Von Moltke, J.; Ji, M.; Liang, H.E.; Locksley, R.M. Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature 2016, 529, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Neill, D.R.; Wong, S.H.; Bellosi, A.; Flynn, R.J.; Daly, M.; Langford, T.K.; Bucks, C.; Kane, C.M.; Fallon, P.G.; Pannell, R.; et al. Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature 2010, 464, 1367–1370. [Google Scholar] [CrossRef] [PubMed]
- Moro, K.; Yamada, T.; Tanabe, M.; Takeuchi, T.; Ikawa, T.; Kawamoto, H.; Furusawa, J.; Ohtani, M.; Fujii, H.; Koyasu, S. Innate production of T(H)2 cytokines by adipose tissue-associated c-Kit(+)Sca-1(+) lymphoid cells. Nature 2010, 463, 540–544. [Google Scholar] [CrossRef]
- Kim, B.S.; Artis, D. Group 2 innate lymphoid cells in health and disease. Cold Spring Harb. Perspect. Biol. 2015, 7. [Google Scholar] [CrossRef]
- Longman, R.S.; Diehl, G.E.; Victorio, D.A.; Huh, J.R.; Galan, C.; Miraldi, E.R.; Swaminath, A.; Bonneau, R.; Scherl, E.J.; Littman, D.R. CX(3)CR1(+) mononuclear phagocytes support colitis-associated innate lymphoid cell production of IL-22. J. Exp. Med. 2014, 211, 1571–1583. [Google Scholar] [CrossRef]
- Kim, B.S.; Siracusa, M.C.; Saenz, S.A.; Noti, M.; Monticelli, L.A.; Sonnenberg, G.F.; Hepworth, M.R.; van Voorhees, A.S.; Comeau, M.R.; Artis, D. TSLP elicits IL-33-independent innate lymphoid cell responses to promote skin inflammation. Sci. Transl. Med. 2013, 5. [Google Scholar] [CrossRef] [PubMed]
- Zook, E.C.; Kee, B.L. Development of innate lymphoid cells. Nat. Immunol. 2016, 17, 775–782. [Google Scholar] [CrossRef] [PubMed]
- Klose, C.S.; Flach, M.; Mohle, L.; Rogell, L.; Hoyler, T.; Ebert, K.; Fabiunke, C.; Pfeifer, D.; Sexl, V.; Fonseca-Pereira, D.; et al. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell 2014, 157, 340–356. [Google Scholar] [CrossRef] [PubMed]
- Daussy, C.; Faure, F.; Mayol, K.; Viel, S.; Gasteiger, G.; Charrier, E.; Bienvenu, J.; Henry, T.; Debien, E.; Hasan, U.A.; et al. T-bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow. J. Exp. Med. 2014, 211, 563–577. [Google Scholar] [CrossRef] [PubMed]
- Yagi, R.; Zhong, C.; Northrup, D.L.; Yu, F.; Bouladoux, N.; Spencer, S.; Hu, G.; Barron, L.; Sharma, S.; Nakayama, T.; et al. The transcription factor GATA3 is critical for the development of all IL-7Ralpha-expressing innate lymphoid cells. Immunity 2014, 40, 378–388. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Cella, M.; McDonald, K.G.; Garlanda, C.; Kennedy, G.D.; Nukaya, M.; Mantovani, A.; Kopan, R.; Bradfield, C.A.; Newberry, R.D.; et al. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat. Immunol. 2012, 13, 144–151. [Google Scholar] [CrossRef]
- Kiss, E.A.; Diefenbach, A. Role of the Aryl Hydrocarbon Receptor in Controlling Maintenance and Functional Programs of RORgammat(+) Innate Lymphoid Cells and Intraepithelial Lymphocytes. Front. Immunol. 2012, 3, 124. [Google Scholar] [CrossRef]
- Sawa, S.; Cherrier, M.; Lochner, M.; Satoh-Takayama, N.; Fehling, H.J.; Langa, F.; di Santo, J.P.; Eberl, G. Lineage relationship analysis of RORgammat+ innate lymphoid cells. Science 2010, 330, 665–669. [Google Scholar] [CrossRef]
- O’Sullivan, T.E.; Rapp, M.; Fan, X.; Weizman, O.E.; Bhardwaj, P.; Adams, N.M.; Walzer, T.; Dannenberg, A.J.; Sun, J.C. Adipose-Resident Group 1 Innate Lymphoid Cells Promote Obesity-Associated Insulin Resistance. Immunity 2016, 45, 428–441. [Google Scholar] [CrossRef]
- Sojka, D.K.; Plougastel-Douglas, B.; Yang, L.; Pak-Wittel, M.A.; Artyomov, M.N.; Ivanova, Y.; Zhong, C.; Chase, J.M.; Rothman, P.B.; Yu, J.; et al. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. eLife 2014, 3, e01659. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, A.; Vermi, W.; Lee, J.S.; Lonardi, S.; Gilfillan, S.; Newberry, R.D.; Cella, M.; Colonna, M. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-gamma-producing cells. Immunity 2013, 38, 769–781. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.W.; Odegaard, J.I.; Mukundan, L.; Qiu, Y.; Molofsky, A.B.; Nussbaum, J.C.; Yun, K.; Locksley, R.M.; Chawla, A. Activated type 2 innate lymphoid cells regulate beige fat biogenesis. Cell 2015, 160, 74–87. [Google Scholar] [CrossRef] [PubMed]
- Pantelyushin, S.; Haak, S.; Ingold, B.; Kulig, P.; Heppner, F.L.; Navarini, A.A.; Becher, B. Rorgammat+ innate lymphocytes and gammadelta T cells initiate psoriasiform plaque formation in mice. J. Clin. Investig. 2012, 122, 2252–2256. [Google Scholar] [CrossRef] [PubMed]
- Constantinides, M.G.; McDonald, B.D.; Verhoef, P.A.; Bendelac, A. A committed precursor to innate lymphoid cells. Nature 2014, 508, 397–401. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Li, F.; Harly, C.; Xing, S.; Ye, L.; Xia, X.; Wang, H.; Wang, X.; Yu, S.; Zhou, X.; et al. TCF-1 upregulation identifies early innate lymphoid progenitors in the bone marrow. Nat. Immunol. 2015, 16, 1044–1050. [Google Scholar] [CrossRef] [PubMed]
- Seehus, C.R.; Kadavallore, A.; Torre, B.; Yeckes, A.R.; Wang, Y.; Tang, J.; Kaye, J. Alternative activation generates IL-10 producing type 2 innate lymphoid cells. Nat. Commun. 2017, 8, 1900. [Google Scholar] [CrossRef]
- Yu, X.; Wang, Y.; Deng, M.; Li, Y.; Ruhn, K.A.; Zhang, C.C.; Hooper, L.V. The basic leucine zipper transcription factor NFIL3 directs the development of a common innate lymphoid cell precursor. eLife 2014, 3. [Google Scholar] [CrossRef] [PubMed]
- Monticelli, L.A.; Sonnenberg, G.F.; Abt, M.C.; Alenghat, T.; Ziegler, C.G.; Doering, T.A.; Angelosanto, J.M.; Laidlaw, B.J.; Yang, C.Y.; Sathaliyawala, T.; et al. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat. Immunol. 2011, 12, 1045–1054. [Google Scholar] [CrossRef]
- Chang, Y.J.; Kim, H.Y.; Albacker, L.A.; Baumgarth, N.; McKenzie, A.N.; Smith, D.E.; Dekruyff, R.H.; Umetsu, D.T. Innate lymphoid cells mediate influenza-induced airway hyper-reactivity independently of adaptive immunity. Nat. Immunol. 2011, 12, 631–638. [Google Scholar] [CrossRef]
- Nussbaum, J.C.; van Dyken, S.J.; von Moltke, J.; Cheng, L.E.; Mohapatra, A.; Molofsky, A.B.; Thornton, E.E.; Krummel, M.F.; Chawla, A.; Liang, H.E.; et al. Type 2 innate lymphoid cells control eosinophil homeostasis. Nature 2013, 502, 245–248. [Google Scholar] [CrossRef]
- Halim, T.Y. Group 2 innate lymphoid cells in disease. Int. Immunol. 2016, 28, 13–22. [Google Scholar] [CrossRef]
- Wilhelm, C.; Hirota, K.; Stieglitz, B.; van Snick, J.; Tolaini, M.; Lahl, K.; Sparwasser, T.; Helmby, H.; Stockinger, B. An IL-9 fate reporter demonstrates the induction of an innate IL-9 response in lung inflammation. Nat. Immunol. 2011, 12, 1071–1077. [Google Scholar] [CrossRef]
- Halim, T.Y.; Hwang, Y.Y.; Scanlon, S.T.; Zaghouani, H.; Garbi, N.; Fallon, P.G.; McKenzie, A.N. Group 2 innate lymphoid cells license dendritic cells to potentiate memory TH2 cell responses. Nat. Immunol. 2016, 17, 57–64. [Google Scholar] [CrossRef]
- Silver, J.S.; Kearley, J.; Copenhaver, A.M.; Sanden, C.; Mori, M.; Yu, L.; Pritchard, G.H.; Berlin, A.A.; Hunter, C.A.; Bowler, R.; et al. Inflammatory triggers associated with exacerbations of COPD orchestrate plasticity of group 2 innate lymphoid cells in the lungs. Nat. Immunol. 2016, 17, 626–635. [Google Scholar] [CrossRef]
- Ohne, Y.; Silver, J.S.; Thompson-Snipes, L.; Collet, M.A.; Blanck, J.P.; Cantarel, B.L.; Copenhaver, A.M.; Humbles, A.A.; Liu, Y.J. IL-1 is a critical regulator of group 2 innate lymphoid cell function and plasticity. Nat. Immunol. 2016, 17, 646–655. [Google Scholar] [CrossRef]
- Martinez-Gonzalez, I.; Matha, L.; Steer, C.A.; Ghaedi, M.; Poon, G.F.; Takei, F. Allergen-Experienced Group 2 Innate Lymphoid Cells Acquire Memory-like Properties and Enhance Allergic Lung Inflammation. Immunity 2016, 45, 198–208. [Google Scholar] [CrossRef]
- Huang, Y.; Guo, L.; Qiu, J.; Chen, X.; Hu-Li, J.; Siebenlist, U.; Williamson, P.R.; Urban, J.F., Jr.; Paul, W.E. IL-25-responsive, lineage-negative KLRG1(hi) cells are multipotential ‘inflammatory’ type 2 innate lymphoid cells. Nat. Immunol. 2015, 16, 161–169. [Google Scholar] [CrossRef]
- Huang, Y.; Mao, K.; Chen, X.; Sun, M.A.; Kawabe, T.; Li, W.; Usher, N.; Zhu, J.; Urban, J.F., Jr.; Paul, W.E.; et al. S1P-dependent interorgan trafficking of group 2 innate lymphoid cells supports host defense. Science 2018, 359, 114–119. [Google Scholar] [CrossRef]
- Klein Wolterink, R.G.; Serafini, N.; van Nimwegen, M.; Vosshenrich, C.A.; de Bruijn, M.J.; Fonseca Pereira, D.; Veiga Fernandes, H.; Hendriks, R.W.; di Santo, J.P. Essential, dose-dependent role for the transcription factor Gata3 in the development of IL-5+ and IL-13+ type 2 innate lymphoid cells. Proc. Natl. Acad. Sci. USA 2013, 110, 10240–10245. [Google Scholar] [CrossRef]
- Hoyler, T.; Klose, C.S.; Souabni, A.; Turqueti-Neves, A.; Pfeifer, D.; Rawlins, E.L.; Voehringer, D.; Busslinger, M.; Diefenbach, A. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity 2012, 37, 634–648. [Google Scholar] [CrossRef]
- Califano, D.; Cho, J.J.; Uddin, M.N.; Lorentsen, K.J.; Yang, Q.; Bhandoola, A.; Li, H.; Avram, D. Transcription Factor Bcl11b Controls Identity and Function of Mature Type 2 Innate Lymphoid Cells. Immunity 2015, 43, 354–368. [Google Scholar] [CrossRef]
- Walker, J.A.; Oliphant, C.J.; Englezakis, A.; Yu, Y.; Clare, S.; Rodewald, H.R.; Belz, G.; Liu, P.; Fallon, P.G.; McKenzie, A.N. Bcl11b is essential for group 2 innate lymphoid cell development. J. Exp. Med. 2015, 212, 875–882. [Google Scholar] [CrossRef]
- Yu, Y.; Wang, C.; Clare, S.; Wang, J.; Lee, S.C.; Brandt, C.; Burke, S.; Lu, L.; He, D.; Jenkins, N.A.; et al. The transcription factor Bcl11b is specifically expressed in group 2 innate lymphoid cells and is essential for their development. J. Exp. Med. 2015, 212, 865–874. [Google Scholar] [CrossRef]
- Spooner, C.J.; Lesch, J.; Yan, D.; Khan, A.A.; Abbas, A.; Ramirez-Carrozzi, V.; Zhou, M.; Soriano, R.; Eastham-Anderson, J.; Diehl, L.; et al. Specification of type 2 innate lymphocytes by the transcriptional determinant Gfi1. Nat. Immunol. 2013, 14, 1229–1236. [Google Scholar] [CrossRef]
- Halim, T.Y.; MacLaren, A.; Romanish, M.T.; Gold, M.J.; McNagny, K.M.; Takei, F. Retinoic-acid-receptor-related orphan nuclear receptor alpha is required for natural helper cell development and allergic inflammation. Immunity 2012, 37, 463–474. [Google Scholar] [CrossRef]
- Zook, E.C.; Ramirez, K.; Guo, X.; van der Voort, G.; Sigvardsson, M.; Svensson, E.C.; Fu, Y.X.; Kee, B.L. The ETS1 transcription factor is required for the development and cytokine-induced expansion of ILC2. J. Exp. Med. 2016, 213, 687–696. [Google Scholar] [CrossRef]
- Robinette, M.L.; Fuchs, A.; Cortez, V.S.; Lee, J.S.; Wang, Y.; Durum, S.K.; Gilfillan, S.; Colonna, M.; Immunological Genome, C. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat. Immunol. 2015, 16, 306–317. [Google Scholar] [CrossRef]
- Malhotra, N.; Leyva-Castillo, J.M.; Jadhav, U.; Barreiro, O.; Kam, C.; O’Neill, N.K.; Meylan, F.; Chambon, P.; von Andrian, U.H.; Siegel, R.M.; et al. RORalpha-expressing T regulatory cells restrain allergic skin inflammation. Sci. Immunol. 2018, 3. [Google Scholar] [CrossRef]
- Lo, B.C.; Gold, M.J.; Hughes, M.R.; Antignano, F.; Valdez, Y.; Zaph, C.; Harder, K.W.; McNagny, K.M. The orphan nuclear receptor ROR alpha and group 3 innate lymphoid cells drive fibrosis in a mouse model of Crohn’s disease. Sci. Immunol. 2016, 1. [Google Scholar] [CrossRef]
- Voon, D.C.; Hor, Y.T.; Ito, Y. The RUNX complex: reaching beyond haematopoiesis into immunity. Immunology 2015, 146, 523–536. [Google Scholar] [CrossRef]
- Collins, A.; Littman, D.R.; Taniuchi, I. RUNX proteins in transcription factor networks that regulate T-cell lineage choice. Nat. Rev. Immunol. 2009, 9, 106–115. [Google Scholar] [CrossRef]
- Bone, K.R.; Gruper, Y.; Goldenberg, D.; Levanon, D.; Groner, Y. Translation regulation of Runx3. Blood Cells Mol. Dis. 2010, 45, 112–116. [Google Scholar] [CrossRef]
- Okuda, T.; van Deursen, J.; Hiebert, S.W.; Grosveld, G.; Downing, J.R. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 1996, 84, 321–330. [Google Scholar] [CrossRef]
- Wang, Q.; Stacy, T.; Miller, J.D.; Lewis, A.F.; Gu, T.L.; Huang, X.; Bushweller, J.H.; Bories, J.C.; Alt, F.W.; Ryan, G.; et al. The CBFbeta subunit is essential for CBFalpha2 (AML1) function in vivo. Cell 1996, 87, 697–708. [Google Scholar] [CrossRef]
- Taniuchi, I.; Osato, M.; Egawa, T.; Sunshine, M.J.; Bae, S.C.; Komori, T.; Ito, Y.; Littman, D.R. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell 2002, 111, 621–633. [Google Scholar] [CrossRef]
- Egawa, T.; Tillman, R.E.; Naoe, Y.; Taniuchi, I.; Littman, D.R. The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J. Exp. Med. 2007, 204, 1945–1957. [Google Scholar] [CrossRef]
- Kitoh, A.; Ono, M.; Naoe, Y.; Ohkura, N.; Yamaguchi, T.; Yaguchi, H.; Kitabayashi, I.; Tsukada, T.; Nomura, T.; Miyachi, Y.; et al. Indispensable role of the Runx1-Cbfbeta transcription complex for in vivo-suppressive function of FoxP3+ regulatory T cells. Immunity 2009, 31, 609–620. [Google Scholar] [CrossRef]
- Zhang, F.; Meng, G.; Strober, W. Interactions among the transcription factors Runx1, RORgammat and Foxp3 regulate the differentiation of interleukin 17-producing T cells. Nat. Immunol. 2008, 9, 1297–1306. [Google Scholar] [CrossRef]
- Naoe, Y.; Setoguchi, R.; Akiyama, K.; Muroi, S.; Kuroda, M.; Hatam, F.; Littman, D.R.; Taniuchi, I. Repression of interleukin-4 in T helper type 1 cells by Runx/Cbf beta binding to the Il4 silencer. J. Exp. Med. 2007, 204, 1749–1755. [Google Scholar] [CrossRef]
- Kim, B.; Sasaki, Y.; Egawa, T. Restriction of Nonpermissive RUNX3 Protein Expression in T Lymphocytes by the Kozak Sequence. J. Immunol. 2015, 195, 1517–1523. [Google Scholar] [CrossRef]
- Komori, T.; Yagi, H.; Nomura, S.; Yamaguchi, A.; Sasaki, K.; Deguchi, K.; Shimizu, Y.; Bronson, R.T.; Gao, Y.H.; Inada, M.; et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 1997, 89, 755–764. [Google Scholar] [CrossRef]
- Otto, F.; Thornell, A.P.; Crompton, T.; Denzel, A.; Gilmour, K.C.; Rosewell, I.R.; Stamp, G.W.; Beddington, R.S.; Mundlos, S.; Olsen, B.R.; et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 1997, 89, 765–771. [Google Scholar] [CrossRef]
- Sawai, C.M.; Sisirak, V.; Ghosh, H.S.; Hou, E.Z.; Ceribelli, M.; Staudt, L.M.; Reizis, B. Transcription factor Runx2 controls the development and migration of plasmacytoid dendritic cells. J. Exp. Med. 2013, 210, 2151–2159. [Google Scholar] [CrossRef]
- Ebihara, T.; Song, C.; Ryu, S.H.; Plougastel-Douglas, B.; Yang, L.; Levanon, D.; Groner, Y.; Bern, M.D.; Stappenbeck, T.S.; Colonna, M.; et al. Runx3 specifies lineage commitment of innate lymphoid cells. Nat. Immunol. 2015, 16, 1124–1133. [Google Scholar] [CrossRef]
- Levanon, D.; Negreanu, V.; Lotem, J.; Bone, K.R.; Brenner, O.; Leshkowitz, D.; Groner, Y. Transcription factor Runx3 regulates interleukin-15-dependent natural killer cell activation. Mol. Cell. Biol. 2014, 34, 1158–1169. [Google Scholar] [CrossRef]
- Tenno, M.; Kojo, S.; Lawir, D.F.; Hess, I.; Shiroguchi, K.; Ebihara, T.; Endo, T.A.; Muroi, S.; Satoh, R.; Kawamoto, H.; et al. Cbfbeta2 controls differentiation of and confers homing capacity to prethymic progenitors. J. Exp. Med. 2018, 215, 595–610. [Google Scholar] [CrossRef]
- Tachibana, M.; Tenno, M.; Tezuka, C.; Sugiyama, M.; Yoshida, H.; Taniuchi, I. Runx1/Cbfbeta2 complexes are required for lymphoid tissue inducer cell differentiation at two developmental stages. J. Immunol. 2011, 186, 1450–1457. [Google Scholar] [CrossRef]
- Mao, A.P.; Ishizuka, I.E.; Kasal, D.N.; Mandal, M.; Bendelac, A. A shared Runx1-bound Zbtb16 enhancer directs innate and innate-like lymphoid lineage development. Nat. Commun. 2017, 8, 863. [Google Scholar] [CrossRef]
- Miyamoto, C.; Kojo, S.; Yamashita, M.; Moro, K.; Lacaud, G.; Shiroguchi, K.; Taniuchi, I.; Ebihara, T. Runx/Cbfbeta complexes protect group 2 innate lymphoid cells from exhausted-like hyporesponsiveness during allergic airway inflammation. Nat. Commun. 2019, 10, 447. [Google Scholar] [CrossRef]
- Yagi, R.; Junttila, I.S.; Wei, G.; Urban, J.F., Jr.; Zhao, K.; Paul, W.E.; Zhu, J. The transcription factor GATA3 actively represses RUNX3 protein-regulated production of interferon-gamma. Immunity 2010, 32, 507–517. [Google Scholar] [CrossRef]
- Wallrapp, A.; Riesenfeld, S.J.; Burkett, P.R.; Abdulnour, R.E.; Nyman, J.; Dionne, D.; Hofree, M.; Cuoco, M.S.; Rodman, C.; Farouq, D.; et al. The neuropeptide NMU amplifies ILC2-driven allergic lung inflammation. Nature 2017, 549, 351–356. [Google Scholar] [CrossRef]
- Cardoso, V.; Chesne, J.; Ribeiro, H.; Garcia-Cassani, B.; Carvalho, T.; Bouchery, T.; Shah, K.; Barbosa-Morais, N.L.; Harris, N.; Veiga-Fernandes, H. Neuronal regulation of type 2 innate lymphoid cells via neuromedin U. Nature 2017, 549, 277–281. [Google Scholar] [CrossRef]
- Klose, C.S.N.; Mahlakoiv, T.; Moeller, J.B.; Rankin, L.C.; Flamar, A.L.; Kabata, H.; Monticelli, L.A.; Moriyama, S.; Putzel, G.G.; Rakhilin, N.; et al. The neuropeptide neuromedin U stimulates innate lymphoid cells and type 2 inflammation. Nature 2017, 549, 282–286. [Google Scholar] [CrossRef]
- Maazi, H.; Patel, N.; Sankaranarayanan, I.; Suzuki, Y.; Rigas, D.; Soroosh, P.; Freeman, G.J.; Sharpe, A.H.; Akbari, O. ICOS:ICOS-ligand interaction is required for type 2 innate lymphoid cell function, homeostasis, and induction of airway hyperreactivity. Immunity 2015, 42, 538–551. [Google Scholar] [CrossRef]
- Wherry, E.J.; Kurachi, M. Molecular and cellular insights into T cell exhaustion. Nat. Rev. Immunol. 2015, 15, 486–499. [Google Scholar] [CrossRef]
- Crespo, J.; Sun, H.; Welling, T.H.; Tian, Z.; Zou, W. T cell anergy, exhaustion, senescence, and stemness in the tumor microenvironment. Curr. Opin. Immunol. 2013, 25, 214–221. [Google Scholar] [CrossRef]
- Yi, J.S.; Cox, M.A.; Zajac, A.J. T-cell exhaustion: characteristics, causes and conversion. Immunology 2010, 129, 474–481. [Google Scholar] [CrossRef]
- Taylor, S.; Huang, Y.; Mallett, G.; Stathopoulou, C.; Felizardo, T.C.; Sun, M.A.; Martin, E.L.; Zhu, N.; Woodward, E.L.; Elias, M.S.; et al. PD-1 regulates KLRG1(+) group 2 innate lymphoid cells. J. Exp. Med. 2017, 214, 1663–1678. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ebihara, T.; Taniuchi, I. Transcription Factors in the Development and Function of Group 2 Innate Lymphoid Cells. Int. J. Mol. Sci. 2019, 20, 1377. https://doi.org/10.3390/ijms20061377
Ebihara T, Taniuchi I. Transcription Factors in the Development and Function of Group 2 Innate Lymphoid Cells. International Journal of Molecular Sciences. 2019; 20(6):1377. https://doi.org/10.3390/ijms20061377
Chicago/Turabian StyleEbihara, Takashi, and Ichiro Taniuchi. 2019. "Transcription Factors in the Development and Function of Group 2 Innate Lymphoid Cells" International Journal of Molecular Sciences 20, no. 6: 1377. https://doi.org/10.3390/ijms20061377
APA StyleEbihara, T., & Taniuchi, I. (2019). Transcription Factors in the Development and Function of Group 2 Innate Lymphoid Cells. International Journal of Molecular Sciences, 20(6), 1377. https://doi.org/10.3390/ijms20061377




