Genomic Analysis of Shewanella sp. O23S—The Natural Host of the pSheB Plasmid Carrying Genes for Arsenic Resistance and Dissimilatory Reduction
Abstract
1. Introduction
2. Results and Discussion
2.1. General Features of the Shewanella sp. O23S Genome
2.2. Mobilome of O23S
2.2.1. Plasmid pSheA
2.2.2. Plasmid pSheB
2.2.3. Prophages and Plasmid-Like Phages
2.2.4. Other Mobile Genetic Elements
2.3. Other Heavy Metal and Antibiotic Resistance Genes
2.4. Antibiotic Susceptibility and Putative Antimicrobial Drugs Resistance Mechanisms of O23S
2.5. Adherence and Biofilm Formation Analysis
2.6. Metabolic Preferences of O23S
3. Materials and Methods
3.1. Strains, Vectors, Media, and Growth Conditions
3.2. DNA Manipulations
3.2.1. DNA Isolation
3.2.2. Genome Sequencing, Assembly and Analysis
3.2.3. Phage Induction, Annotation and Analysis
3.3. Physiological Analyses
3.3.1. Resistance to Phosphate, Glycerol, and Heavy Metal Ions
3.3.2. Antibiotic Susceptibility Tests
3.3.3. Metabolic Substrate Preferences (Biolog™) Test
3.4. Functionality of Arsenic Resistance and Arsenic Respiration Modules
3.5. Microplate Adherence Assay
3.6. Phylogenetic Analysis
3.7. Nucleotide Sequence Accession Numbers
4. Conclusions
5. Patents
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| DARB | Dissimilatory arsenate reducing bacterium/bacteria |
| HGT | Horizontal Gene Transfer |
| hmr | Heavy metal resistance |
| MIC | Minimal Inhibitory Concentration |
| ORF | Open Reading Frame |
References
- Fashola, M.O.; Ngole-Jeme, V.M.; Babalola, O.O. Heavy metal pollution from gold mines: Environmental effects and bacterial strategies for resistance. Int. J. Environ. Res. Public Health 2016, 13. [Google Scholar] [CrossRef] [PubMed]
- Drewniak, L.; Styczek, A.; Majder-Lopatka, M.; Sklodowska, A. Bacteria, hypertolerant to arsenic in the rocks of an ancient gold mine, and their potential role in dissemination of arsenic pollution. Environ. Pollut. 2008, 156, 1069–1074. [Google Scholar] [CrossRef] [PubMed]
- diCenzo, G.C.; Debiec, K.; Krzysztoforski, J.; Uhrynowski, W.; Mengoni, A.; Fagorzi, C.; Gorecki, A.; Dziewit, L.; Bajda, T.; Rzepa, G.; et al. Genomic and biotechnological characterization of the heavy-metal resistant, arsenic-oxidizing bacterium Ensifer sp. M14. Genes 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Uhrynowski, W.; Debiec, K.; Sklodowska, A.; Drewniak, L. The role of dissimilatory arsenate reducing bacteria in the biogeochemical cycle of arsenic based on the physiological and functional analysis of Aeromonas sp. O23A. Sci. Total Environ. 2017, 598, 680–689. [Google Scholar] [CrossRef] [PubMed]
- Gnanaprakasam, E.T.; Lloyd, J.R.; Boothman, C.; Ahmed, K.M.; Choudhury, I.; Bostick, B.C.; van Geen, A.; Mailloux, B.J. Microbial community structure and arsenic biogeochemistry in two arsenic-impacted aquifers in Bangladesh. MBio 2017, 8, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Héry, M.; Gault, A.G.; Rowland, H.A.L.; Lear, G.; Polya, D.A.; Lloyd, J.R. Molecular and cultivation-dependent analysis of metal-reducing bacteria implicated in arsenic mobilisation in south-east asian aquifers. Appl. Geochem. 2008, 23, 3215–3223. [Google Scholar] [CrossRef]
- Drewniak, L.; Rajpert, L.; Mantur, A.; Sklodowska, A. Dissolution of arsenic minerals mediated by dissimilatory arsenate reducing bacteria: Estimation of the physiological potential for arsenic mobilization. BioMed Res. Int. 2014, 2014. [Google Scholar] [CrossRef]
- Das, S.; Liu, C.C.; Jean, J.S.; Liu, T. Dissimilatory Arsenate Reduction and In Situ Microbial Activities and Diversity in Arsenic-rich Groundwater of Chianan Plain, Southwestern Taiwan. Microb. Ecol. 2016, 71, 365–374. [Google Scholar] [CrossRef] [PubMed]
- Maizel, D.; Balverdi, P.; Rosen, B.; Sales, A.M.; Ferrero, M.A. Arsenic-hypertolerant and arsenic-reducing bacteria isolated from wells in Tucuman, Argentina. Can. J. Microbiol. 2018, 64, 876–886. [Google Scholar] [CrossRef] [PubMed]
- Drewniak, L.; Stasiuk, R.; Uhrynowski, W.; Sklodowska, A. Shewanella sp. O23S as a driving agent of a system utilizing dissimilatory arsenate-reducing bacteria responsible for self-cleaning of water contaminated with arsenic. Int. J. Mol. Sci. 2015, 16, 14409. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Shi, Q.; Jing, C. Arsenic biotransformation in solid waste residue: Comparison of contributions from bacteria with arsenate and iron reducing pathways. Environ. Sci. Technol. 2015, 49, 2140–2146. [Google Scholar] [CrossRef] [PubMed]
- Kudo, K.; Yamaguchi, N.; Makino, T.; Ohtsuka, T.; Kimura, K.; Dong, D.T.; Amachi, S. Release of arsenic from soil by a novel dissimilatory arsenate-reducing bacterium, Anaeromyxobacter sp. strain PSR-1. Appl. Environ. Microbiol. 2013, 79, 4635–4642. [Google Scholar] [CrossRef] [PubMed]
- Rajpert, L.; Schäffer, A.; Lenz, M. Redox-stat bioreactors for elucidating mobilisation mechanisms of trace elements: An example of As-contaminated mining soils. Appl. Microbiol. Biotechnol. 2018, 102, 7635–7641. [Google Scholar] [CrossRef] [PubMed]
- Uhrynowski, W.; Decewicz, P.; Dziewit, L.; Radlinska, M.; Krawczyk, P.S.; Lipinski, L.; Adamska, D.; Drewniak, L. Analysis of the genome and mobilome of a dissimilatory arsenate reducing Aeromonas sp. O23A reveals multiple mechanisms for heavy metal resistance and metabolism. Front. Microbiol. 2017, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Hou, Q.; Wang, Y.; Li, J.; Li, W.; Kwok, L.Y.; Sun, Z.; Zhang, H.; Zhong, Z. Comparative genomic analysis of Enterococcus faecalis: Insights into their environmental adaptations. BMC Genom. 2018, 19, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Carlin, A.; Shi, W.; Dey, S.; Rosen, B.P. The ars Operon of Escherichia coli Confers Arsenical and Antimonial Resistance. J. Bacteriol. 1995, 177, 981–986. [Google Scholar] [CrossRef] [PubMed]
- Freel, K.C.; Krueger, M.C.; Farasin, J.; Brochier-Armanet, C.; Barbe, V.; Andres, J.; Cholley, P.-E.; Dillies, M.-A.; Jagla, B.; Koechler, S.; et al. Adaptation in Toxic Environments: Arsenic Genomic Islands in the Bacterial Genus Thiomonas. PLoS ONE 2015, 10, e0139011. [Google Scholar] [CrossRef] [PubMed]
- Bruhn, D.F.; Li, J.; Silver, S.; Roberto, F.; Rosen, B.P. The arsenical resistance operon of IncN plasmid R46. FEMS Microbiol. Lett. 1996, 139, 149–153. [Google Scholar] [CrossRef] [PubMed]
- Saltikov, C.W.; Newman, D.K. Genetic identification of a respiratory arsenate reductase. Proc. Natl. Acad. Sci. USA 2003, 100, 10983–10988. [Google Scholar] [CrossRef] [PubMed]
- Silver, S.; Phung, L.T. Genes and enzymes involved in bacterial oxidation and reduction of inorganic arsenic. Appl. Environ. Microbiol. 2005, 71, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Glasser, N.R.; Oyala, P.H.; Osborne, T.H.; Santini, J.M.; Newman, D.K. Structural and mechanistic analysis of the arsenate respiratory reductase provides insight into environmental arsenic transformations. Proc. Natl. Acad. Sci. USA 2018, 115, E8614–E8623. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.L.; Wang, N.; Wang, H.M.; Deng, Y.M.; Ma, T.; Wu, M.X.J.; Zhang, Y.N. Molecular Characterization of the Total Bacteria and Dissimilatory Arsenate-Reducing Bacteria in Core Sediments of the Jianghan Plain, Central China. Geomicrobiol. J. 2017, 34, 467–479. [Google Scholar] [CrossRef]
- Suhadolnik, M.L.S.; Salgado, A.P.C.; Scholte, L.L.S.; Bleicher, L.; Costa, P.S.; Reis, M.P.; Dias, M.F.; Ávila, M.P.; Barbosa, F.A.R.; Chartone-Souza, E.; et al. Novel arsenic-transforming bacteria and the diversity of their arsenic-related genes and enzymes arising from arsenic-polluted freshwater sediment. Sci. Rep. 2017, 7, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Muller, D.; Médigue, C.; Koechler, S.; Barbe, V.; Barakat, M.; Talla, E.; Bonnefoy, V.; Krin, E.; Arsène-Ploetze, F.; Carapito, C.; et al. A tale of two oxidation states: Bacterial colonization of arsenic-rich environments. PLoS Genet. 2007, 3, 0518–0530. [Google Scholar] [CrossRef] [PubMed]
- Copeland, A.; Lucas, S.; Lapidus, A.; Barry, K.; Glavina del Rio, T.; Dalin, E.; Tice, H.; Pitluck, S.; Chain, P.; Malfatti, S.; et al. Complete Sequence of Pyrobaculum arsenaticum DSM 13514; US DOE Joint Genome Institute: Walnut Creek, CA, USA, 2007; p. CP000660.1. [Google Scholar]
- Wattam, A.R.; Davis, J.J.; Assaf, R.; Boisvert, S.; Brettin, T.; Bun, C.; Conrad, N.; Dietrich, E.M.; Disz, T.; Gabbard, J.L.; et al. Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. 2017, 45, D535–D542. [Google Scholar] [CrossRef] [PubMed]
- Caro-Quintero, A.; Auchtung, J.; Deng, J.; Brettar, I.; Höfle, M.; Tiedje, J.M.; Konstantinidis, K.T. Genome sequencing of five Shewanella baltica strains recovered from the oxic-anoxic interface of the baltic sea. J. Bacteriol. 2012, 194, 1236. [Google Scholar] [CrossRef] [PubMed]
- Caro-Quintero, A.; Deng, J.; Auchtung, J.; Brettar, I.; Ho, M.G.; Hofle, M.G.; Klappenbach, J.; Konstantinidis, K.T. Unprecedented levels of horizontal gene transfer among spatially co-occurring Shewanella bacteria from the Baltic Sea. ISME J. 2011, 5, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Belogurov, A.A.; Delver, E.P.; Rodzevich, O. V Plasmid pKM101 encodes two nonhomologous antirestriction proteins (ArdA and ArdB) whose expression is controlled by homologous regulatory sequences. J. Bacteriol. 1993, 175, 4843–4850. [Google Scholar] [CrossRef] [PubMed]
- Korotkov, K.V.; Sandkvist, M.; Hol, W.G. The type II secretion system: Biogenesis, molecular architecture and mechanism. Nat. Rev. Microbiol. 2012, 10, 336–351. [Google Scholar] [CrossRef] [PubMed]
- Sandkvist, M. Type II Secretion and Pathogenesis MINIREVIEW Type II Secretion and Pathogenesis. Infect. Immun. 2001, 69, 3523–3535. [Google Scholar] [CrossRef] [PubMed]
- Hirano, N.; Muroi, T.; Takahashi, H.; Haruki, M. Site-specific recombinases as tools for heterologous gene integration. Appl. Microbiol. Biotechnol. 2011, 92, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Castillo, F.; Benmohamed, A.; Szatmari, G. Xer site specific recombination: Double and single recombinase systems. Front. Microbiol. 2017, 8, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Bartosik, D.; Szymanik, M.; Wysocka, E. Identification of the partitioning site within the repABC-type replicon of the composite Paracoccus versutus plasmid pTAV1. J. Bacteriol. 2001, 183, 6234–6243. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Drewniak, L.; Dziewit, L.; Ciezkowska, M.; Gawor, J.; Gromadka, R.; Sklodowska, A. Structural and functional genomics of plasmid pSinA of Sinorhizobium sp. M14 encoding genes for the arsenite oxidation and arsenic resistance. J. Biotechnol. 2013, 164, 479–488. [Google Scholar] [CrossRef] [PubMed]
- Gödeke, J.; Paul, K.; Lassak, J.; Thormann, K.M. Phage-induced lysis enhances biofilm formation in Shewanella oneidensis MR-1. ISME J. 2011, 5, 613–626. [Google Scholar] [CrossRef] [PubMed]
- Harshey, R.M. Transposable Phage Mu. Microbiol. Spectr. 2014, 2. [Google Scholar] [CrossRef]
- Canchaya, C.; Proux, C.; Fournous, G.; Bruttin, A.; Brussow, H. Prophage Genomics. Microbiol. Mol. Biol. Rev. 2003, 67, 238–276. [Google Scholar] [CrossRef] [PubMed]
- Mai-Prochnow, A.; Hui, J.G.K.; Kjelleberg, S.; Rakonjac, J.; McDougald, D.; Rice, S.A. Big things in small packages: The genetics of filamentous phage and effects on fitness of their host. FEMS Microbiol. Rev. 2015, 39, 465–487. [Google Scholar] [CrossRef] [PubMed]
- Matsushita, O.; Okabe, A. Clostridial hydrolytic enzymes degrading extracellular components. Toxicon 2001, 39, 1769–1780. [Google Scholar] [CrossRef]
- Drewniak, L.; Krawczyk, P.S.; Mielnicki, S.; Adamska, D.; Sobczak, A.; Lipinski, L.; Burec-Drewniak, W.; Sklodowska, A. Physiological and metagenomic analyses of microbial mats involved in self-purification of mine waters contaminated with heavy metals. Front. Microbiol. 2016, 7, 1252. [Google Scholar] [CrossRef] [PubMed]
- Drewniak, L.; Matlakowska, R.; Rewerski, B.; Sklodowska, A. Arsenic release from gold mine rocks mediated by the activity of indigenous bacteria. Hydrometallurgy 2010, 104, 437–442. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001; ISBN 9780879695774. [Google Scholar]
- Tuovinen, O.H.; Kelly, D.P. Studies on the Growth of Thiobacillus ferrooxidans. Arch. Mikrobiol. 1973, 88, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Carver, T.; Berriman, M.; Tivey, A.; Patel, C.; Böhme, U.; Barrell, B.G.; Parkhill, J.; Rajandream, M.A. Artemis and ACT: Viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 2008, 24, 2672–2676. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.J.; Vincze, T.; Posfai, J.; Macelis, D. REBASE-a database for DNA restriction and modification: Enzymes, genes and genomes. Nucleic Acids Res. 2015, 43, D298–D299. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liang, Y.; Lynch, K.H.; Dennis, J.J.; Wishart, D.S. PHAST: A Fast Phage Search Tool. Nucleic Acids Res. 2011, 39, 347–352. [Google Scholar] [CrossRef] [PubMed]
- Lopes, A.; Tavares, P.; Petit, M.-A.; Guérois, R.; Zinn-Justin, S. Automated classification of tailed bacteriophages according to their neck organization. BMC Genomics 2014, 15, 1027. [Google Scholar] [CrossRef] [PubMed]
- Malasarn, D.; Saltikov, C.W.; Campbell, K.M.; Santini, J.M.; Hering, J.G.; Newman, D.K. arrA Is a Reliable Marker for As(V) Respiration. Science 2004, 306, 455. [Google Scholar] [CrossRef] [PubMed]
- Kovach, M.E.; Elzer, P.H.; Hill, D.S.; Robertson, G.T.; Farris, M.A.; Roop, R.M.; Peterson, K.M. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 1995, 166, 175–176. [Google Scholar] [CrossRef]
- Ondov, B.D.; Treangen, T.J.; Melsted, P.; Mallonee, A.B.; Bergman, N.H.; Koren, S.; Phillippy, A.M. Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016, 17, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 2008, 57, 758–771. [Google Scholar] [CrossRef] [PubMed]
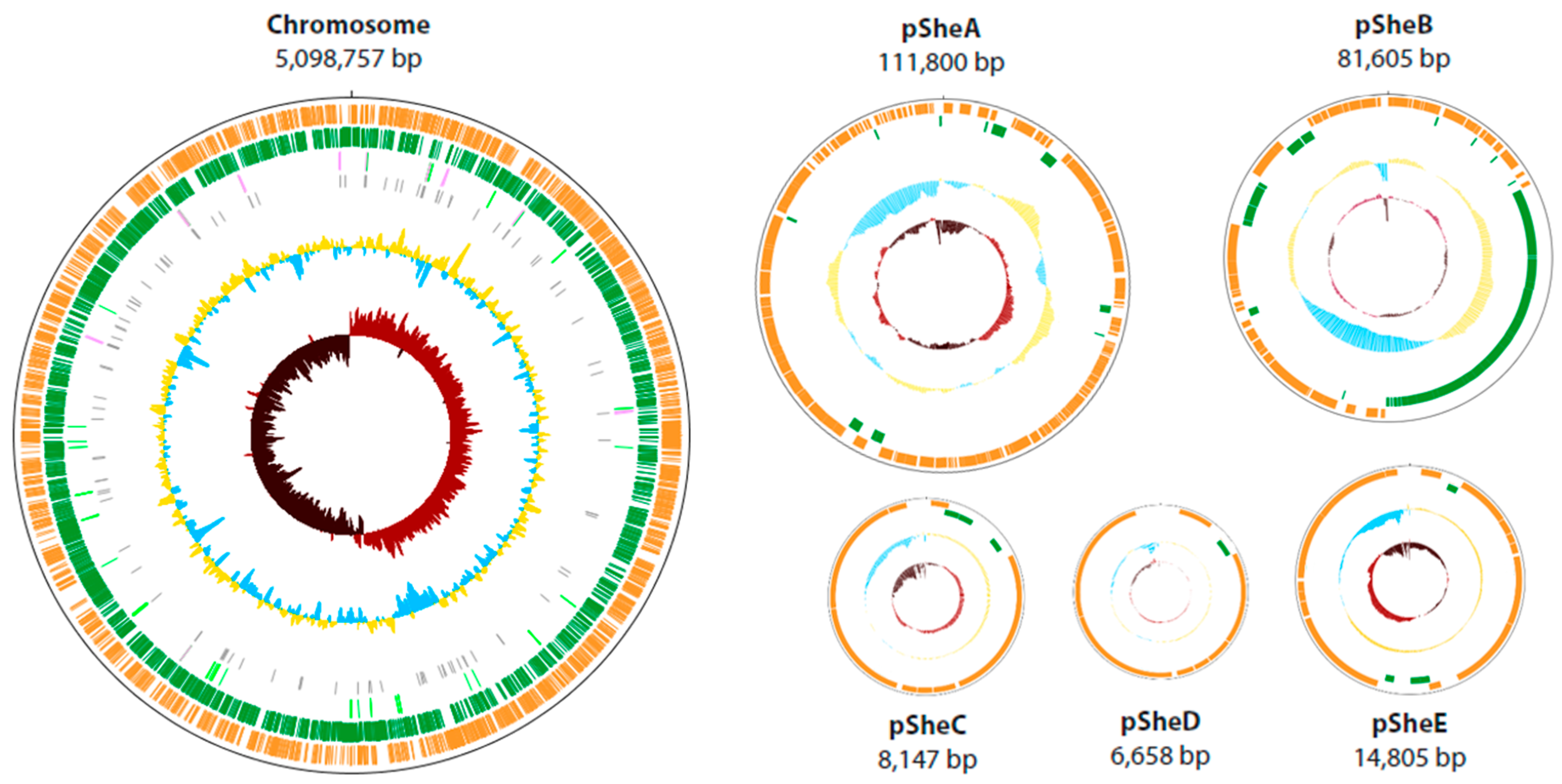
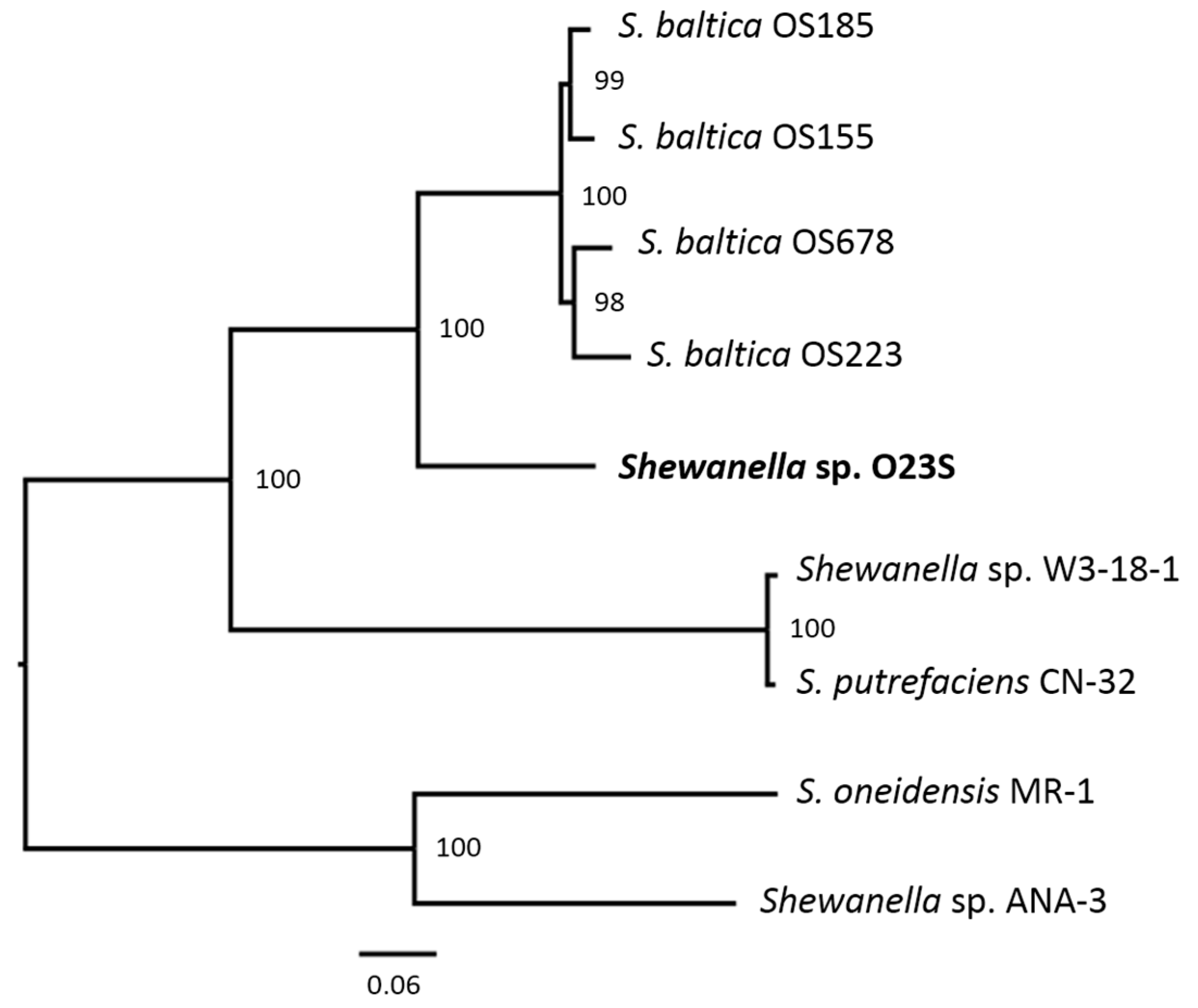
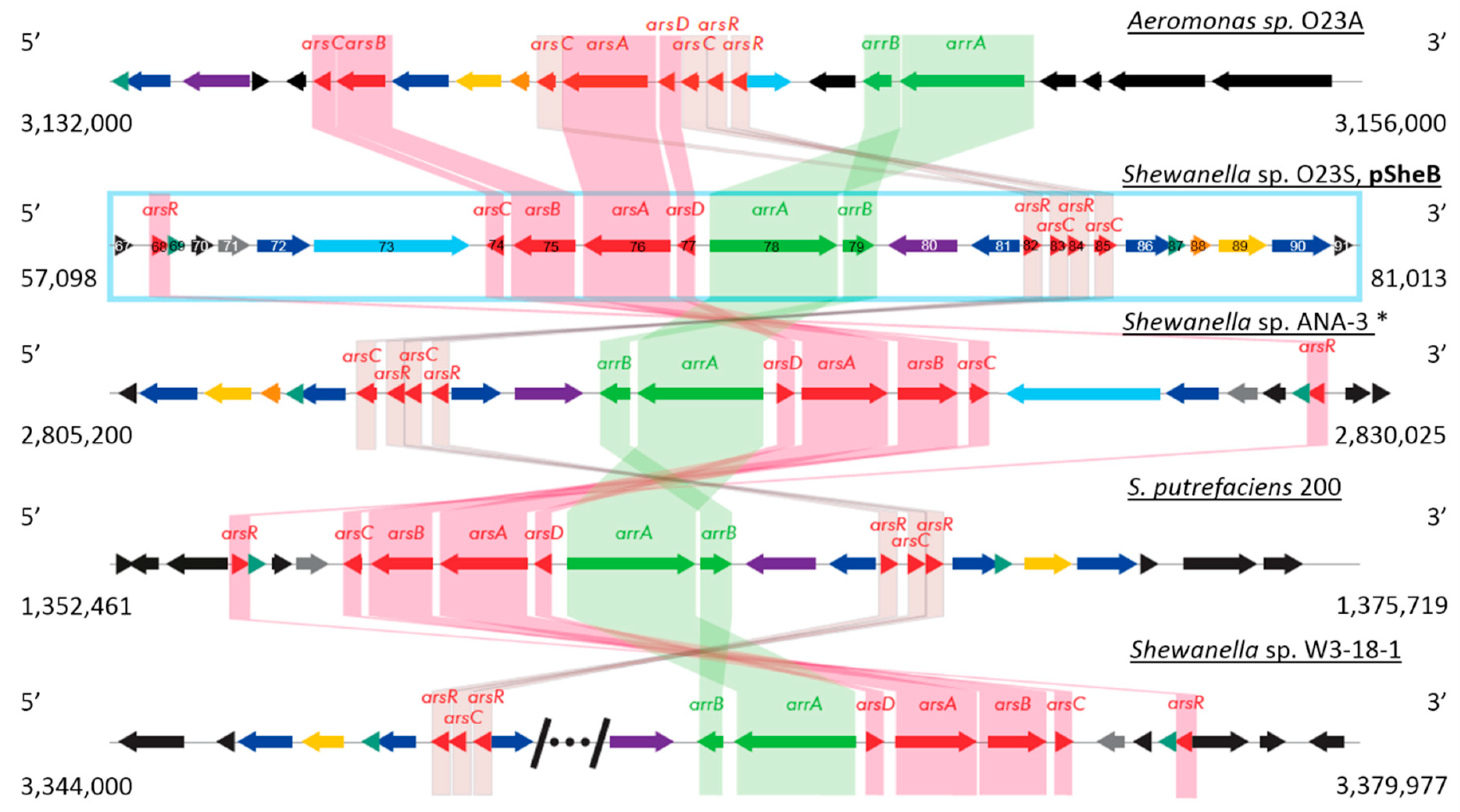
| General Features | Chromosome | pSheA | pSheB | pSheC | pSheD | pSheE |
|---|---|---|---|---|---|---|
| size (bp) | 5,098,757 | 111,800 | 81,605 | 8147 | 6658 | 14,805 |
| GC content (%) | 45.34 | 39.4 | 44.0 | 40.7 | 40.0 | 40.4 |
| coding density (%) | 85.40 | 86.3 | 89.4 | 87.7 | 90.7 | 89.0 |
| number of genes | 4654 | 157 | 91 | 12 | 11 | 23 |
| number of tRNA genes | 106 | 0 | 0 | 0 | 0 | 0 |
| number of 16S-23S-5S rRNA gene clusters | 10 (plus one additional gene for 5S rRNA) | 0 | 0 | 0 | 0 | 0 |
| phage regions | 3 | 0 | 0 | 1 | 1 | 2 |
| Strain | As(III) Concentration [mM] | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |
| E. coli F96401-1 (FOS 41C with arr/ars) | ||||||||||||
| E. coli F96401-1 (no fosmid) | ||||||||||||
| E. coli F96401-1 (fosmid without arr/ars) | ||||||||||||
| Shewanella sp. O23S | ||||||||||||
| Uninoculated medium (control) | ||||||||||||
| OD600 nm | 0 | 0.2 | 0.4 | 0.6 | 0.8 | 1 | 1.2 | 1.4 | 1.6 |
| Strain | As(V) Concentration [mM] | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 25 | 50 | 100 | 150 | 200 | 250 | 300 | 350 | 400 | 450 | 500 | |
| E. coli F96401-1 (FOS 41C with arr/ars) | ||||||||||||
| E. coli F96401-1 (no fosmid) | ||||||||||||
| E. coli F96401-1 (fosmid without arr/ars) | ||||||||||||
| Shewanella sp. O23S | ||||||||||||
| Uninoculated medium (control) | ||||||||||||
| OD600 nm | 0 | 0.2 | 0.4 | 0.6 | 0.8 | 1 | 1.2 | 1.4 | 1.6 |
| (A) | MS | MS + As(III) [mM] | MS + As(V) [mM] | (B) | MS | MS + As(III) [mM] | MS + As(V) [mM] | ||||||
| - | 2.5 | 5 | 5 | 10 | - | 2.5 | 5 | 5 | 10 | ||||
| - | - | ||||||||||||
| Cu(II) | Cu(II) | ||||||||||||
| Fe(III) | Fe(III) | ||||||||||||
| Ni(II) | Ni(II) | ||||||||||||
| Zn(II) | Zn(II) | ||||||||||||
| OD600 nm | 0.00–0.05 | 0.05–0.10 | 0.10–0.15 | >0.15 |
| No. | ORF no. (Chromosome) | Gene Name | Putative Product | Predicted Function | Result, MIC Value (mg/L) |
|---|---|---|---|---|---|
| 1 2 | 2288 2289 | mex | MexPQ-OpmE multidrug efflux pump | carbapenem, acridine dye, phenicol antibiotic, diaminopyrimidine, tetracycline, macrolide resistance | n–f., O23S is susceptible to: AMP—1.0; CFM—0.38; CTX—0.19; CRO—0.19; TE—0.25; E—0.75 |
| 3 4 5 6 | 2298 2517 3381 4409 | bcr | Drug resistance transporter Bcr/CmlA | bicyclomycin, chloramphenicol and florfenicol resistance | n–f., O23S is susceptible to: C—0.75 |
| 7 | 2321 | mdtK/norM | Multidrug efflux transporter MdtK/NorM (MATE family) | fluoroquinolone, tetraphenylphosphonium ion, deoxycholate, doxorubicin, trimethoprim, fosfomycin, ethidium bromide, benzalkonium, kanamycin and streptomycin resistance | n–f., O23S is susceptible to: CIP—0.016; MXF—0.004; TM—0.25; C—0.75 |
| 8 | 2377 | emrD | EmrD (Multidrug resistance efflux pump) | aminoglycoside resistance | n–f., O23S is susceptible to: CN—0.75 |
| 9 10 11 12 13 | 880 2919 3979 3980 3132 | acrA acrB | AcrA-B (Multidrug resistance efflux pump) | acriflavin, aminoglycoside, β-lactam, glycylcycline, macrolide resistance | n–f., O23S is susceptible to: AMP—1.0; CFM—0.38; CTX—0.19;CRO—0.19; E—0.75; CN—0.75 |
| 14 15 | 569 3371 | Metallo-beta-lactamase superfamily protein | resistance to almost all clinically-available β-lactam antibiotics including carbapenems | n–f., O23S is susceptible to: AMP—1.0; CFM—0.38; CTX—0.19; CRO—0.19 | |
| 16 17 | 3390 3391 | mexF mexE | MexE/F-OprN | resistance to chloramphenicol and fluoroquinolones | n–f., O23S is susceptible to: CIP—0.016; MXF—0.004; C—0.75 |
| 18 19 | 3753 3752 | macB macA | MacA/B (Macrolide-specific efflux system) | Macrolide resistance | n–f., O23S is susceptible to: E—0.75 |
| 20 | 4001 | mdtL | MdtL. (Multidrug resistance efflux pump) | Chloramphenicol resistance | n–f., O23S is susceptible to: C—0.75 |
| 21 22 23 | 4552 4553 4554 | fus | Fus | Fusaric acid resistance | - |
| 24 25 | 847 2376 | Beta-lactamase class C-like and penicillin binding proteins (PBPs) | Resistance to β-lactam antibiotics | n–f., O23S is susceptible to: AMP—1.0; CFM—0.38; CTX—0.19; CRO—0.19 | |
| 26 | 3739 | Class D beta-lactamase, OXA-48 family | Resistance to β-lactam antibiotics including carbapenems | n–f., O23S is susceptible to: AMP—1.0; CFM—0.38; CTX—0.19; —0.19 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uhrynowski, W.; Radlinska, M.; Drewniak, L. Genomic Analysis of Shewanella sp. O23S—The Natural Host of the pSheB Plasmid Carrying Genes for Arsenic Resistance and Dissimilatory Reduction. Int. J. Mol. Sci. 2019, 20, 1018. https://doi.org/10.3390/ijms20051018
Uhrynowski W, Radlinska M, Drewniak L. Genomic Analysis of Shewanella sp. O23S—The Natural Host of the pSheB Plasmid Carrying Genes for Arsenic Resistance and Dissimilatory Reduction. International Journal of Molecular Sciences. 2019; 20(5):1018. https://doi.org/10.3390/ijms20051018
Chicago/Turabian StyleUhrynowski, Witold, Monika Radlinska, and Lukasz Drewniak. 2019. "Genomic Analysis of Shewanella sp. O23S—The Natural Host of the pSheB Plasmid Carrying Genes for Arsenic Resistance and Dissimilatory Reduction" International Journal of Molecular Sciences 20, no. 5: 1018. https://doi.org/10.3390/ijms20051018
APA StyleUhrynowski, W., Radlinska, M., & Drewniak, L. (2019). Genomic Analysis of Shewanella sp. O23S—The Natural Host of the pSheB Plasmid Carrying Genes for Arsenic Resistance and Dissimilatory Reduction. International Journal of Molecular Sciences, 20(5), 1018. https://doi.org/10.3390/ijms20051018






