FGF23 and Fetuin-A Interaction and Mesenchymal Osteogenic Transformation
Abstract
1. Introduction
2. Results
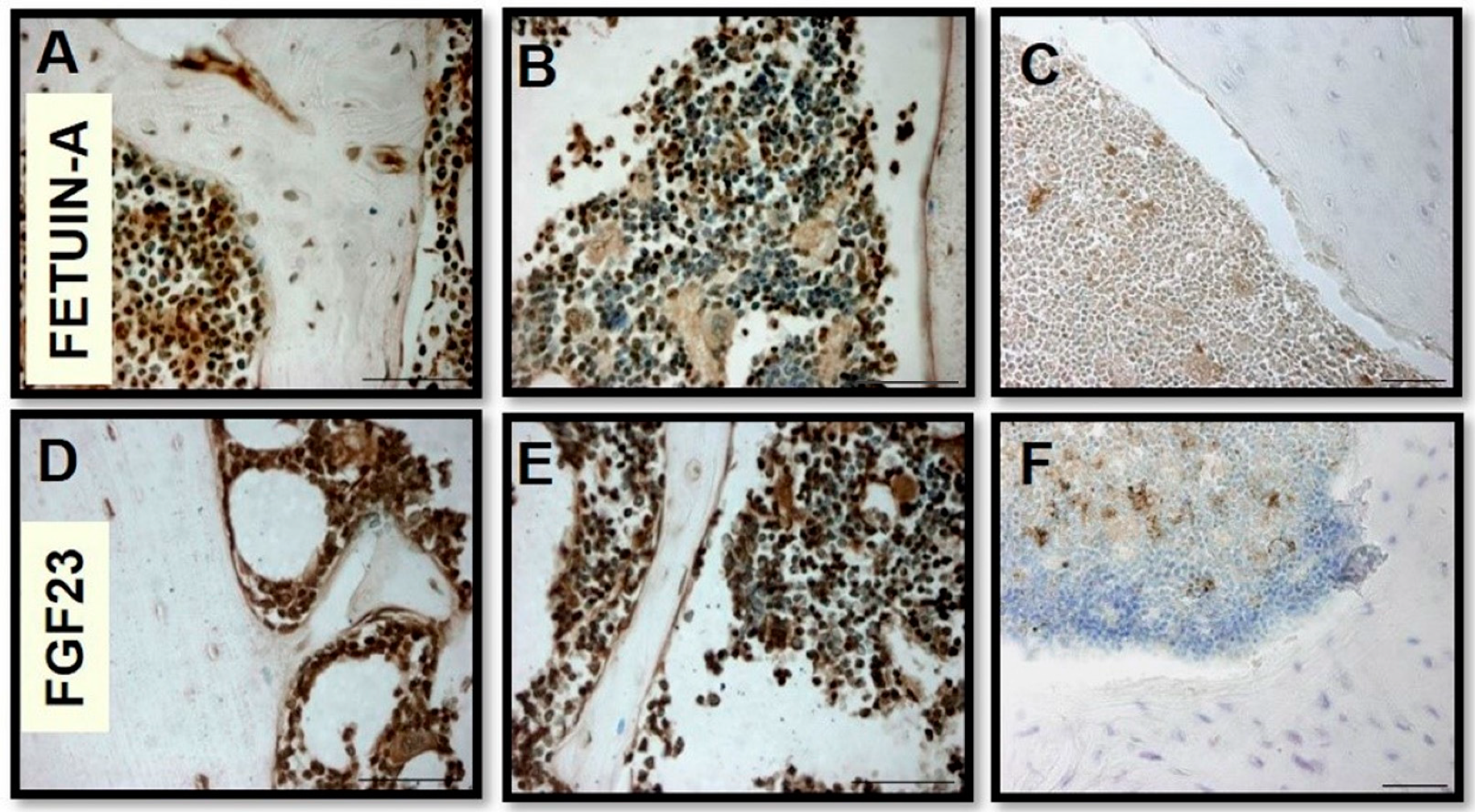
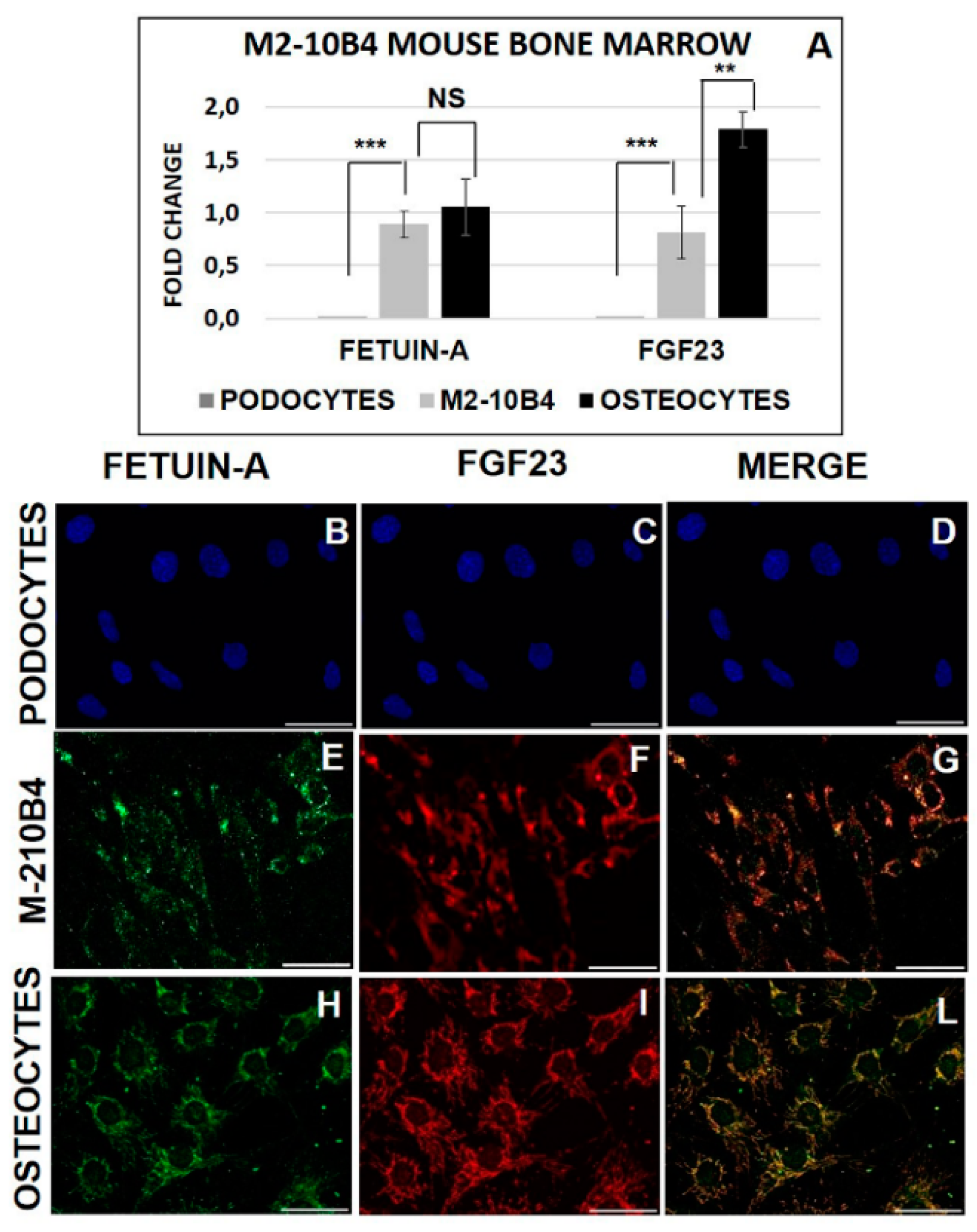
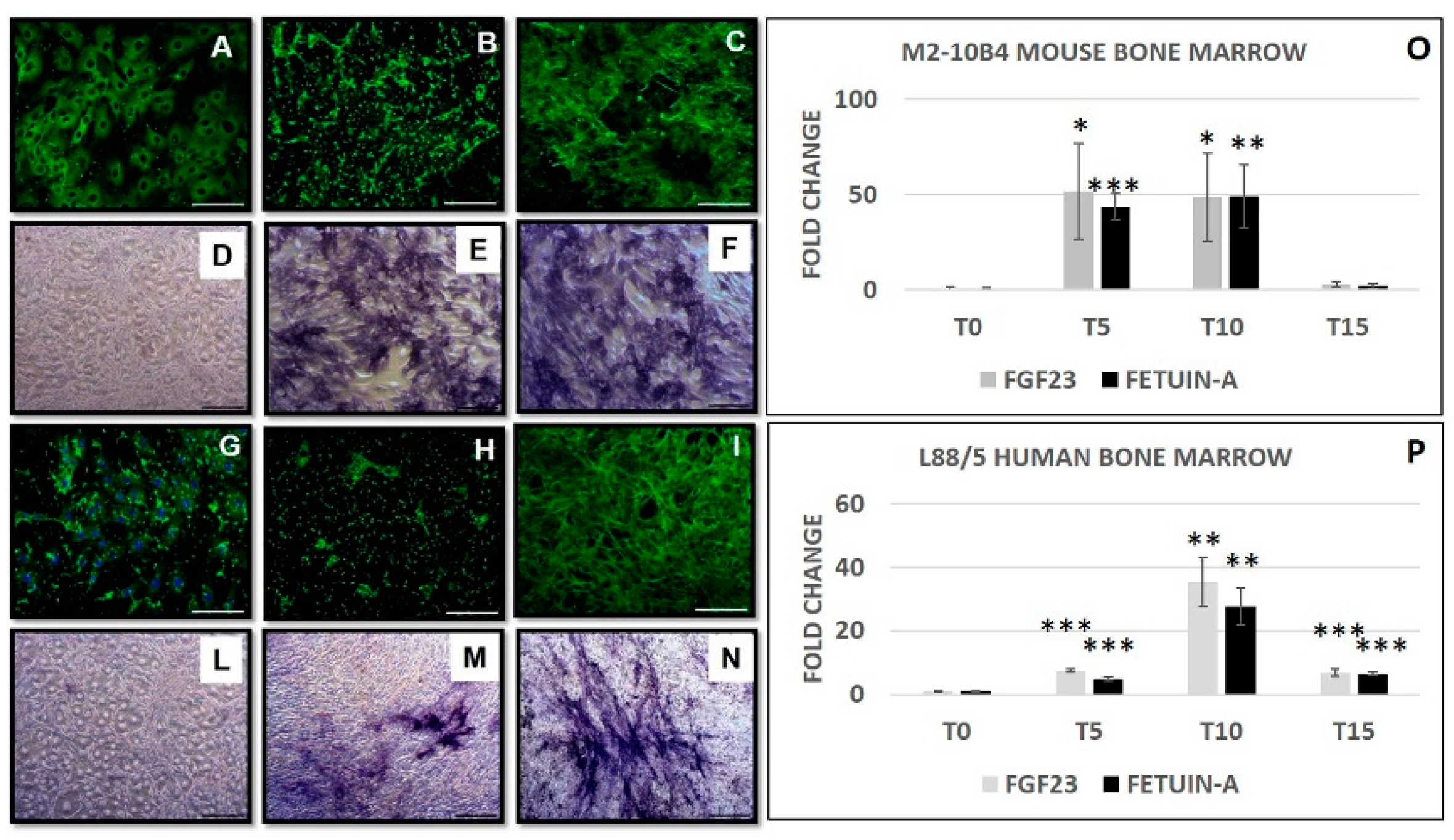
2.1. FGF23 and Fetuin-A Expression in Tissue and Cells Mice Bone Marrow
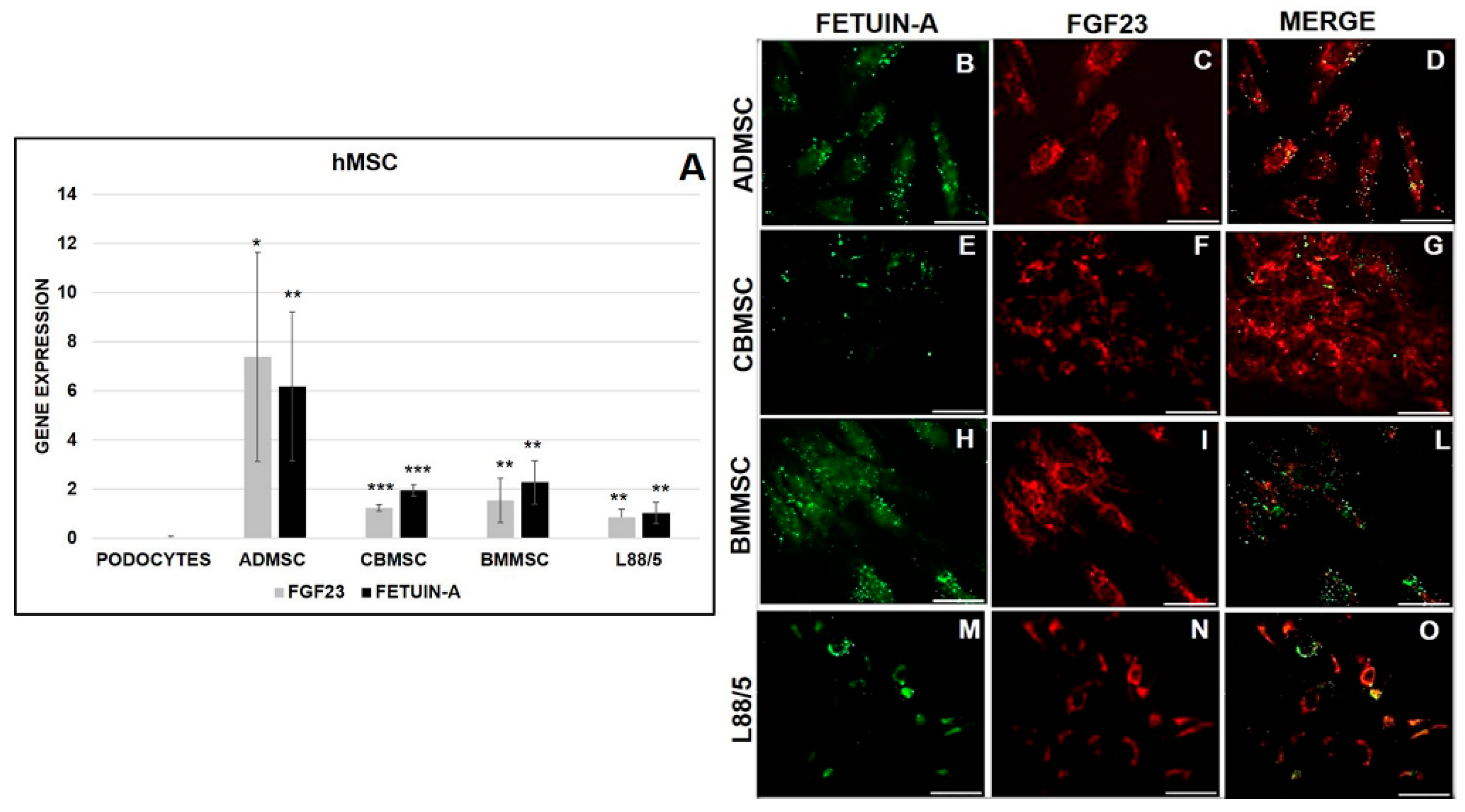
2.2. FGF23 and Fetuin-A Expression in Primary and Human MSCs
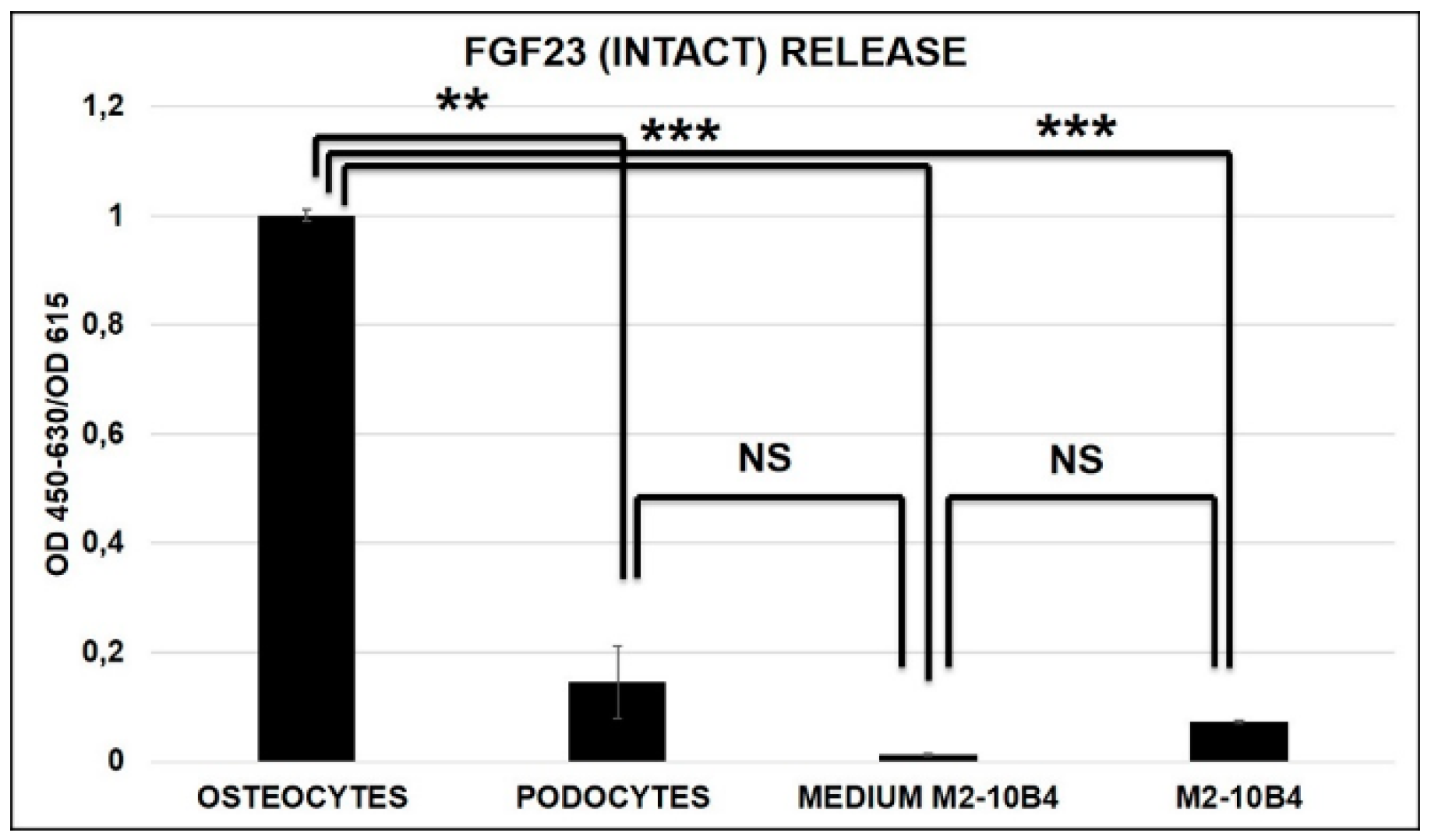
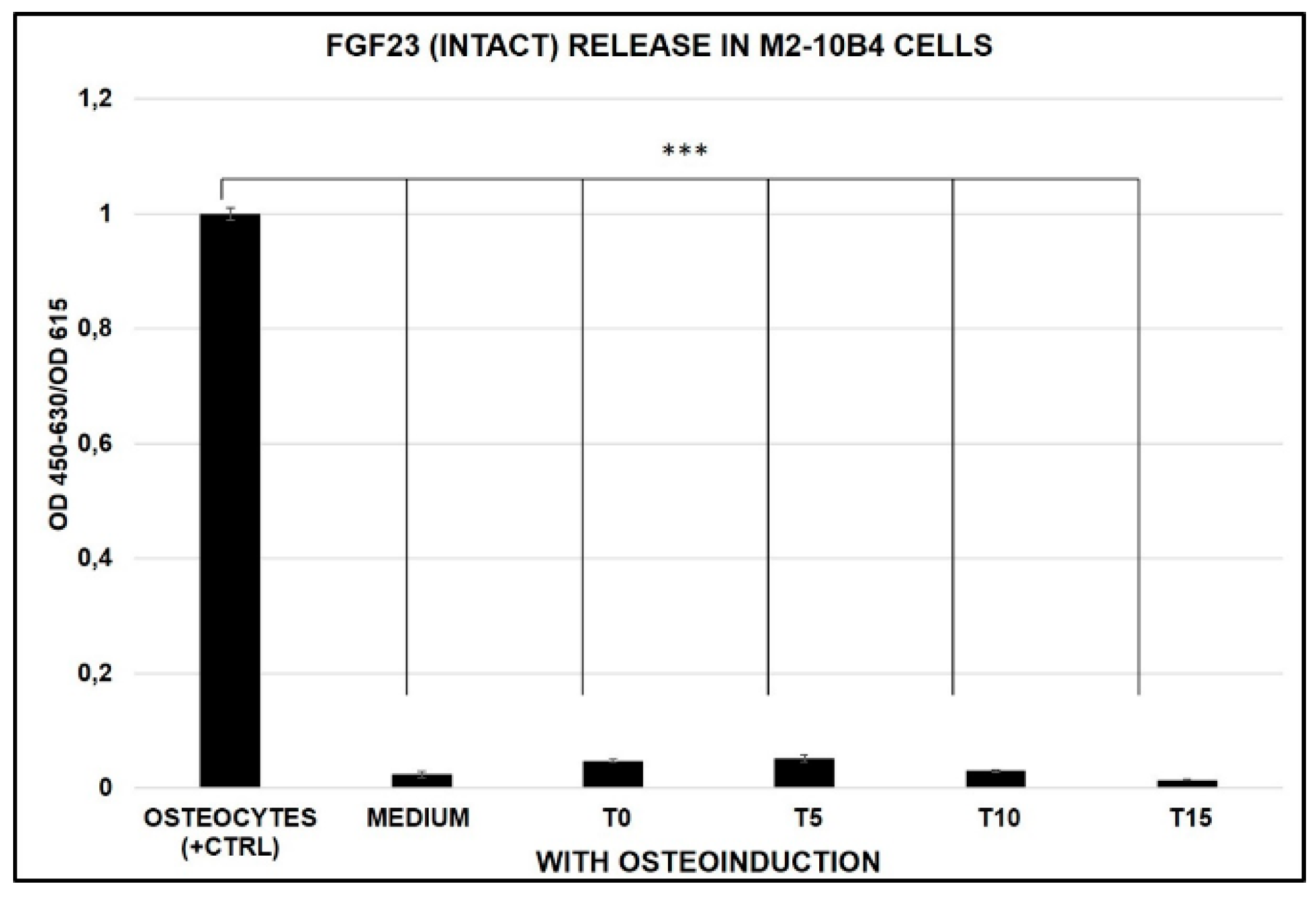
2.3. FGF23 Release
2.4. Effect of Osteogenic Differentiation on FGF23 and Fetuin-A Expression
2.5. FGF23 Release during MSCs Transformation in OB
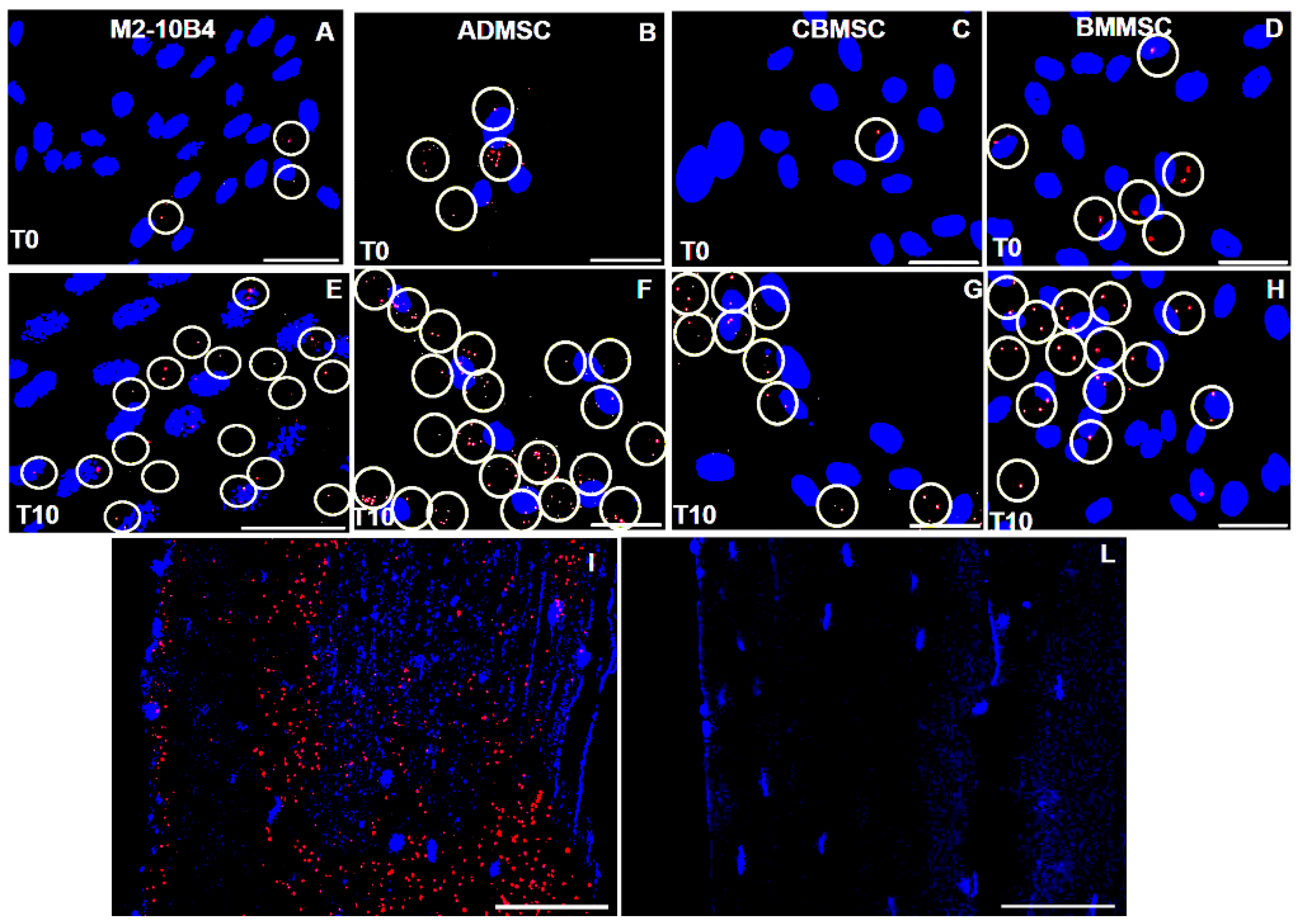
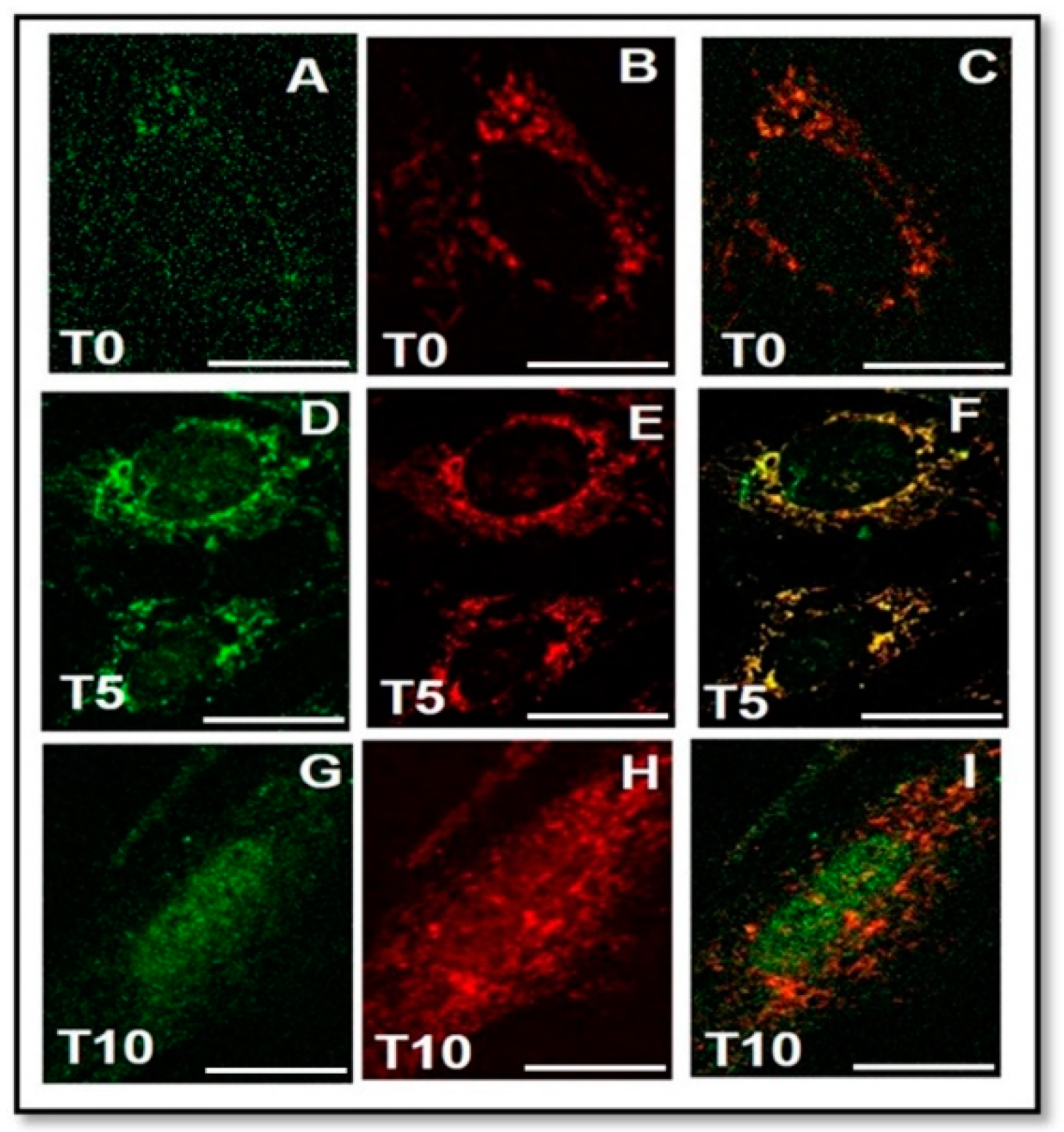
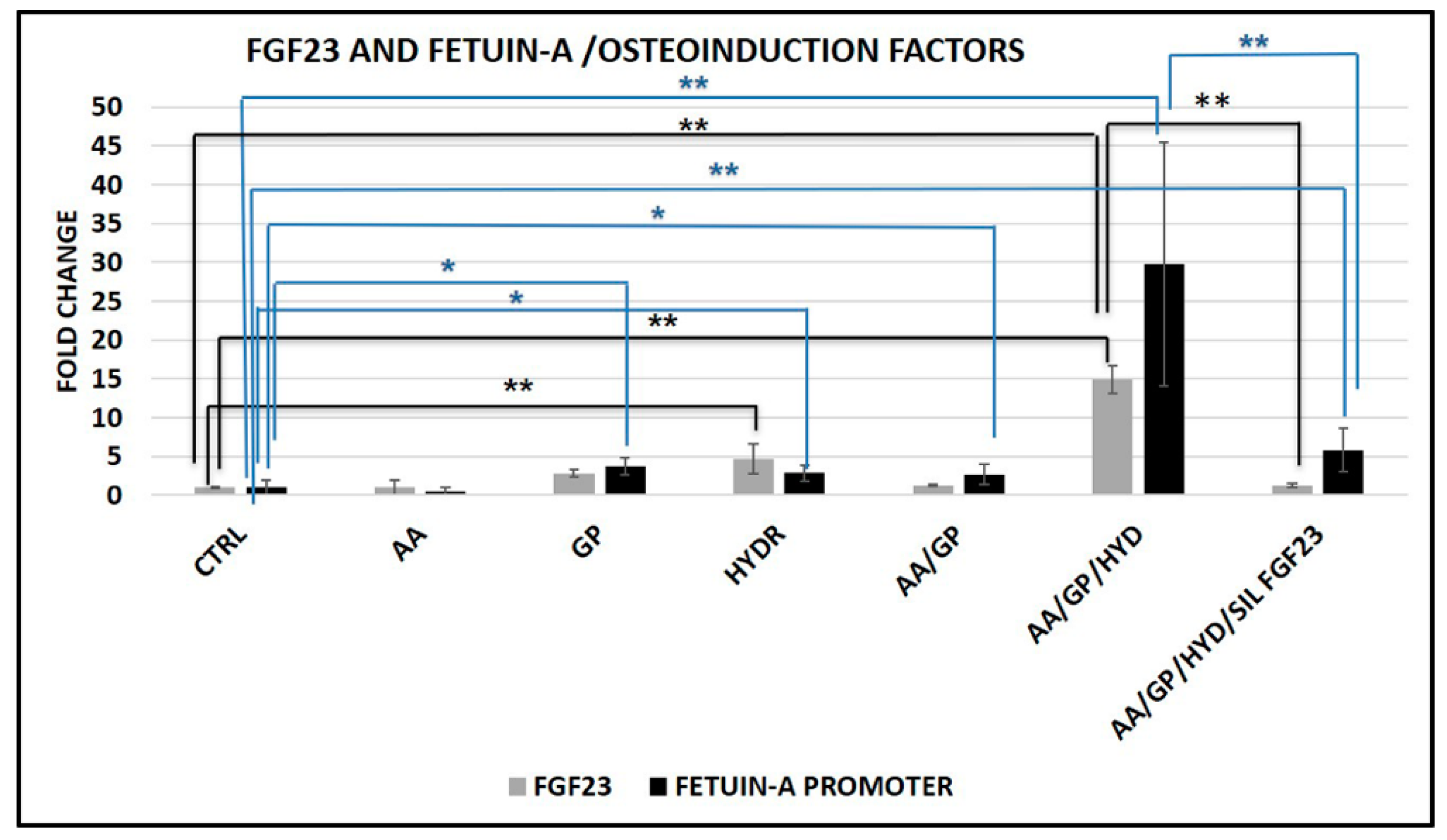
2.6. Interaction between FGF23 and Fetuin-A
2.6.1. Fetuin-A and FGF23 Interaction in Mouse, in Primary Human Cells and Tibial Mouse Tissue
2.6.2. Nuclear Biochemical Interaction between FGF23 and Fetuin-A promoter
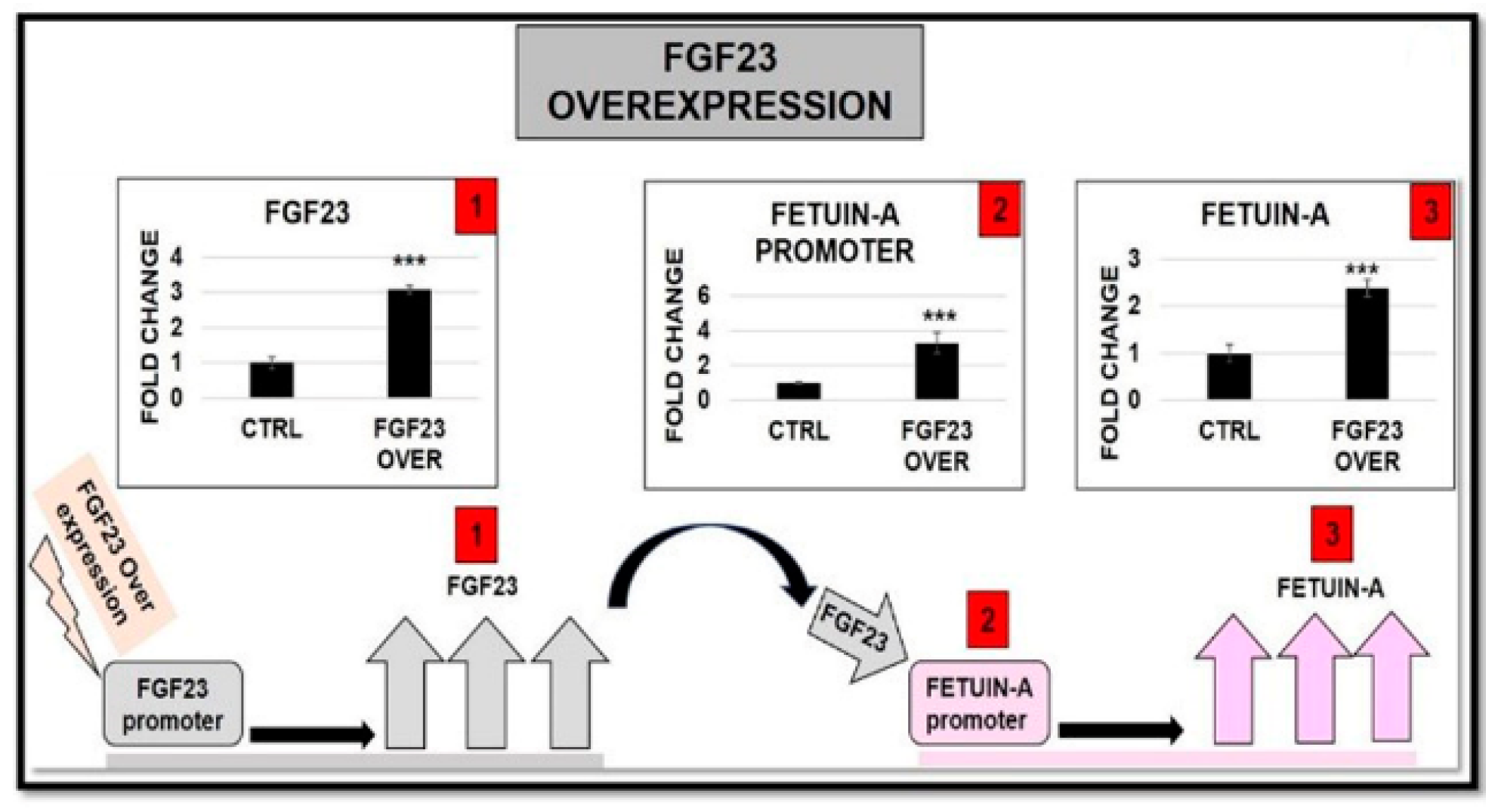
2.6.3. FGF23 Overexpression and Addition on both FGF23 Expression and Fetuin-A Promoter Activation
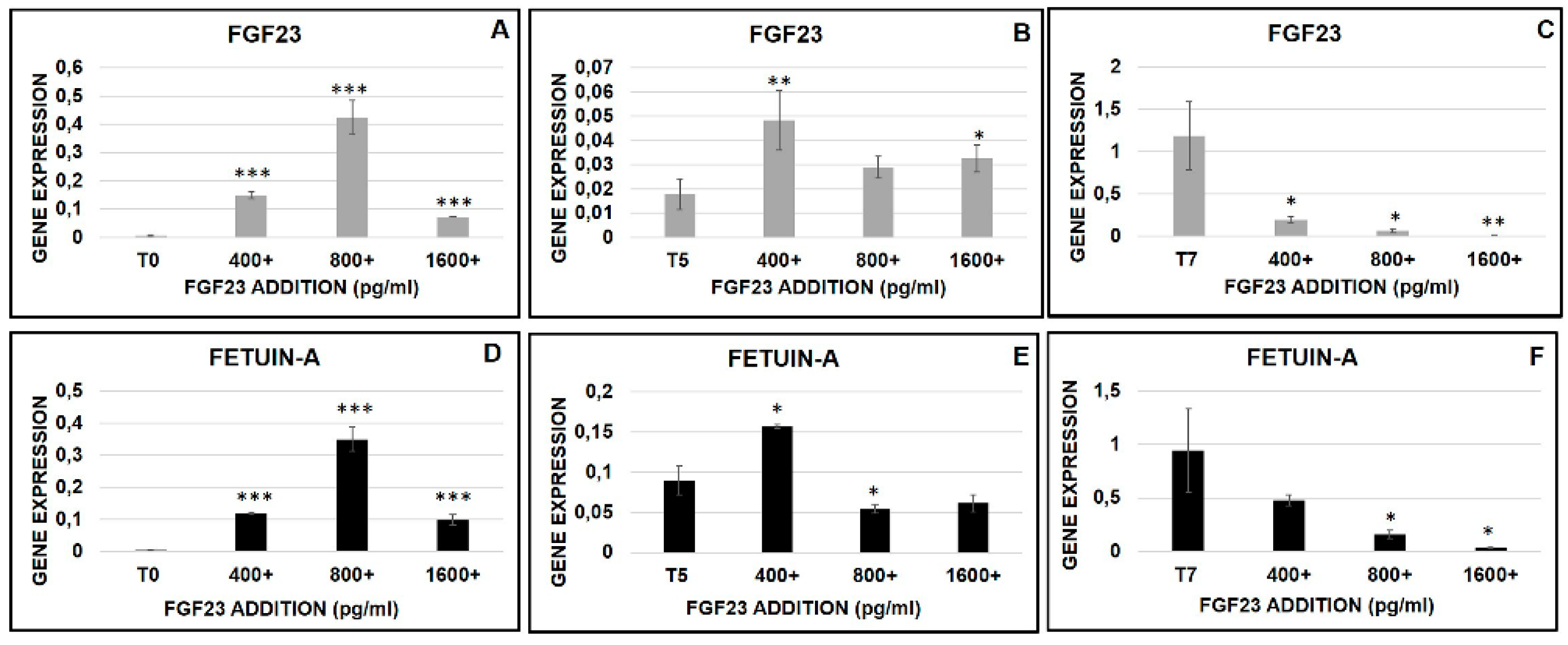
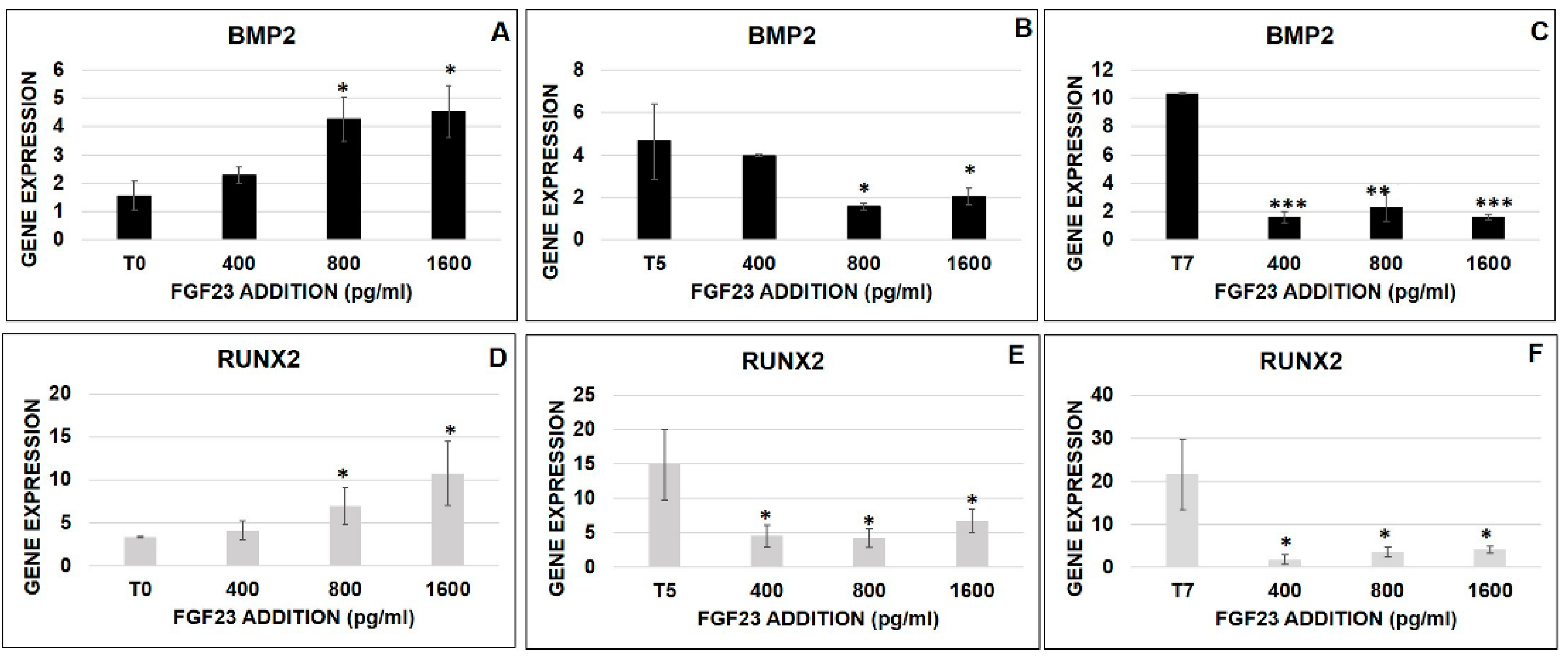
2.6.4. FGF23 Influence on Osteo-Differentiation Markers during the OB Transformation
2.6.5. FGF23 Influence on Fetuin-A Promoter Expression during the OB Transformation
3. Discussion
4. Material and Methods
4.1. Tissues
4.2. Human and Mouse Bone Marrow, Osteocytes (OS), and Podocytes (PODO) Cell Line Culture
4.3. Primary MSCs Isolation and Culture
4.4. Alkaline Phosphatase (ALP) and Collagen Type I Stain
4.5. mRNA Extraction and qRT-PCR
4.6. Immunohistochemistry (IHC) and IF
4.7. FGF23 and Fetuin-A Overexpression and Silencing
4.8. Protein Interaction
4.9. Chromatine Immunoprecipitation (CHIP) and Semi-Quantitative PCR
4.10. Enzyme-Linked Immunosorbent (ELISA) Tests
4.11. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| MSCs | Mesenchymal stem cells |
| OB | Osteoblasts |
| FGF | Fibroblast growth factor |
| OS | Osteocytes |
| POD | Podocytes |
| HYD | Hydrocortisone |
| COLIαI | Collagen type i |
| ALP | Alkaline phosphatase |
| Rb | Rabbit |
| Hu | Human |
| BMMSCs | Bone marrows mesenchymal stem cells |
| ADMSCs | Adipose derived mesenchymal stem cells |
| CBMSCs | Cord blood mesenchymal cells |
| Ms | Mouse |
| AHSG/Fetuin-A | Alpha 2-hs glycoprotein |
| GP | Glycerol phosphate |
| AA | Ascorbic acid |
| CVD | Cardio vascular disease |
| MBD | Mineral bone disorder |
| CKD | Chronic kidney disease |
References
- Pittenger, M.F.; Mackay, A.M.; Beck, S.C.; Jaiswal, R.K.; Douglas, R.; Mosca, J.D.; Moorman, M.A.; Simonetti, D.W.; Craig, S.; Marshak, D.R. Multilineage potential of adult human mesenchymal stem cells. Science 1999, 284, 143–147. [Google Scholar] [CrossRef] [PubMed]
- De Kok, I.J.; Drapeau, S.J.; Young, R.; Cooper, L.F. Evaluation of mesenchymal stem cells following implantation in alveolar sockets: A canine safety study. Int. J. Oral Maxillofac. Implants 2005, 20, 511–518. [Google Scholar] [PubMed]
- Dallari, D.; Fini, M.; Stagni, C.; Torricelli, P.; Nicoli Aldini, N.; Giavaresi, G.; Cenni, E.; Baldini, N.; Cenacchi, A.; Bassi, A.; et al. In vivo study on the healing of bone defects treated with bone marrow stromal cells, platelet-rich plasma, and freeze-dried bone allografts, alone and in combination. J. Orthop. Res. 2006, 24, 877–888. [Google Scholar] [CrossRef] [PubMed]
- Perrotta, F.M.; Guerra, G.; De Socio, A.; Scriffignano, S.; Lubrano, E. Mesenchymal stem cells: A possible role in the pathogenesis and treatment of spondyloarthritis. Reumatismo 2017, 69, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Miana, V.V.; González, E.A.P. Adipose tissue stem cells in regenerative medicine. Ecancermedicalscience 2018, 12, 822. [Google Scholar] [CrossRef] [PubMed]
- Komori, T. Regulation of osteoblast differentiation by Runx2. Adv. Exp. Med. Biol. 2010, 658, 43–49. [Google Scholar] [PubMed]
- Tsutsumi, S.; Shimazu, A.; Miyazaki, K.; Pan, H.; Koike, C.; Yoshida, E.; Takagishi, K.; Kato, Y. Retention of multilineage differentiation potential of mesenchymal cells during proliferationin response to FGF. Biochem. Biophys. Res. Commun. 2001, 288, 413–419. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, R.; Kato, Y.; Chen, D.; Harris, S.E.; Mundy, G.R.; Yoneda, T. Smad5 and DPC4 are key molecules in mediating BMP-2-induced osteoblastic differentiation of the pluripotent mesenchymal precursor cell line C2C12. J. Biol. Chem. 1998, 273, 1872–1879. [Google Scholar] [CrossRef] [PubMed]
- Bonewald, L.F. The amazing osteocyte. J. Bone Miner. Res. 2011, 26, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Capulli, M.; Paone, R.; Rucci, N. Osteoblast and osteocyte: Games without frontiers. Arch. Biochem. Biophys. 2014, 561, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Palumbo, C.; Palazzini, S.; Marotti, G. Morphological study of intercellular junctions during osteocyte differentiation. Bone 1990, 11, 401–406. [Google Scholar] [CrossRef]
- Skerry, T.M.; Bitensky, L.; Chayen, J.; Lanyon, L.E. Early strain-related changes in enzyme activity in osteocytes following bone loading in vivo. J. Bone Miner. Res. 1989, 4, 783–788. [Google Scholar] [CrossRef] [PubMed]
- Gittens, S.A.; Uludag, H. Growth factor delivery for bone tissue engineering. J. Drug Target. 2001, 9, 407–429. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.E.; Eom, Y.W. Maintenance of Proliferation and Adipogenic Differentiation by Fibroblast Growth Factor-2 and Dexamethasone Through Expression of Hepatocyte Growth Factor in Bone Marrow-derived Mesenchymal Stem Cells. Biomed. Sci. Lett. 2016, 22, 1–8. [Google Scholar] [CrossRef][Green Version]
- Yun, Y.R.; Won, J.E.; Jeon, E.; Lee, S.; Kang, W.; Jo, H.; Jang, J.H.; Shin, U.S.; Kim, H.W. Fibroblast Growth Factors: Biology, Function, and Application for Tissue Regeneration. J. Tissue Eng. 2010, 2010, 218142. [Google Scholar] [CrossRef] [PubMed]
- Eom, Y.W.; Oh, J.E.; Lee, J.I.; Baik, S.K.; Rhee, K.J.; Shin, H.C.; Kim, Y.M.; Ahn, C.M.; Kong, J.H.; Kim, H.S.; et al. The role of growth factors in maintenance of stemness in bone marrow-derived mesenchymal stem cells. Biochem. Biophys. Res. Commun. 2014, 445, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Xie, N.; Li, W.; Yuan, B.; Shi, Y.; Wang, Y. Immunobiology of mesenchymal stem cells. Cell Death Differ. 2014, 21, 216–225. [Google Scholar] [CrossRef] [PubMed]
- Uccelli, A.; Moretta, L.; Pistoia, V. Mesenchymal stem cells in health and disease. Nat. Rev. Immunol. 2008, 8, 726–762. [Google Scholar] [CrossRef]
- Lu, X.; Hu, M.C. Klotho/FGF23 Axis in Chronic Kidney Disease and Cardiovascular Disease. Kidney Dis. 2017, 3, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.C.; Shiizaki, K.; Kuro-o, M. Fibroblast growth factor 23 and Klotho: Physiology and pathophysiology of an endocrine network of mineral metabolism. Annu. Rev. Physiol. 2013, 75, 503–505. [Google Scholar] [CrossRef]
- Shalhoub, V.; Ward, S.C.; Sun, B.; Stevens, J.; Renshaw, L.; Hawkins, N.; Richards, W.G. Fibroblast growth factor 23 (FGF23) and alpha-klotho stimulate osteoblastic MC3T3.E1 cell proliferation and inhibit mineralization. Calcif. Tissue Int. 2011, 89, 140–150. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; He, X.; Olauson, H.; Larsson, T.E.; Lindgren, U. FGF23 affects the lineage fate determination of mesenchymal stem cells. Calcif. Tissue Int. 2013, 93, 556–564. [Google Scholar] [CrossRef] [PubMed]
- Mattinzoli, D.; Rastaldi, M.P.; Ikehata, M.; Armelloni, S.; Pignatari, C.; Giardino, L.A.; Li, M.; Alfieri, C.M.; Regalia, A.; Riccardi, D.; et al. FGF23-regulated production of Fetuin-A (AHSG) in osteocytes. Bone 2016, 83, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Mattinzoli, D.; Ikehata, M.; Alfieri, C.M.; Messa, P. Authors’ reply to the comments on the paper: “FGF23-regulated production of Fetuin A (AHSG) in osteocytes”. Bone 2016, 93, 225–229. [Google Scholar] [CrossRef] [PubMed]
- Mattinzoli, D.; Ikehata, M.; Tsugawa, K.; Alfieri, C.M.; Dongiovanni, P.; Trombetta, E.; Valenti, L.; Puliti, A.; Lazzari, L.; Messa, P. FGF23 and Fetuin-A interaction in the liver and in the circulation. Int. J. Biol. Sci. 2018, 14, 586–598. [Google Scholar] [CrossRef] [PubMed]
- Mori, K.; Emoto, M.; Inaba, M. Fetuin-A: A multifunctional protein. Recent Pat. Endocr. Metab. Immune Drug Discov. 2011, 5, 124–146. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Sama, A.E. Anti-inflammatory role of fetuin-A in injury and infection. Curr. Mol. Med. 2012, 12, 625–633. [Google Scholar] [CrossRef] [PubMed]
- Szweras, M.; Liu, D.; Partridge, E.A.; Pawling, J.; Sukhu, B.; Clokie, C.; Jahnen-Dechent, W.; Tenenbaum, H.C.; Swallow, C.J.; Grynpas, M.D.; et al. Alpha 2-HS glycoprotein/fetuin, a transforming growth factor-beta/bone morphogenetic protein antagonist, regulates postnatal bone growth and remodeling. J. Biol. Chem. 2002, 277, 19991–19997. [Google Scholar] [CrossRef]
- Bussolati, G.; Radulescu, R.T. Blocking endogenous peroxidases in immunohistochemistry: A mandatory, yet also subtle measure. Appl. Immunohistochem. Mol. Morphol. 2011, 19, 484. [Google Scholar] [CrossRef]
- Mundel, P.; Shankland, S.J. Podocyte Biology and Response to Injury. J. Am. Soc. Nephrol. 2002, 13, 3005–3015. [Google Scholar] [CrossRef]
- Li, M.; Armelloni, S.; Ikehata, M.; Corbelli, A.; Pesaresi, M.; Calvaresi, N.; Giardino, L.; Mattinzoli, D.; Nisticò, F.; Andreoni, S.; et al. Nephrin expression in adult rodent central nervous system and its interaction with glutamate receptors. J. Pathol. 2011, 225, 118–128. [Google Scholar] [CrossRef]
- Giardino, L.; Armelloni, S.; Corbelli, A.; Mattinzoli, D.; Zennaro, C.; Guerrot, D.; Tourrel, F.; Ikehata, M.; Li, M.; Berra, S.; et al. Podocyte glutamatergic signaling contributes to the function of the glomerular filtration barrier. J. Am. Soc. Nephrol. 2009, 20, 1929–1940. [Google Scholar] [CrossRef] [PubMed]
- Armelloni, S.; Ikehata, M.; Mattinzoli, D.; Li, M.; Alfieri, C.M.; Rastaldi, M.; Messa, P.; Neuro, D. Expression in Podocytes and Interrelationships with Nephrin at Both Nuclear and Cytoplasmic Sites. Cell. Physiol. Biochem. 2018, 46, 873–889. [Google Scholar] [CrossRef] [PubMed]
- Rittenberg, B.; Partridge, E.; Baker, G.; Clokie, C.; Zohar, R.; Dennis, J.W.; Tenenbaum, H.C. Regulation of BMP-induced ectopic bone formation by Ahsg. J. Orthop. Res. 2005, 23, 653–662. [Google Scholar] [CrossRef] [PubMed]
- Sato, C.; Iso, Y.; Mizukami, T.; Otabe, K.; Sasai, M.; Kurata, M.; Sanbe, T.; Sekiya, I.; Miyazaki, A.; Suzuki, H. Fibroblast growth factor-23 induces cellular senescence in human mesenchymal stem cells from skeletal muscle. Biochem. Biophys. Res. Commun. 2016, 470, 657–662. [Google Scholar] [CrossRef]
- Papierska, L.; Ćwikła, J.B.; Misiorowski, W.; Rabijewski, M.; Sikora, K.; Wanyura, H. FGF23 Producing Mesenchymal Tumor. Case Rep. Endocrinol. 2014, 2014, 492789. [Google Scholar] [CrossRef] [PubMed]
- Heo, J.S.; Choi, Y.; Kim, H.S.; Kim, H.O. Comparison of molecular profiles of human MSCs stem cells derived from bone marrow, umbilical cord blood, placenta and adipose tissue. Int. J. Mol. Med. 2016, 37, 115–125. [Google Scholar] [CrossRef]
- Wagner, W.; Wein, F.; Seckinger, A.; Frankhauser, M.; Wirkner, U.; Krause, U.; Blake, J.; Schwager, C.; Eckstein, V.; Ansorge, W.; et al. Comparative characteristics of mesenchymal stem cells from human bone marrow, adipose tissue, and umbilical cord blood. Exp. Hematol. 2005, 33, 1402–1416. [Google Scholar] [CrossRef]
- Ren, H.; Sang, Y.; Zhang, F.; Liu, Z.; Qi, N.; Chen, Y. Comparative Analysis of Human Mesenchymal Stem Cells from Umbilical Cord, Dental Pulp, and Menstrual Blood as Sources for Cell Therapy. Stem Cells Int. 2016, 2016, 3516574. [Google Scholar] [CrossRef]
- Pal, D.; Dasgupta, S.; Kundu, R.; Maitra, S.; Das, G.; Mukhopadhyay, S.; Ray, S.; Majumdar, S.S.; Bhattacharya, S. Fetuin-A acts as an endogenous ligand of TLR4 to promote lipid-induced insulin resistance. Nat. Med. 2012, 18, 1279–1285. [Google Scholar] [CrossRef]
- Stefan, N.; Häring, H. Circulating fetuin-A and free fatty acids interact to predict insulin resistance in humans. Nat. Med. 2013, 19, 394–395. [Google Scholar] [CrossRef] [PubMed]
- Trepanowski, J.; Mey, J.; Varady, K. Fetuin-A: A novel link between obesity and related complications. Int. J. Obes. 2015, 39, 734–741. [Google Scholar] [CrossRef] [PubMed]
- Hoemann, C.D.; El-Gabalawy, H.; McKee, M.D. In vitro osteogenesis assays: Influence of the primary cell source on alkaline phosphatase activity and mineralization. Pathol. Biol. 2009, 57, 318–323. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Blaber, M. Structural basis of conserved cysteine in the fibroblast growth factor family: Evidence for a vestigial half-cystine. J. Mol. Biol. 2009, 393, 128–139. [Google Scholar] [CrossRef] [PubMed]
- Xie, B.; Tassi, E.; Swift, M.R.; McDonnell, K.; Bowden, E.T.; Wang, S.; Ueda, Y.; Tomita, Y.; Riegel, A.T.; Wellstein, A. Identification of the fibroblast growth factor (FGF)-interacting domain in a secreted FGF-binding protein by phage display. J. Biol. Chem. 2006, 281, 1137–1144. [Google Scholar] [CrossRef] [PubMed]
- Birmingham, E.; Niebur, G.L.; McHugh, P.E.; Shaw, G.; Barry, F.P.; McNamara, L.M. Osteogenic differentiation of mesenchymal stem cells is regulated by osteocyte and osteoblast cells in a simplified bone niche. Eur. Cells Mater. 2012, 23, 13–27. [Google Scholar] [CrossRef]
- Fujii, H.; Joki, N. Mineral metabolism and cardiovascular disease in CKD. Clin. Exp. Nephrol. 2017, 21 (Suppl. 1), 53–63. [Google Scholar] [CrossRef] [PubMed]
- Tsuruya, K.; Eriguchi, M. Cardiorenal syndrome in chronic kidney disease. Curr. Opin. Nephrol. Hypertens. 2015, 24, 154–162. [Google Scholar] [CrossRef]
- Wöltje, M.; Tschöke, B.; von Bülow, V.; Westenfeld, R.; Denecke, B.; Gräber, S.; Jahnen-Dechent, W. CCAAT enhancer binding protein beta and hepatocyte nuclear factor 3beta are necessary and sufficient to mediate dexamethasone-induced up-regulation of alpha2HS-glycoprotein/fetuin-A gene expression. J. Mol. Endocrinol. 2006, 36, 261–277. [Google Scholar] [CrossRef]
- Beyer, N.; da Silva, M.L.; Hand, B. Mesenchymal stem cells: Isolation in vitro expansion and characterization. Exp. Pharmacol. 2006, 174, 249–282. [Google Scholar]
- Langenbach, F.; Handschel, J. Effects of dexamethasone, ascorbic acid and β-glycerophosphate on the osteogenic differentiation of stem cells in vitro. Stem Cell Res. Ther. 2013, 4, 117. [Google Scholar] [CrossRef] [PubMed]
- Mattinzoli, D.; Messa, P.; Corbelli, A.; Ikehata, M.; Zennaro, C.; Armelloni, S.; Li, M.; Giardino, L.; Rastaldi, M.P. A novel model of in vitro osteogenesis induced by retinoic acid treatment. Eur. Cells Mater. 2012, 24, 403–425. [Google Scholar] [CrossRef]
- Mattinzoli, D.; Messa, P.; Corbelli, A.; Ikehata, M.; Mondini, A.; Zennaro, C.; Armelloni, S.; Li, M.; Giardino, L.; Rastaldi, M.P. Application of retinoic acid to obtain osteocytes cultures from primary mouse osteoblasts. J. Vis. Exp. 2014. [Google Scholar] [CrossRef] [PubMed]
- Canesi, M.; Girdano, R.; Lazzari, L.; Isalberti, M.; Isaia, I.U.; Benti, R.; Rampini, P.; Marotta, G.; Colombo, A.; Cereda, E.; et al. Finding a new therapeutic approach for no-option Parkinsonisms: Mesenchimal stromal cells for progressive suprenuclear palsy. J. Transl. Med. 2016, 14, 127. [Google Scholar] [CrossRef] [PubMed]
- Montelatici, E.; Baluce, B.; Ragni, E.; Lavazza, C.; Parazzi, V.; Mazzola, R.; Cantarella, G.; Brambilla, M.; Giordano, R.; Lazzari, L. Defining the identity of human adipose-derived mesenchymal stem cells. Biochem. Cell Biol. 2015, 93, 74–82. [Google Scholar] [CrossRef]
- Barilani, M.; Lavazza, C.; Viganò, M.; Montemurro, T.; Boldrin, V.; Parazzi, V.; Montelatici, E.; Crosti, M.; Moro, M.; Giordano, R.; et al. Dissection of the cord blood stromal component reveals predictive parameters for culture outcome. Stem Cells Dev. 2015, 24, 104–114. [Google Scholar] [CrossRef] [PubMed]
- Barilani, M.; Lavazza, C.; Boldrin, V.; Ragni, E.; Parazzi, V.; Crosti, M.; Montelatici, E.; Giordano, R.; Lazzari, L. A Chemically Defined Medium-Based Strategy to Efficiently Generate Clinically Relevant Cord Blood Mesenchymal Stromal Colonies. Cell Transplant. 2016, 25, 1501–1514. [Google Scholar] [CrossRef]
- Jahnen-Dechent, W.; Schinke, T.; Trindl, A.; Müller-Esterel, W.; Sablitzky, F.; Kaiser, S.; Blessing, M. Cloning and targeted Deletion of the Mouse Fetuin Gene. J. Biol. Chem. 1997, 272, 31496–31503. [Google Scholar] [CrossRef]
- Banine, F.; Gangneux, C.; Mercier, L.; Le Cam, A.; Salier, J.P. Positive and negative elements modulate the promoter of the human liver-specific a2-HS-glycoprotein gene. Eur. J. Biochem. 2000, 267, 1214–1222. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(− delta delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mattinzoli, D.; Ikehata, M.; Tsugawa, K.; Alfieri, C.M.; Barilani, M.; Lazzari, L.; Andreetta, P.; Elli, F.M.; Mantovani, G.; Messa, P. FGF23 and Fetuin-A Interaction and Mesenchymal Osteogenic Transformation. Int. J. Mol. Sci. 2019, 20, 915. https://doi.org/10.3390/ijms20040915
Mattinzoli D, Ikehata M, Tsugawa K, Alfieri CM, Barilani M, Lazzari L, Andreetta P, Elli FM, Mantovani G, Messa P. FGF23 and Fetuin-A Interaction and Mesenchymal Osteogenic Transformation. International Journal of Molecular Sciences. 2019; 20(4):915. https://doi.org/10.3390/ijms20040915
Chicago/Turabian StyleMattinzoli, Deborah, Masami Ikehata, Koji Tsugawa, Carlo M. Alfieri, Mario Barilani, Lorenza Lazzari, Paola Andreetta, Francesca M. Elli, Giovanna Mantovani, and Piergiorgio Messa. 2019. "FGF23 and Fetuin-A Interaction and Mesenchymal Osteogenic Transformation" International Journal of Molecular Sciences 20, no. 4: 915. https://doi.org/10.3390/ijms20040915
APA StyleMattinzoli, D., Ikehata, M., Tsugawa, K., Alfieri, C. M., Barilani, M., Lazzari, L., Andreetta, P., Elli, F. M., Mantovani, G., & Messa, P. (2019). FGF23 and Fetuin-A Interaction and Mesenchymal Osteogenic Transformation. International Journal of Molecular Sciences, 20(4), 915. https://doi.org/10.3390/ijms20040915







