Proteins and Signaling Pathways Response to Dry Needling Combined with Static Stretching Treatment for Chronic Myofascial Pain in a RAT Model: An Explorative Proteomic Study
Abstract
1. Introduction
2. Results
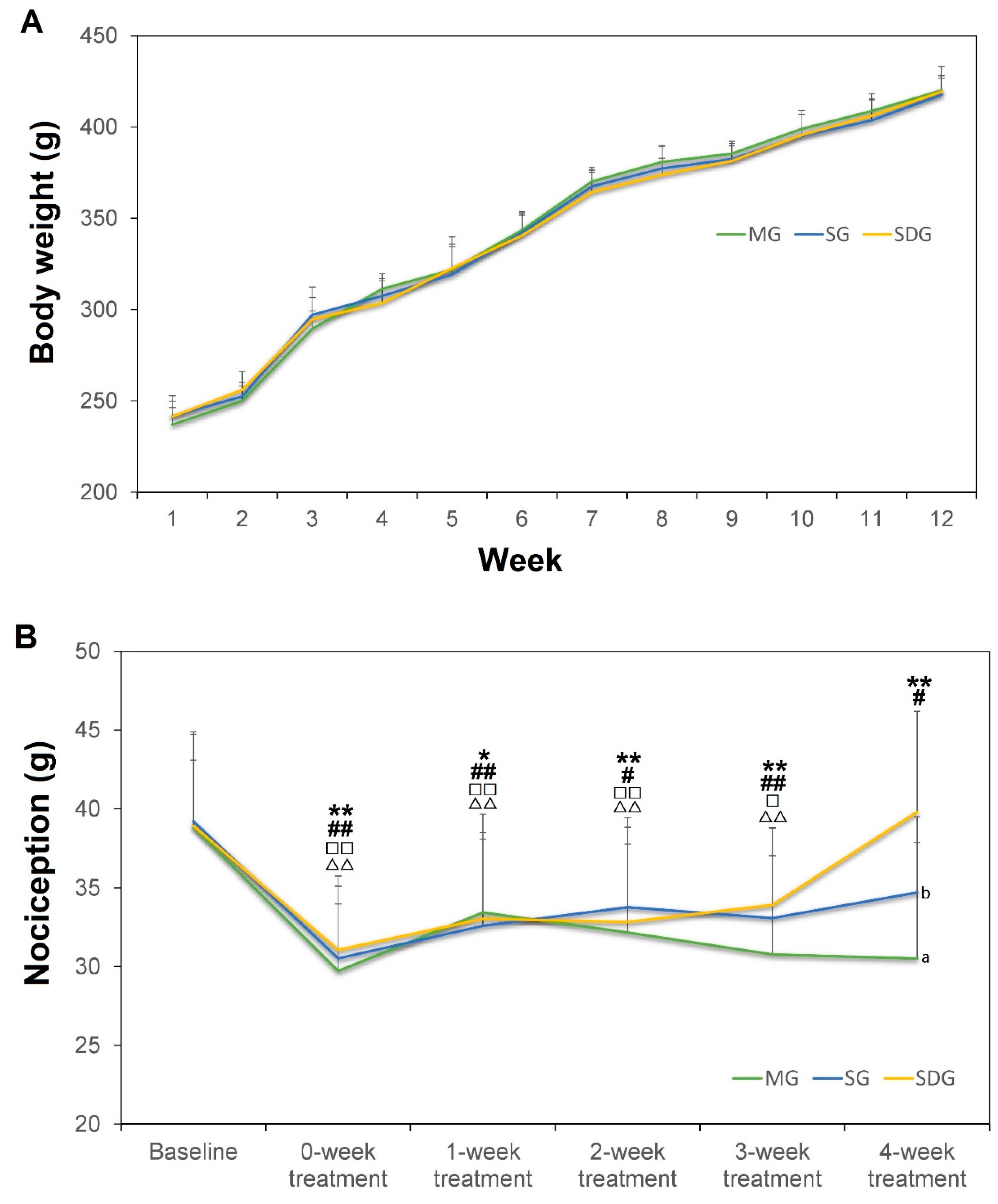
2.1. Body Weight and the Nociception Value
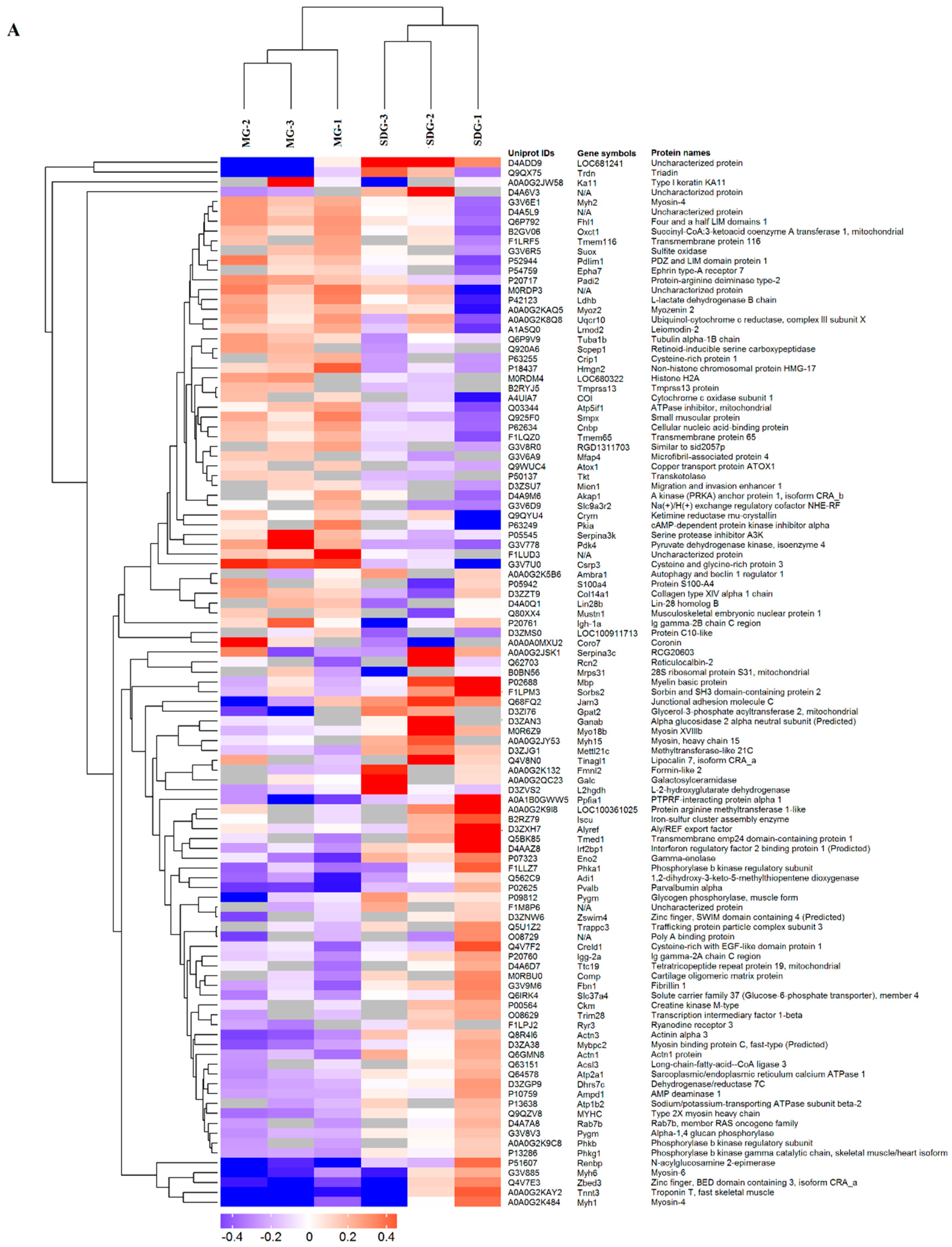
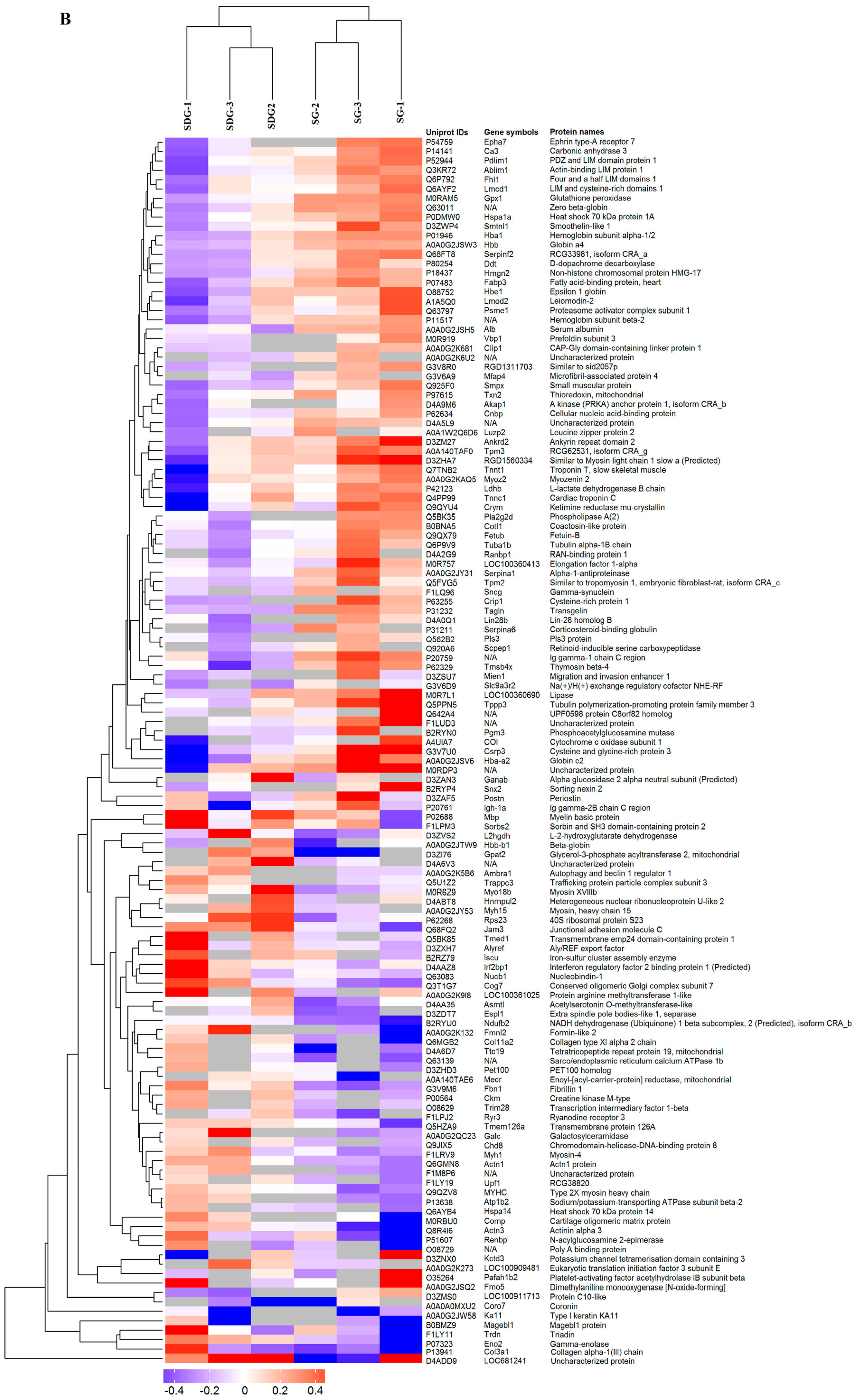
2.2. Hierarchical Cluster
2.3. GO and KEGG Pathway Enrichment
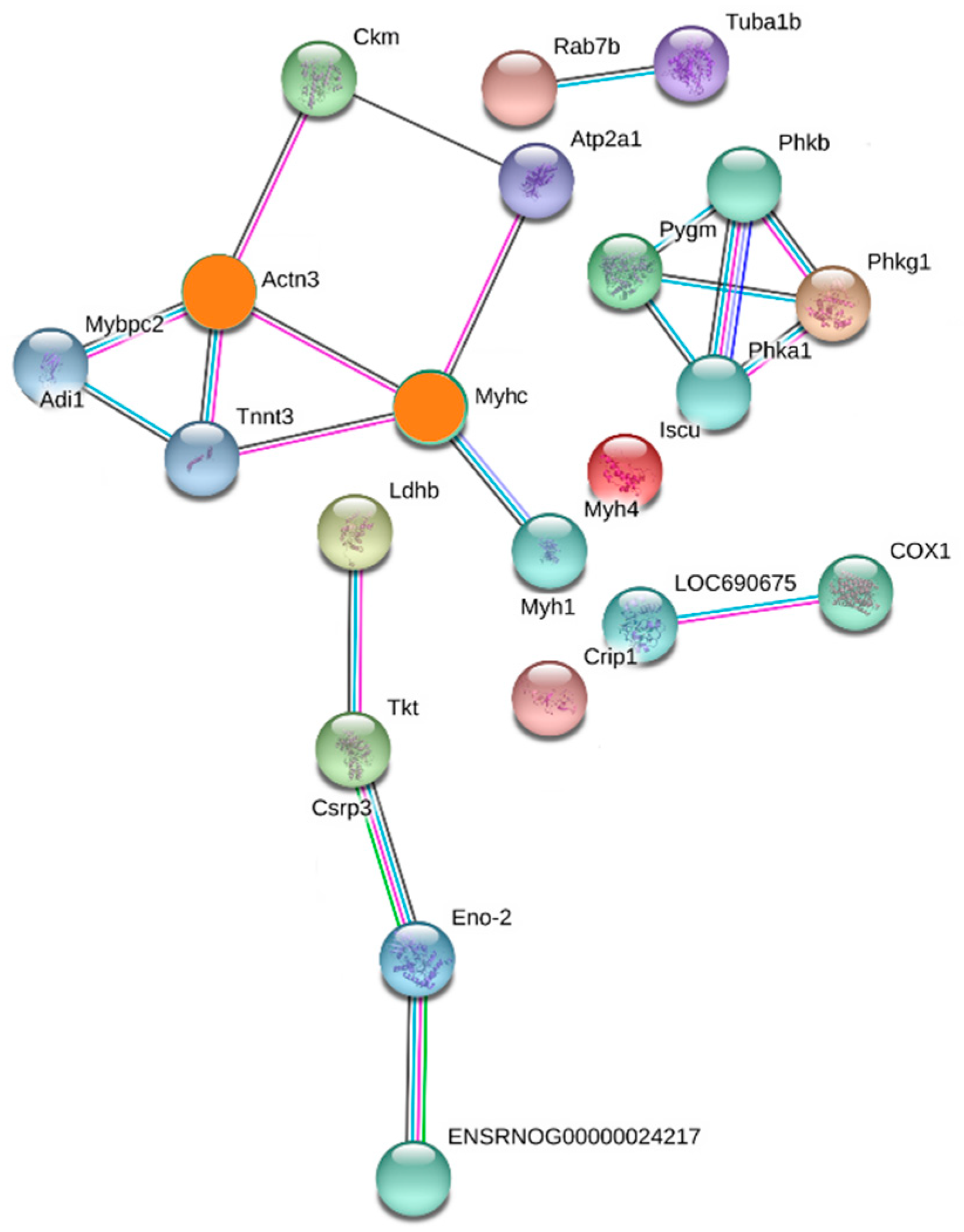
2.4. PPI Analysis
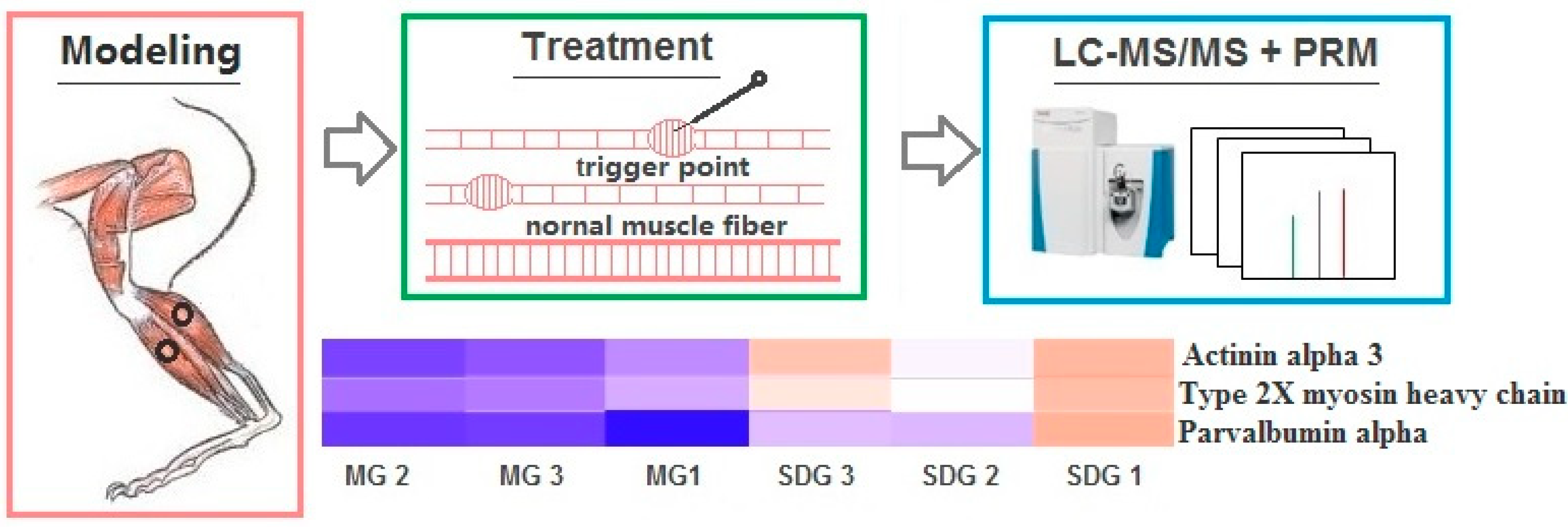
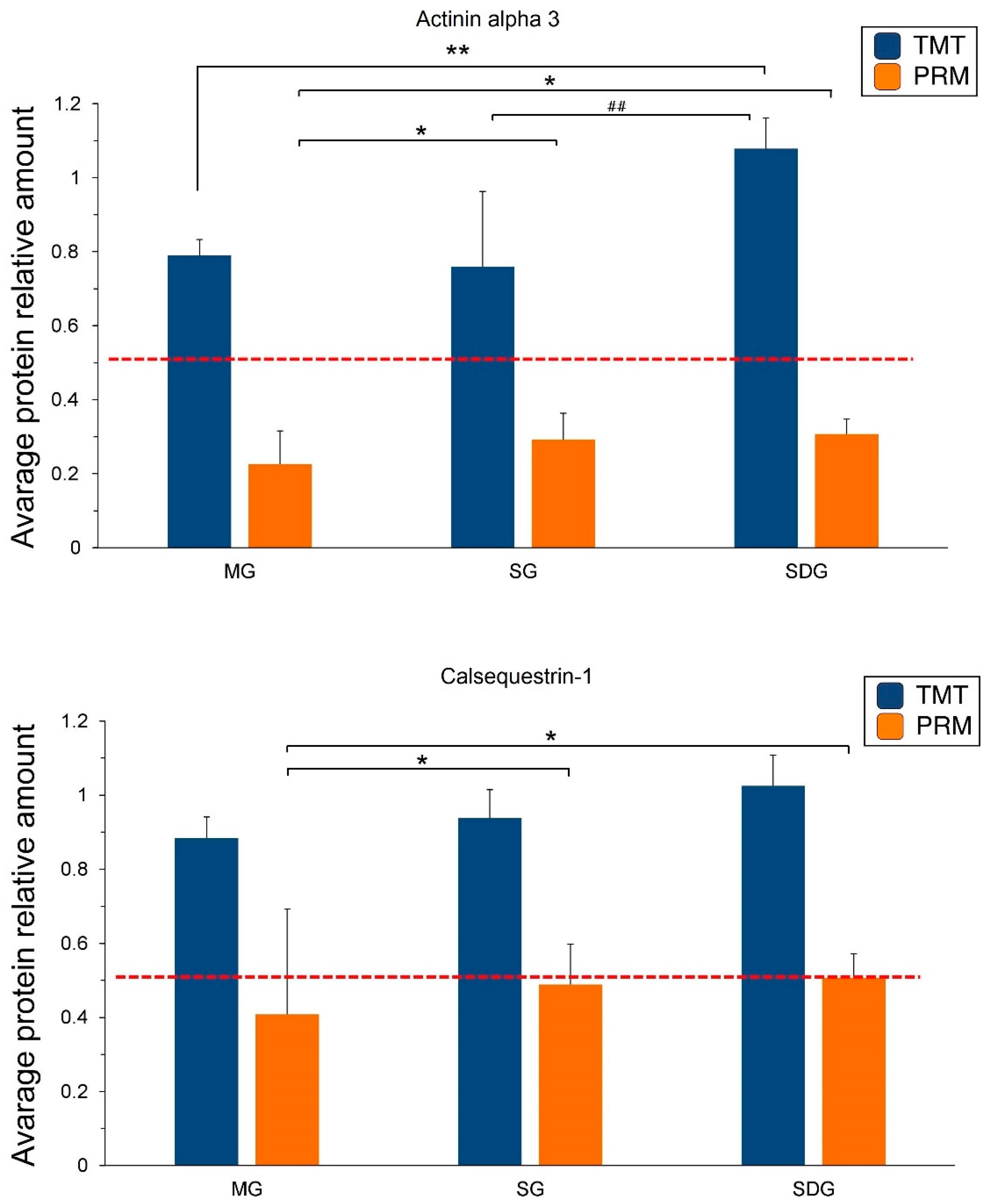
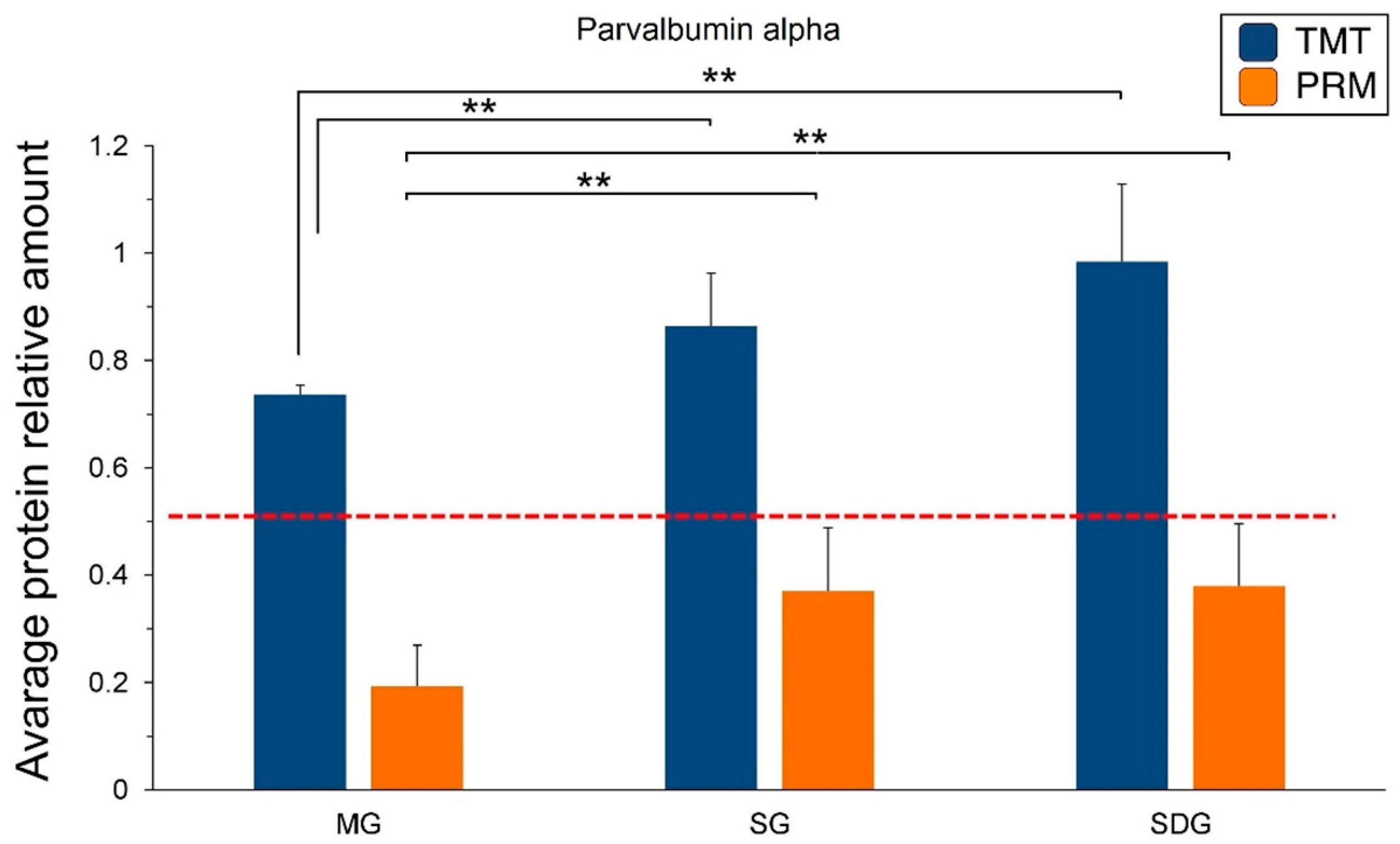
2.5. PRM Validation of TMT-Based Results
3. Discussion
4. Materials and Methods
4.1. MTrPs Modeling
4.2. Static Stretching and Dry Needling Treatment
4.3. Proteomics Analysis
4.3.1. Tandem Mass Tag (TMT) Sample Preparation
4.3.2. LC-MS/MS Analysis
4.3.3. Bioinformatics Analysis
4.4. Parallel Reaction Monitoring (PRM) Validation
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Barbero, M.; Fernández-de-Las-Peñas, C.; Palacios-Ceña, M.; Cescon, C.; Falla, D. Pain extent is associated with pain intensity but not with widespread pressure or thermal pain sensitivity in women with fibromyalgia syndrome. Clin. Rheumatol. 2017, 36, 1427–1432. [Google Scholar] [CrossRef] [PubMed]
- Ris, I.; Barbero, M.; Falla, D.; Larsen, M.H.; Kraft, M.N.; Søgaard, K.; Juul-Kristensen, B. Pain extent is more strongly associated with disability, psychological factors, and neck muscle function in people with non-traumatic versus traumatic chronic neck pain: A cross sectional study. Eur. J. Phys. Rehabil. Med. 2018. [Google Scholar] [CrossRef]
- Zhang, H.; Lü, J.J.; Huang, Q.M.; Liu, L.; Liu, Q.G.; Eric, O.A. Histopathological nature of myofascial trigger points at different stages of recovery from injury in a rat model. Acupunct. Med. 2017, 35, 445–451. [Google Scholar] [CrossRef] [PubMed]
- Tuzun, E.H.; Gildir, S.; Angin, E.; Tecer, B.H.; Dana, K.Ö.; Malkoç, M. Effectiveness of dry needling versus a classical physiotherapy program in patients with chronic low-back pain: A single-blind, randomized, controlled trial. J. Phys. Ther. Sci. 2017, 29, 1502–1509. [Google Scholar] [CrossRef] [PubMed]
- Jafari, M.; Bahrpeyma, F.; Togha, M. Effect of ischemic compression for cervicogenic headache and elastic behavior of active trigger point in the sternocleidomastoid muscle using ultrasound imaging. J. Bodyw. Mov. Ther. 2017, 21, 933–939. [Google Scholar] [CrossRef] [PubMed]
- Bron, C.; Dommerholt, J.D. Etiology of myofascial trigger points. Curr. Pain Headache Rep. 2012, 16, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Mense, S.; Simons, D.G.; Hoheisel, U.; Quenzer, B. Lesions of rat skeletal muscle after local block of acetylcholinesterase and neuromuscular stimulation. J. Appl. Physiol. 2003, 94, 2494–2501. [Google Scholar] [CrossRef] [PubMed]
- Akamatsu, F.E.; Ayres, B.R.; Saleh, S.O.; Hojaij, F.; Andrade, M.; Hsing, W.T.; Jacomo, A.L. Trigger points: An anatomical substratum. BioMed Res. Int. 2015, 2015, 623287. [Google Scholar] [CrossRef]
- Chen, Q.; Wang, H.J.; Gay, R.E.; Thompson, J.M.; Manduca, A.; An, K.N.; Ehman, R.E.; Basford, J.R. Quantification of myofascial taut bands. Arch. Phys. Med. Rehabil. 2016, 97, 67–73. [Google Scholar] [CrossRef]
- Akamatsu, F.E.; Saleh, S.; Pinesi, H.T.; Itezerote, A.M.; Hojaij, F.; Andrade, M.; Hsing, W.T.; Jacomo, A.L. Anatomical basis of the myofascial trigger points of the gluteus maximus muscle. BioMed Res. Int. 2017, 2017, 4821968. [Google Scholar] [CrossRef]
- Liu, L.; Huang, Q.M.; Liu, Q.G.; Thitham, N.; Li, L.H.; Ma, Y.T.; Zhao, J.M. Evidence for dry needling in the management of myofascial trigger points associated with low back pain: A systematic review and meta-analysis. Arch. Phys. Med. Rehabil. 2018, 99, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Cerezotéllez, E.; Torreslacomba, M.; Fuentesgallardo, I.; Perez-Muñoz, M.; Mayoral-Del-Moral, O.; Lluch-Girbés, E.; Prieto-Valiente, L.; Falla, D. Effectiveness of dry needling for chronic nonspecific neck pain: A randomized, single-blinded, clinical trial. Pain 2016, 157, 1905–1917. [Google Scholar] [CrossRef] [PubMed]
- Stoop, R.; Clijsen, R.; Leoni, D.; Soldini, E.; Castellini, G.; Redaelli, V.; Barbero, M. Evolution of the methodological quality of controlled clinical trials for myofascial trigger point treatments for the period 1978–2015: A systematic review. Musculoskelet. Sci. Pract. 2017, 30, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Arias-Buría, J.L.; Fernández-De-Las-Peñas, C.; Palacios-Ceña, M.; Koppenhaver, S.L.; Salom-Moreno, J. Exercises and dry needling for subacromial pain syndrome: A randomized parallel-group trial. J. Pain 2017, 18, 11–18. [Google Scholar] [CrossRef]
- Martínpintado, Z.A.; Rodríguezfernández, Á.L.; Garcíamuro, F.; López-López, A.; Mayoral, O.; Mesa-Jiménez, J.; Fernández-Carnero, J. Effects of spray and stretch on postneedling soreness and sensitivity after dry needling of a latent myofascial trigger point. Arch. Phys. Med. Rehabil. 2014, 195, 1925–1932. [Google Scholar] [CrossRef]
- Budini, F.; Gallasch, E.; Christova, M.; Rafolt, D.; Rauscher, A.B.; Tilp, M. One minute static stretch of plantar flexors transiently increases H reflex excitability and exerts no effect on corticospinal pathways. Exp. Physiol. 2017, 102, 901–910. [Google Scholar] [CrossRef]
- Simons, D.G. New views of myofascial trigger points: Etiology and diagnosis. Arch. Phys. Med. Rehabil. 2008, 89, 157–159. [Google Scholar] [CrossRef]
- Gerwin, R.D.; Dommerholt, J.; Shah, J.P. An expansion of Simons’ integrated hypothesis of trigger point formation. Curr. Pain Headache Rep. 2004, 8, 468–475. [Google Scholar] [CrossRef]
- Hsieh, Y.L.; Yang, C.C.; Liu, S.Y.; Chou, L.W.; Hong, C.Z. Remote dose-dependent effects of dry needling at distant myofascial trigger spots of rabbit skeletal muscles on reduction of substance p levels of proximal muscle and spinal cords. BioMed Res. Int. 2014, 2014, 982121. [Google Scholar] [CrossRef]
- Shah, J.P.; Danoff, J.V.; Desai, M.J.; Parikh, S.; Nakamura, L.Y.; Phillips, T.M.; Gerber, L.H. Biochemicals associated with pain and inflammation are elevated in sites near to and remote from active myofascial trigger points. Arch. Phys. Med. Rehabil. 2008, 89, 157–159. [Google Scholar] [CrossRef]
- Liu, Q.G.; Liu, L.; Huang, Q.M.; Nguyen, T.T.; Ma, Y.T.; Zhao, J.M. Decreased spontaneous electrical activity and acetylcholine at myofascial trigger spots after dry needling treatment: A Pilot Study. Evid. Based Complement. Alternat. Med. 2017, 2017, 3938191. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.M.; Liu, L. Wet needling of myofascial trigger points in abdominal muscles for treatment of primary dysmenorrhoea. Acupunct. Med. 2014, 32, 346–349. [Google Scholar] [CrossRef]
- Ge, H.Y.; Serrao, M.; Andersen, O.K.; Graven-Nielsen, T.; Arendt-Nielsen, L. Increased H-reflex response induced by intramuscular electrical stimulation of latent myofascial trigger points. Acupunct. Med. 2009, 27, 150–154. [Google Scholar] [CrossRef] [PubMed]
- Catherman, A.D.; Skinner, O.S.; Kelleher, N.L. Top Down proteomics: Facts and perspectives. Biochem. Biophys. Res. Commun. 2014, 445, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Varela, D.; Barry, A.M.; Schmidt, M. Proteome-based systems biology in chronic pain. J. Proteom. 2018, 190, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Overall, C.M. Can proteomics fill the gap between genomics and phenotypes? J. Proteom. 2014, 100, 1–2. [Google Scholar] [CrossRef]
- Li, L.H.; Huang, Q.M.; Barbero, M.; Liu, L.; Nguyen, T.T.; Beretta-Piccoli, M.; Xu, A.L.; Ji, L.J. Quantitative proteomics analysis to identify biomarkers of chronic myofascial pain and therapeutic targets of dry needling in a rat model of myofascial trigger points. J. Pain Res. 2019, 12, 283–298. [Google Scholar] [CrossRef]
- Ruedi, A.; Burlingame, A.L.; Bradshaw, R.A. Western blots versus selected reaction monitoring assays: Time to turn the tables? Mol. Cell Proteom. 2013, 12, 2381–2382. [Google Scholar]
- Hackel, D.; Brack, A.; Fromm, M.; Rittner, H.L. Modulation of tight junction proteins in the perineurium for regional pain control. Ann. N. Y. Acad. Sci. 2012, 1257, 199–206. [Google Scholar] [CrossRef]
- Rittner, H.L.; Amasheh, S.; Moshourab, R.; Hackel, D.; Yamdeu, R.S.; Mousa, S.A.; Fromm, M.; Stein, C.; Brack, A. Modulation of tight junction proteins in the perineurium to facilitate peripheral opioid analgesia. Anesthesiology 2012, 116, 1323–1334. [Google Scholar] [CrossRef]
- Ronaldson, P.T.; Davis, T.P. Targeting blood-brain barrier changes during inflammatory pain: An opportunity for optimizing CNS drug delivery. Ther. Deliv. 2011, 2, 1015–1041. [Google Scholar] [CrossRef] [PubMed]
- Ronaldson, P.T.; Davis, T.P. Targeted drug delivery to treat pain and cerebral hypoxia. Pharmacol. Rev. 2013, 65, 291–314. [Google Scholar] [CrossRef]
- Feuerecker, M.; van Oosterhout, W.P.; Feuerecker, B.; Matzel, S.; Schelling, G.; Rehm, M.; Vein, A.A.; Choukèr, A. Headache under simulated microgravity is related to endocrine, fluid distribution, and tight junction changes. Pain 2016, 157, 1072–1078. [Google Scholar] [CrossRef] [PubMed]
- Garton, F.C.; Seto, J.T.; Quinlan, K.G.; Yang, N.; Houweling, P.J.; North, K.N. alpha-Actinin-3 deficiency alters muscle adaptation in response to denervation and immobilization. Hum. Mol. Genet. 2014, 23, 1879–1893. [Google Scholar] [CrossRef]
- Ichinoseki-Sekine, N.; Yoshihara, T.; Kakigi, R.; Ogura, Y.; Sugiura, T.; Naito, H. Fiber-type specific expression of alpha-actinin isoforms in rat skeletal muscle. Biochem. Biophys. Res. Commun. 2012, 419, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Paolini, C.; Quarta, M.; Pierre, L.; Michelucci, A.; Nori, A.; Reggiani, C.; Dirksen, R.T.; Protasi, F. Oxidative stress, mitochondrial damage, and cores in muscle from calsequestrin-1 knockout mice. Skelet. Muscle 2015, 5, 10. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, M.Q.; Wu, X.; Lazarus, M.; Cherasse, Y.; Yuan, M.Y.; Huang, Z.L.; Li, R.X. Activation of parvalbumin neurons in the rostro-dorsal sector of the thalamic reticular nucleus promotes sensitivity to pain in mice. Neuroscience 2017, 366, 113–123. [Google Scholar] [CrossRef]
- Petitjean, H.; Pawlowski, S.A.; Fraine, S.L.; Sharif, B.; Hamad, D.; Fatima, T.; Berg, J.; Brown, C.M.; Jan, L.Y.; Ribeiro-da-Silva, A.; et al. Dorsal horn parvalbumin neurons are gate-keepers of touch-evoked pain after nerve injury. Cell Rep. 2015, 13, 1246–1257. [Google Scholar] [CrossRef]
- Fernandez-Carnero, J.; Gilarranz-de-Frutos, L.; Leon-Hernandez, J.V.; Pecos-Martin, D.; Alguacil-Diego, I.; Gallego-Izquierdo, T.; Martín-Pintado-Zugasti, A. Effectiveness of different deep dry needling dosages in the treatment of patients with cervical myofascial pain: A pilot RCT. Am. J. Phys. Med. Rehabil. 2017, 96, 726–733. [Google Scholar] [CrossRef] [PubMed]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- MacLean, B.; Tomazela, D.M.; Shulman, N.; Chambers, M.; Finney, G.L.; Frewen, B.; Kern, R.; Tabb, D.L.; Liebler, D.C.; MacCoss, M.J. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010, 26, 966–968. [Google Scholar] [CrossRef] [PubMed]

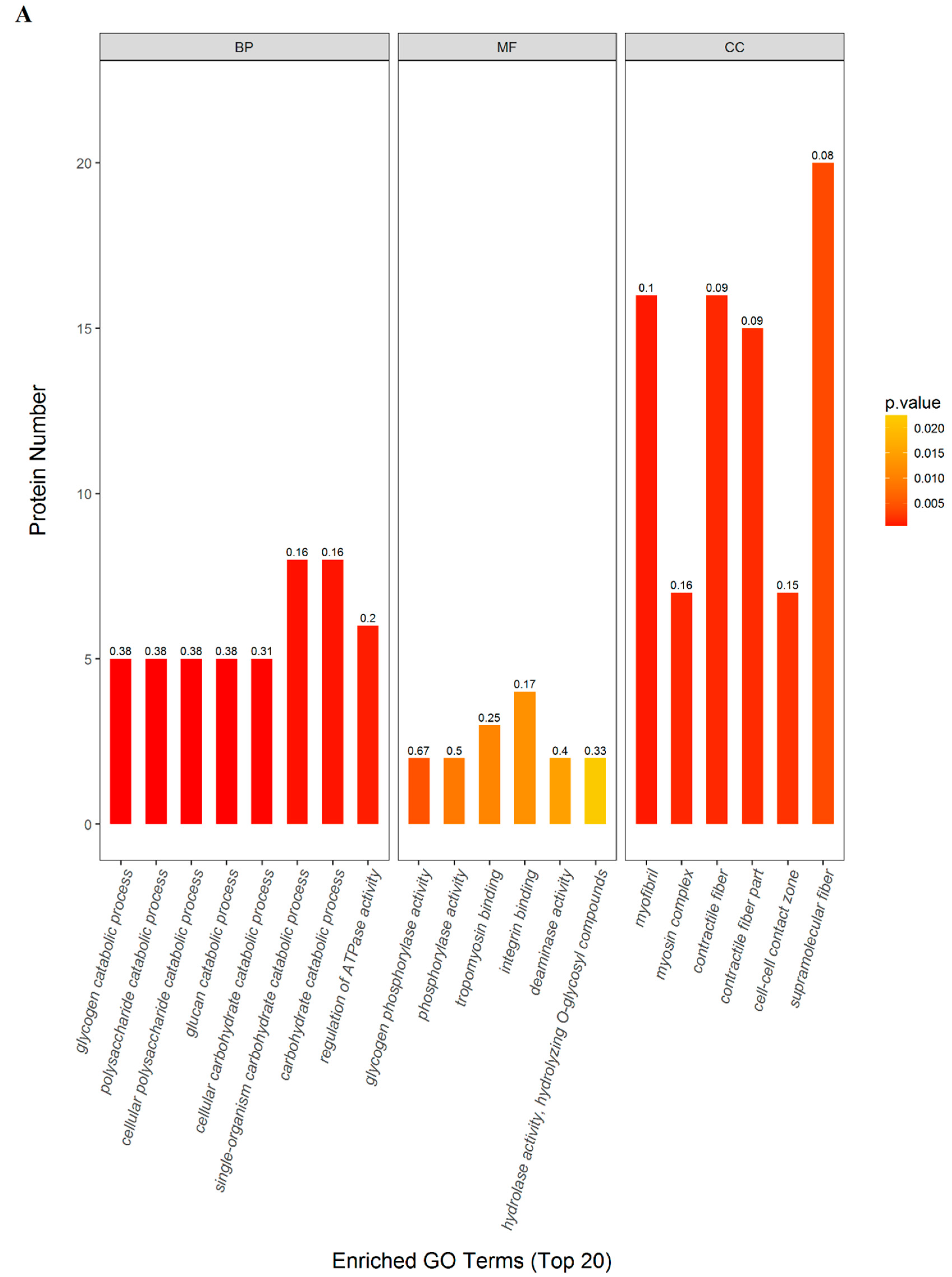
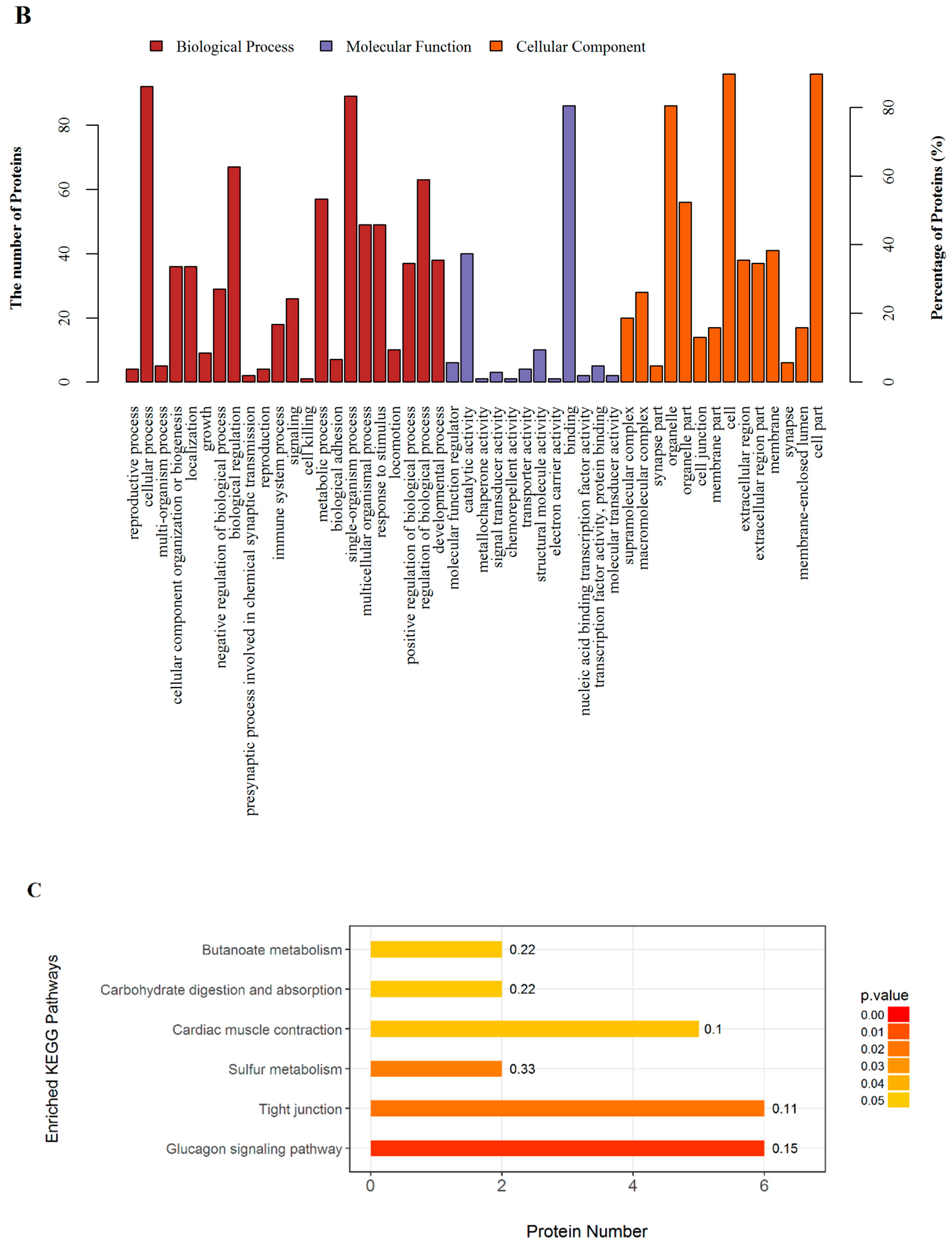
| Terms | Count | P value | FDR (%) | richFactor | Protein Names (Gene Names) |
| GO (gene ontology) | |||||
| (BP) Single-organism carbohydrate catabolic process | 8 | 0.0006 | 0.2982 | 0.1633 | Phosphorylase b kinase gamma catalytic chain, skeletal muscle/heart isoform (Phkg1); Phosphorylase b kinase regulatory subunit (Phka1); Phosphorylase b kinase regulatory subunit (Phkb); Glycogen phosphorylase, muscle form (Pygm); L-lactate dehydrogenase B chain (Ldhb); Actinin alpha 3 (Actn3); Alpha-1,4 glucan phosphorylase (Pygm); Gamma-enolase (Eno2) |
| (MF) Integrin binding | 4 | 0.0125 | 0.4795 | 0.1739 | Actinin alpha 3 (Actn3); Fibrillin 1 (Fbn1); Actn1 protein (Actn1); Junctional adhesion molecule C (Jam3) |
| (CC) Supramolecular fiber | 20 | 0.0040 | 0.4795 | 0.0755 | Type I keratin KA11 (Ka11); Actinin alpha 3 (Actn3); Troponin T, fast skeletal muscle (Tnnt3); Myosin-4 (Myh2); Fibrillin 1 (Fbn1); Small muscular protein (Smpx); Actn1 protein (Actn1); Tubulin alpha-1B chain (Tuba1b); Myosin, heavy chain 15 (Myh15); Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 (Atp2a1); Type 2X myosin heavy chain (Myhc); Myosin-4 (Myh1); Myosin-6 (Myh6); Cysteine and glycine-rich protein 3 (Csrp3); Uncharacterized protein; Leiomodin-2 (Lmod2); Myosin binding protein C, fast-type (Mybpc2); Microfibril-associated protein 4 (Mfap4); Myozenin 2 (Myoz2); Ryanodine receptor 3 (Ryr3) |
| Terms | Count | P value | FDR (%) | richFactor | Protein Names (Gene Names) |
| KEGG (kyoto encyclopedia of genes and genomes) pathways | |||||
| Tight junction | 6 | 0.0189 | 0.7500 | 0.1132 | Myosin-4 (Myh2); Tubulin alpha-1B chain (Tuba1b); Myosin-4 (Myh1); Junctional adhesion molecule C (Jam3); Actn1 protein (Actn1); Myosin, heavy chain 15 (Myh15) |
| Glucagon signaling pathway | 6 | 0.0042 | 0.4642 | 0.1538 | L-lactate dehydrogenase B chain (Ldhb); Protein arginine methyltransferase 1-like (LOC100361025); Glycogen phosphorylase, muscle form (Pygm); Phosphorylase b kinase regulatory subunit (Phka1); Alpha-1,4 glucan phosphorylase (Pygm); Phosphorylase b kinase gamma catalytic chain, skeletal muscle/heart isoform (Phkg1) |
| Sulfur metabolism | 2 | 0.0220 | 0.7501 | 0.3333 | Uncharacterized protein; Sulfite oxidase (Suox) |
| Cardiac muscle contraction | 5 | 0.0464 | 0.7501 | 0.1020 | Cytochrome c oxidase subunit 1 (COI); Ubiquinol-cytochrome c reductase, complex III subunit X (Uqcr10); Myosin-6 (Myh6); Sodium/potassium-transporting ATPase subunit beta-2 (Atp1b2); Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 (Atp2a1) |
| Carbohydratedigestion and absorption | 2 | 0.0488 | 0.7501 | 0.2222 | Sodium/potassium-transporting ATPase subunit beta-2 (Atp1b2); Solute carrier family 37 (Glucose-6-phosphate transporter), member 4 (Slc37a4) |
| Butanoate metabolism | 2 | 0.0488 | 0.7501 | 0.2222 | Succinyl-CoA:3-ketoacid coenzyme A transferase 1, mitochondrial (Oxct1); L-2-hydroxyglutarate dehydrogenase (L2hgdh) |
| Degree | Uniprot ID | Protein Name | Gene Name |
|---|---|---|---|
| Actinin alpha 3 (4-connected) | D3ZA38 | Myosin binding protein C, fast-type | Mybpc2 |
| P00564 | Creatine kinase M-type | Ckm | |
| Q9QZV8 | Type 2X myosin heavy chain | Myhc | |
| A0A0G2KAY2 | Troponin T, fast skeletal muscle | Tnnt3 | |
| Type 2X myosin heavy chain (4-connected) | A0A0G2KAY2 | Troponin T, fast skeletal muscle | Tnnt3 |
| Q8R4I6 | Actinin alpha 3 | Actn3 | |
| Q64578 | ATPase 1 | Atp2a1 | |
| A0A0G2K484 | Myosin-4 | Myh1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.; Huang, Q.; Barbero, M.; Liu, L.; Nguyen, T.; Xu, A.; Ji, L. Proteins and Signaling Pathways Response to Dry Needling Combined with Static Stretching Treatment for Chronic Myofascial Pain in a RAT Model: An Explorative Proteomic Study. Int. J. Mol. Sci. 2019, 20, 564. https://doi.org/10.3390/ijms20030564
Li L, Huang Q, Barbero M, Liu L, Nguyen T, Xu A, Ji L. Proteins and Signaling Pathways Response to Dry Needling Combined with Static Stretching Treatment for Chronic Myofascial Pain in a RAT Model: An Explorative Proteomic Study. International Journal of Molecular Sciences. 2019; 20(3):564. https://doi.org/10.3390/ijms20030564
Chicago/Turabian StyleLi, Lihui, Qiangmin Huang, Marco Barbero, Lin Liu, Thitham Nguyen, Anle Xu, and Lijuan Ji. 2019. "Proteins and Signaling Pathways Response to Dry Needling Combined with Static Stretching Treatment for Chronic Myofascial Pain in a RAT Model: An Explorative Proteomic Study" International Journal of Molecular Sciences 20, no. 3: 564. https://doi.org/10.3390/ijms20030564
APA StyleLi, L., Huang, Q., Barbero, M., Liu, L., Nguyen, T., Xu, A., & Ji, L. (2019). Proteins and Signaling Pathways Response to Dry Needling Combined with Static Stretching Treatment for Chronic Myofascial Pain in a RAT Model: An Explorative Proteomic Study. International Journal of Molecular Sciences, 20(3), 564. https://doi.org/10.3390/ijms20030564





