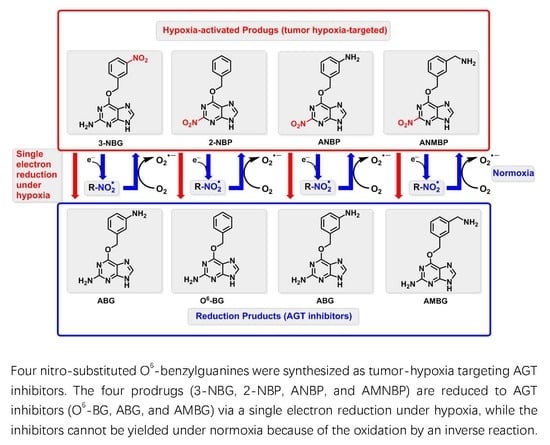
Reductive Activity and Mechanism of Hypoxia- Targeted AGT Inhibitors: An Experimental and Theoretical Investigation
Abstract
1. Introduction
2. Results and Discussion
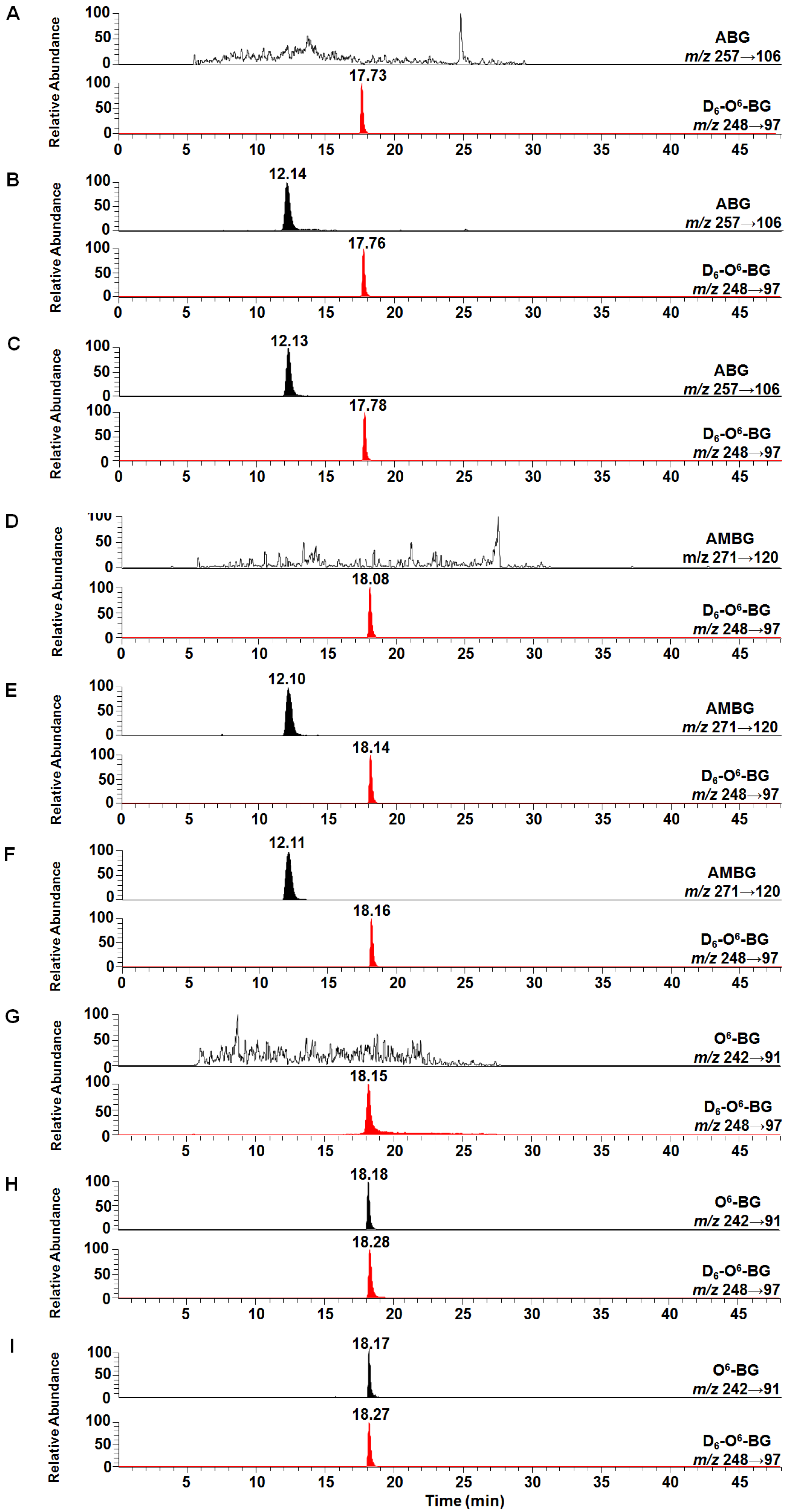
2.1. Reduction of 3-NBG, 2-NBP, ANBP, and AMNBP under Hypoxic and Normoxic Conditions
2.2. Quantum Chemistry Calculations
2.2.1. Mechanism of Single-Electron Reduction Using Nitrobenzene as A Model Compound
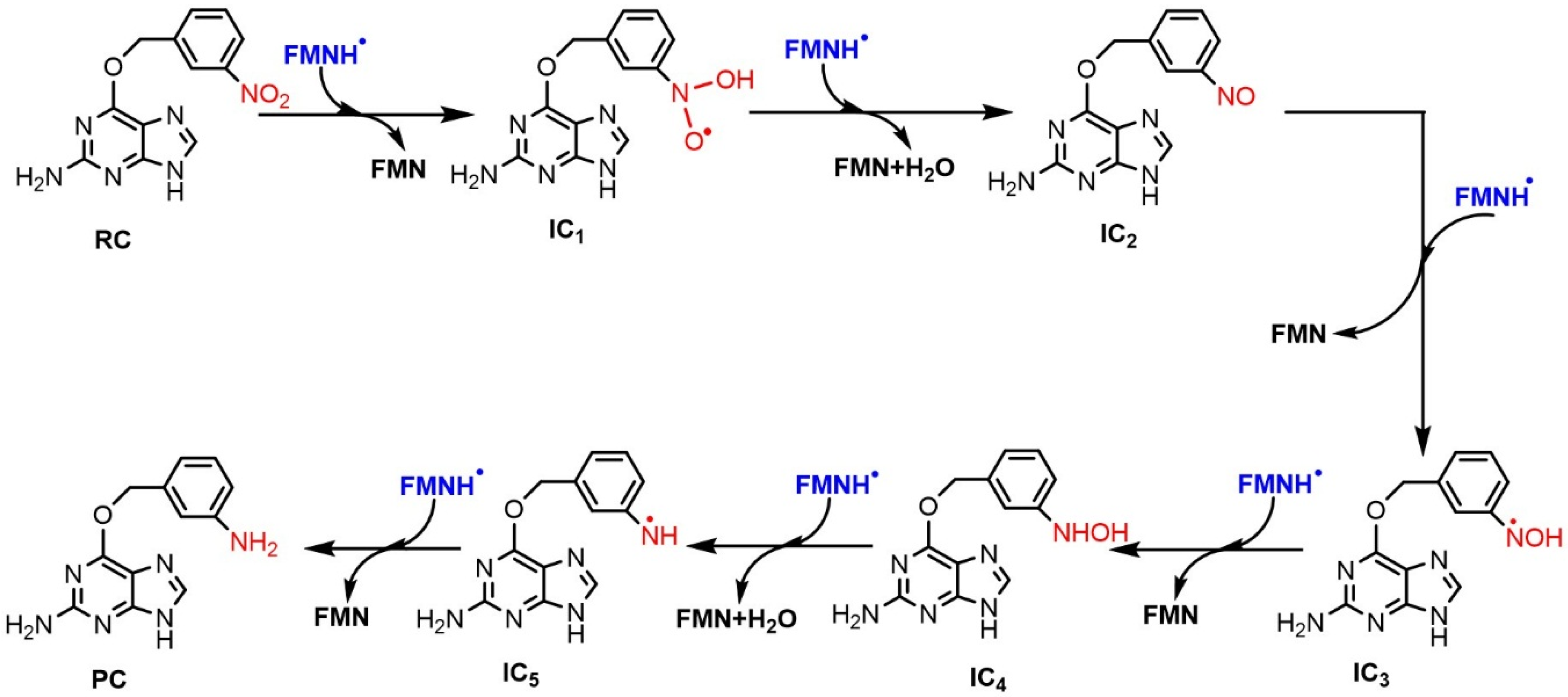
2.2.2. Reduction Mechanisms of 3-NBG, 2-NBP, ANBP, and AMNBP
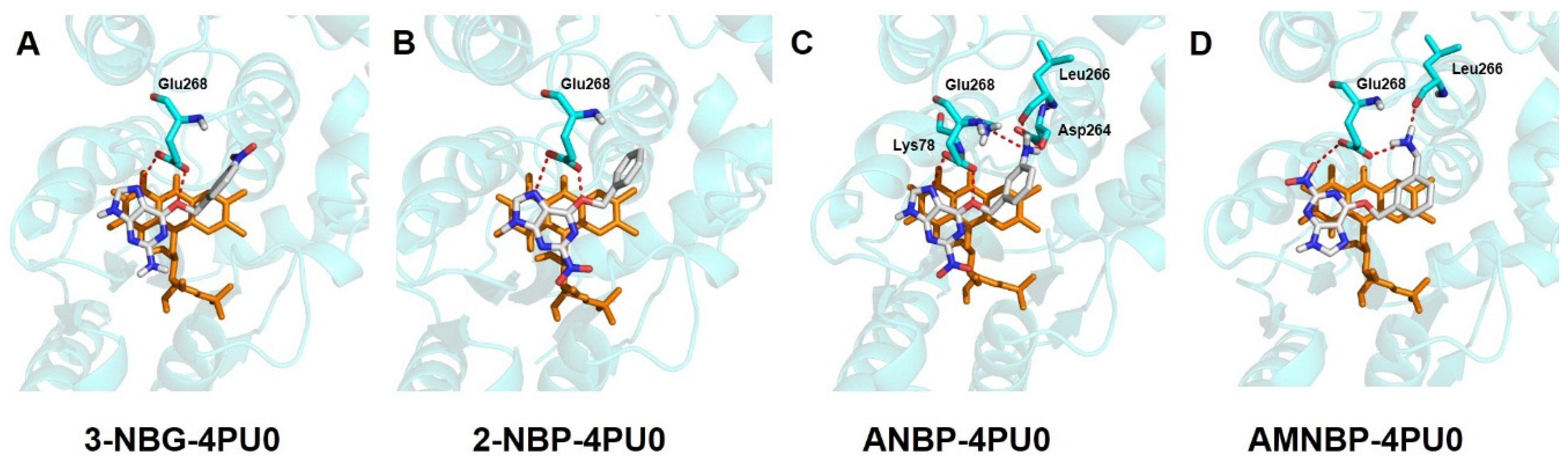
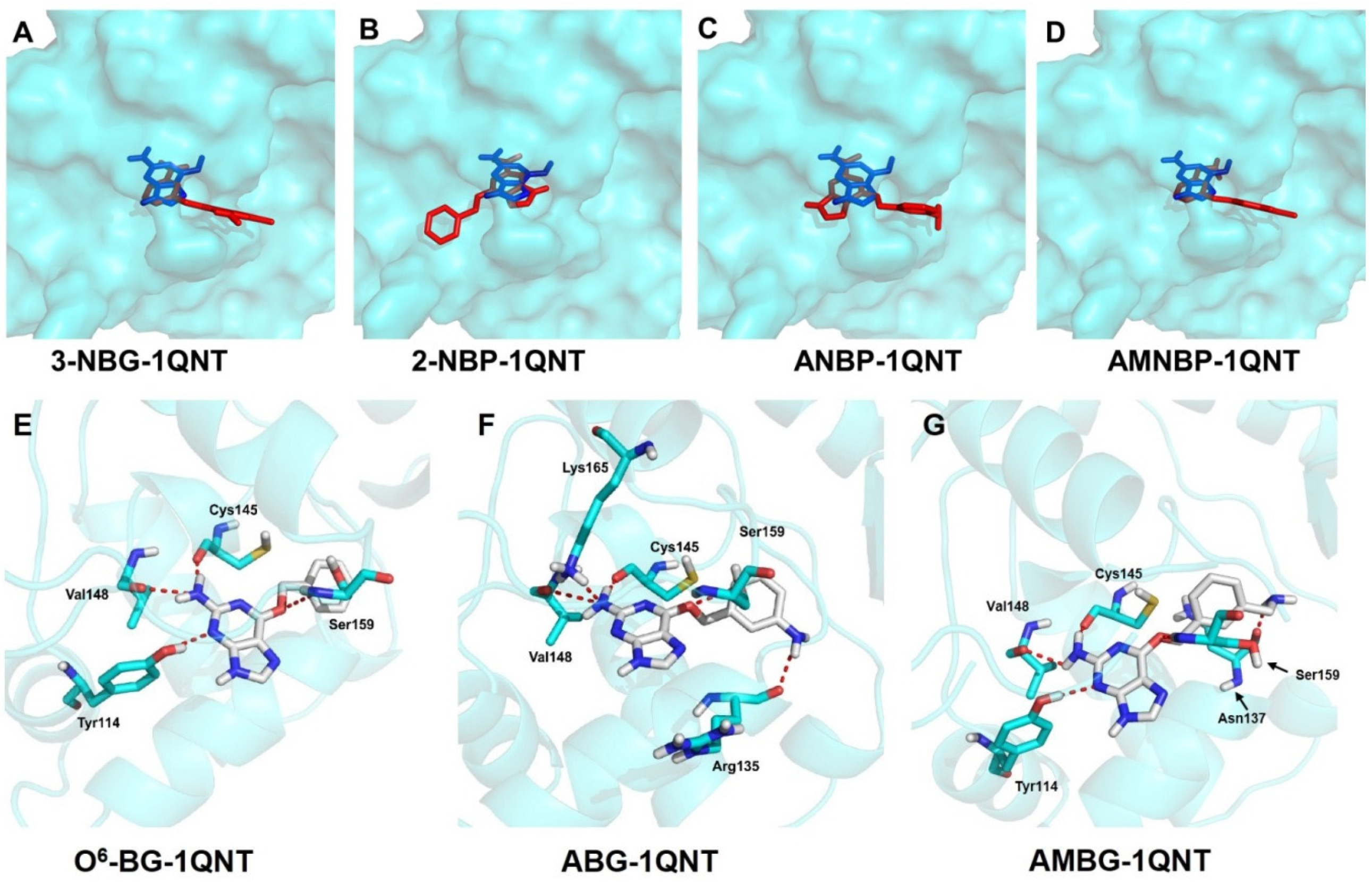
2.3. Docking Results
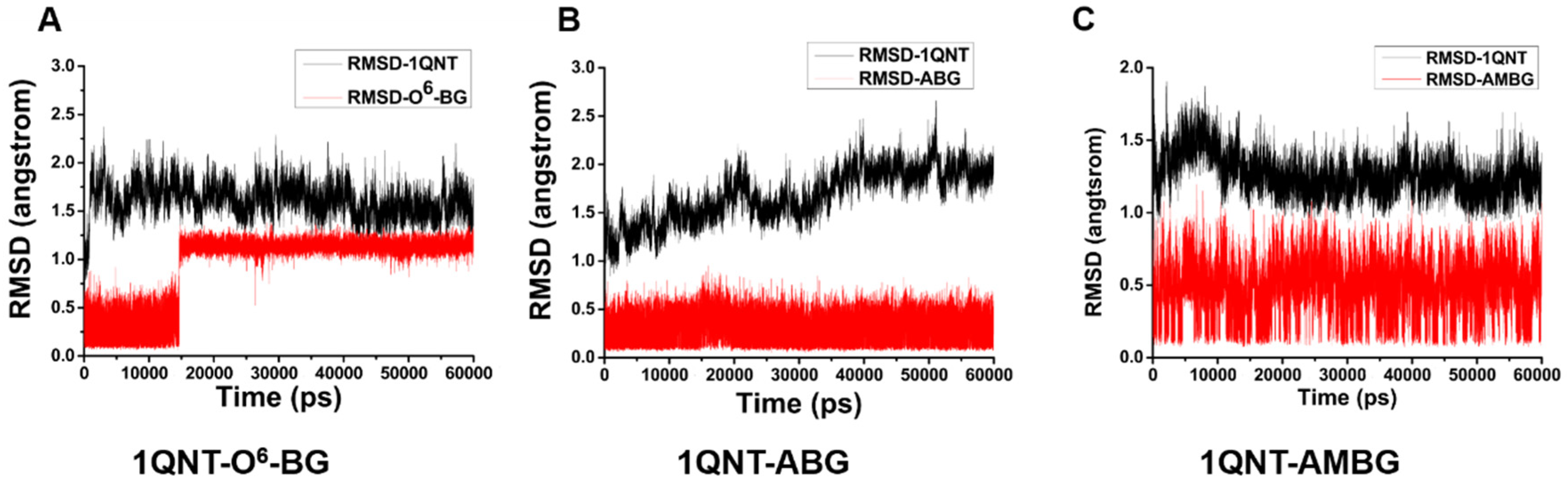
2.4. Molecular Dynamics Simulation and Free Energy Calculation
3. Materials and Methods
3.1. Experimental Study
3.1.1. Chemicals and Reagents
3.1.2. Chemical Syntheses
3.1.3. Reduction of the Four Prodrugs in Normoxic or Hypoxic Conditions
3.1.4. Determination of the Reduction Products by HPLC-ESI-MS/MS
3.2. Theoretical Study
3.2.1. Computational Methods
3.2.2. Molecular Docking Study
3.2.3. Molecular Dynamics Simulations
3.2.4. MM/PBSA Calculations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AGT | O6-alkylguanine-DNA alkyltransferase |
| O6-BG | O6-benzylguanine |
| DFT | Density functional theory |
| FMNH | Flavin mononucleotide |
| 3-NBG | O6-(3-nitro)benzylguanine |
| 2-NBP | 2-nitro-6-benzyloxypurine |
| ANBP | 2-nitro-6-(3-amino)benzyloxypurine |
| AMNBP | 2-nitro-6-(3-aminomethyl)benzyloxypurine |
| ABG | O6-(3-amino)benzylguanine |
| AMBG | O6-(3-aminomethyl)benzylguanine |
| HPLC-ESI-MS/MS | High-performance liquid chromatography electrospray ionization tandem mass spectrometry |
| TMZ | Temozolomide |
| BCNU | Carmustine |
| dG-dC | 1-[N3-deoxycytidyl]-2-[N1-deoxyguanosinyl]ethane |
| RCs | Reactant complexes |
| TSs | Transition states |
| ICs | Intermediate complexes |
| PCs | Product complexes |
References
- Gnewuch, C.T.; Sosnovsky, G.A. Critical appraisal of the evolution of N-nitrosoureas as anticancer drugs. Chem. Rev. 1997, 97, 829–1014. [Google Scholar] [CrossRef] [PubMed]
- Rajski, S.R.; Williams, R.M. DNA cross-linking agents as antitumor drugs. Chem. Rev. 1998, 98, 2723–2796. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.H.; Zhao, L.J.; Zhong, R.G.; Peng, Y.Z. The specific role of O6-methylguanine-DNA methyltransferase inhibitors in cancer chemotherapy. Future Med. Chem. 2018, 10, 1971–1996. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.H.; Zhao, L.J.; Zhong, R.G. The induction and repair of DNA interstrand crosslinks and implications in cancer chemotherapy. Anti Cancer Agents Med. Chem. 2016, 16, 221–246. [Google Scholar]
- Bodell, W.J. DNA alkylation products formed by 1-(2-chloroethyl)-1-nitrosourea as molecular dosimeters of therapeutic response. J. Neuro Oncol. 2009, 91, 257–264. [Google Scholar] [CrossRef]
- Muniandy, P.A.; Liu, J.; Majumdar, A.; Liu, S.; Seidman, M.M. DNA interstrand crosslink repair in mammalian cells: Step by step. Crit. Rev. Biochem. Mol. 2010, 45, 23–49. [Google Scholar] [CrossRef]
- Deans, A.J.; West, S.C. DNA interstrand crosslink repair and cancer. Nat. Rev. Cancer 2011, 11, 467–480. [Google Scholar] [CrossRef]
- Zhao, L.J.; Ma, X.Y.; Zhong, R.G. Comparative theoretical investigation of the formation of DNA interstrand crosslinks induced by two kinds of N-nitroso compounds: Nitrosoureas and nitrosamines. J. Phys. Org. Chem. 2012, 25, 1153–1167. [Google Scholar] [CrossRef]
- Sun, G.H.; Zhao, L.J.; Fan, T.J.; Li, S.S.; Zhong, R.G. Investigations on the effect of O6-benzylguanine on the formation of dG-dC interstrand cross-links induced by chloroethylnitrosoureas in human glioma cells using stable isotope dilution high-performance liquid chromatography electrospray ionization tandem mass spectrometry. Chem. Res. Toxicol. 2014, 27, 1253–1262. [Google Scholar]
- Pegg, A.E. Mammalian O6-alkylguanine-DNA alkyltransferase: Regulation and importance in response to alkylating carcinogenic and therapeutic agents. Cancer Res. 1990, 50, 6119–6129. [Google Scholar]
- Limp-Foster, M.; Kelley, M.R. DNA repair and gene therapy: Implications for translational uses. Environ. Mol. Mutagen. 2000, 35, 71–81. [Google Scholar] [CrossRef]
- Kokkinakis, D.M.; Bocangel, D.B.; Schold, S.C.; Moschel, R.C.; Pegg, A.E. Thresholds of O6-alkylguanine-DNA alkyltransferase which significant resistance of human glial tumor xenografts to treatment with 1,3-bis(2-chloroethyl)-1-nitrosourea or temozolomide. Clin. Cancer Res. 2001, 7, 421–428. [Google Scholar] [PubMed]
- Bacolod, M.D.; Johnson, S.P.; Ali-Osman, F.; Modrich, P.; Bullock, N.S.; Colvin, O.M.; Bigner, D.D.; Friedman, H.S. Mechanisms of resistance to 1, 3-bis(2-chloroethyl)-1-nitrosoureain human medulloblastoma and rhabdomyosarcoma. Mol. Cancer Ther. 2002, 1, 727–736. [Google Scholar] [PubMed]
- Ishiguro, K.; Zhu, Y.; Shyam, K.; Penketh, P.G.; Baumann, R.P.; Sartorelli, A.C. Quantitative relationship between guanine O6-alkyl lesions produced by onrigin™ and tumor resistance by O6-alkylguanine-DNA alkyltransferase. Biochem. Pharmacol. 2010, 80, 1317–1325. [Google Scholar] [CrossRef]
- Pegg, A.E. Multifaceted roles of alkyltransferase and related proteins in DNA repair, DNA damage, resistance to chemotherapy, and research tools. Chem. Res. Toxicol. 2011, 24, 618–639. [Google Scholar] [CrossRef]
- Moschel, R.C.; Mcdougall, M.G.; Dolan, M.E.; Stine, L.; Pegg, A.E. Structural features of substituted purine derivatives compatible with depletion of human O6-alkylguanine-DNA alkyltransferase. J. Med. Chem. 1992, 35, 4486–4491. [Google Scholar] [CrossRef]
- Chae, M.Y.; McDougall, M.G.; Dolan, M.E.; Swebb, K.; Pegg, A.E.; Moschel, R.C. Substituted O6-benzylguanine derivatives and their inactivation of human O-6-alkylguanine-DNA alkyltransferase. J. Med. Chem. 1994, 37, 342–347. [Google Scholar] [CrossRef]
- Chae, M.Y.; Swenn, K.; Kanugula, S.; Dolan, M.E.; Pegg, A.E.; Moschel, R.C. 8-Substituted O6-benzylguanine, substituted 6(4)-(benzyloxy) pyrimidine, and related derivatives as inactivators of human O6-alkylguanine-DNA alkyltransferase. J. Med. Chem. 1995, 38, 359–365. [Google Scholar] [CrossRef]
- McElhinney, R.S.; Donnelly, D.J.; McCormick, J.E.; Kelly, J.; Watson, A.J.; Rafferty, J.A.; Elder, R.H.; Middleton, M.R.; Willington, M.A.; McMurry, T.B.H.; et al. Inactivation of O6-alkylguanine-DNA alkyltransferase. 1. Novel O6-(hetarylmethyl)guanines having basic rings in the side chain. J. Med. Chem. 1998, 41, 5265–5271. [Google Scholar] [CrossRef]
- Griffin, R.J.; Arris, C.E.; Bleasdale, C.; Boyle, F.T.; Calvert, A.H.; Curtin, N.J.; Dalby, C.; Kanugula, S.; Lembicz, N.K.; Newell, D.R.; et al. Resistance-modifying agents. 8. Inhibition of O6-alkylguanine-DNA alkyltransferase by O6-alkenyl-, O6-cycloalkenyl-, and O6-(2-oxoalkyl) guanines and potentiation of temozolomide cytotoxicity in vitro by O6-(1-cyclopentenylmethyl) guanine. J. Med. Chem. 2000, 43, 4071–4083. [Google Scholar] [CrossRef]
- Pauly, G.T.; Loktionova, N.A.; Fang, Q.M.; Vankayala, S.L.; Guida, W.C.; Pegg, A.E. Substitution of aminomethyl at the meta-position enhances the inactivation of O6-alkylguanine-DNA alkyltransferase by O6-benzylguanine. J. Med. Chem. 2008, 51, 7144–7153. [Google Scholar] [CrossRef] [PubMed]
- Lopez, S.; Margison, G.P.; McElhinney, R.S.; Cordeiro, A.; McMurry, T.B.H.; Rozas, I. Towards more specific O6-methylguanine-DNA methyltransferase (MGMT) inactivators. Bioorg. Med. Chem. 2011, 19, 1658–1665. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; La, X.X.; Li, J.T.; Yu, R.; Zhuang, Z.C.; Sun, G.H.; Cui, X.; Zhang, N.; Zhao, L.J.; Upadhyaya, P.; et al. NBGNU: A hypoxia-activated tripartite combi-nitrosourea prodrug overcoming AGT- mediated chemoresistance. Future Med. Chem. 2019, 11, 269–284. [Google Scholar] [CrossRef] [PubMed]
- Apisarnthanarax, N.; Wood, G.S.; Stevens, S.R.; Carlson, S.; Chan, D.V.; Liu, L.L.; Szabo, S.K.; Fu, P.F.; Gilliam, A.C.; Gerson, S.L.; et al. Phase I clinical trial of O6-Benzylguanine and topical carmustine in the treatment of cutaneous t-cell lymphoma, mycosis fungoides type. Arch. Dermatol. 2012, 148, 613–620. [Google Scholar] [CrossRef] [PubMed]
- Wilson, K.A.; Wetmore, S.D. Complex conformational heterogeneity of the highly flexible O6-benzylguanine DNA adduct. Chem. Res. Toxicol. 2014, 27, 1310–1325. [Google Scholar] [CrossRef] [PubMed]
- Osinsky, S.; Zavelevich, M.; Vaupel, P. Tumor hypoxia and malignant progression. Exp. Oncol. 2009, 31, 80–86. [Google Scholar]
- Nichols, A.J.; Roussakis, E.; Klein, O.J.; Evans, C.L. Click-assembled, oxygen-sensing nanoconjugates for depthresolved, near-infrared imaging in a 3D cancer model. Angew. Chem. Int. Ed. 2014, 53, 3671–3674. [Google Scholar] [CrossRef]
- Jian, R.K. Normalization of tumor vasculature: An emerging concept in antiangiogenic therapy. Science 2005, 307, 58–62. [Google Scholar] [CrossRef]
- Pries, A.R.; Annemiek, J.M.C.; Sloot, A.A.; Hinkeldey, M.; Dreher, M.R.; Höpfner, M.; Dewhirst, M.W.; Secomb, T.W. Structural adaptation and heterogeneity of normal and tumor microvascular networks. PLoS Comput. Biol. 2009, 5, e1000394. [Google Scholar] [CrossRef]
- Brahimi-Horn, M.C.; Pouysségur, J. Oxygen, a source of life and stress. FEBS Lett. 2007, 581, 3582–3591. [Google Scholar] [CrossRef]
- Catalano, V.; Turdo, A.; Di Franco, S.; Dieli, F.; Todaro, M.; Stassi, G. Tumor and its microenvironment: A synergistic interplay. Semin. Cancer Biol. 2013, 23, 522–532. [Google Scholar] [CrossRef]
- Patel, A.; Sant, S. Hypoxic tumor microenvironment: Opportunities to develop targeted therapies. Biotechnol. Adv. 2016, 34, 803–812. [Google Scholar] [CrossRef]
- Guise, C.P.; Mowday, A.M.; Ashoorzadeh, A.; Yuan, R.; Lin, W.; Wu, D.; Smaill, J.B.; Patterson, A.V.; Ding, K. Bioreductive prodrugs as cancer therapeutics: Targeting tumor hypoxia. Chin. J. Cancer 2014, 33, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Jaffar, M.; Williams, K.J.; Stratford, I.J. Bioreductive and gene therapy approaches to hypoxic diseases. Adv. Drug Deliver. Rev. 2001, 53, 217–228. [Google Scholar] [CrossRef]
- Jafar, M.; Maylar, M.A.; Robertson, N.; Stratford, I.J. Targeting hypoxia with a new generation of indolequinones. Anti Cancer Drug Des. 1998, 13, 593–609. [Google Scholar]
- Rauth, A.M.; Melo, T.; Misra, V. Bioreductive therapies: An overview of drugs and their mechanisms of action. Int. J. Radiat. Oncol. Biol. Phys. 1998, 42, 755–762. [Google Scholar] [CrossRef]
- Firestone, A.; Mulcahy, R.T.; Borch, R.F. Nitroheterocycle reduction as a paradigm for intramolecular catalysis of drug delivery to hypoxic cells. J. Med. Chem. 1991, 34, 2933–2935. [Google Scholar] [CrossRef]
- Mauger, A.B.; Burke, P.J.; Somani, H.H.; Friedlos, F.; Knox, R.J. Self-Immolative prodrugs: Candidates for antibody-directed enzyme prodrug therapy in conjunction with a nitroreductase enzyme. J. Med. Chem. 1994, 37, 3452–3458. [Google Scholar] [CrossRef]
- Everett, S.A.; Naylor, M.A.; Patel, K.B.; Stratford, M.R.L.; Wardman, P. Bioreductively-activated prodrugs for targeting hypoxic tissues: Elimination of aspirin from 2-nitroimidazole derivatives. Bioorg. Med. Chem. Lett. 1999, 9, 1267–1272. [Google Scholar] [CrossRef]
- Mahmud, N.P.; Garrett, S.W.; Threadgill, M.D. The 5-nitrofuran-2-ylmethylidene group as a potential bioreductively activated prodrug system for diol-containing drugs. Anti Cancer Drug Des. 1998, 13, 655–662. [Google Scholar]
- Zhu, R.; Liu, M.C.; Luo, M.Z.; Penketh, P.G.; Baumann, R.P.; Shyam, K.; Sartorelli, A.C. 4-Nitrobenzyloxycarbonyl derivatives of O6-benzylguanine as hypoxia-activated prodrug inhibitors of O6-alkylguanine-DNA alkyltransferase (AGT), which produces resistance to agents targeting the O-6 position of DNA guanine. J. Med. Chem. 2011, 54, 7720–7728. [Google Scholar] [CrossRef] [PubMed]
- Zhu, R.; Seow, H.A.; Baumann, R.P.; Ishiguro, K.; Penketh, P.G.; Shyam, K.; Sartorelli, A.C. Design of a hypoxia-activated prodrug inhibitor of O6-alkylguanine-DNA alkyltransferase. Bioorg. Med. Chem. Lett. 2012, 22, 6242–6247. [Google Scholar] [CrossRef] [PubMed]
- Penketh, P.G.; Shyam, K.; Baumann, R.P.; Ishiguro, K.; Patridge, E.V.; Zhu, R.; Sartorelli, A.C. A strategy for selective O6-alkylguanine-DNA alkyltransferase depletion under hypoxic conditions. Chem. Biol. Drug Des. 2012, 80, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Zhu, R.; Baumann, R.P.; Penketh, P.G.; Shyam, K.; Sartorelli, A.C. Hypoxia-selective O6-alkylguanine-DNA alkyltransferase inhibitors: Design, synthesis, and evaluation of 6-(Benzyloxy)-2-(aryldiazenyl)-9H-purines as prodrugs of O6-benzylguanine. J. Med. Chem. 2013, 56, 1355–1359. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chua, C.K.; Pumera, M.; Rulíšek, L. Reduction pathways of 2, 4, 6-trinitrotoluene: An electrochemical and theoretical study. J. Phys. Chem. C 2012, 116, 4243–4251. [Google Scholar] [CrossRef]
- Christofferson, A.; Wilkie, J. Mechanism of CB1954 reduction by Escherichia coli nitroreductase. Biochem. Soc. Trans. 2009, 37, 413–418. [Google Scholar] [CrossRef]
- Morrissey, C.; He, H.Y. Silicene catalyzed reduction of nitrobenzene to aniline: A mechanistic study. Chem. Phys. Lett. 2018, 695, 228–234. [Google Scholar] [CrossRef]
- Sheng, T.; Qi, Y.J.; Lin, X.; Hu, P.; Sun, S.G.; Lin, W.F. Insights into the mechanism of nitrobenzene reduction to aniline over Pt catalyst and the significance of the adsorption of phenyl group on kinetics. Chem. Eng. J. 2016, 293, 337–344. [Google Scholar] [CrossRef]
- Corma, A.; Concepción, P.; Serna, P. A Different reaction pathway for the reduction of aromatic nitro compounds on gold catalysts. Angew. Chem. Int. Ed. 2007, 46, 7266–7269. [Google Scholar] [CrossRef]
- Liang, C.J.; Lin, Y.T.; Shiu, J.W. Reduction of nitrobenzene with alkaline ascorbic acid: Kinetics and pathways. J. Hazard. Mater. 2016, 302, 137–143. [Google Scholar] [CrossRef]
- Ciou, C.; Liang, C. 1, 3-Dinitrobenzene reductive degradation by alkaline ascorbic acid—Reaction mechanisms, degradation pathways and reagent optimization. Chemosphere 2017, 166, 482–488. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.H.; Zhang, N.; Zhao, L.J.; Fan, T.J.; Zhang, S.F.; Zhong, R.G. Synthesis and antitumor activity evaluation of a novel combi-nitrosourea prodrug: Designed to release a DNA cross-linking agent and an inhibitor of O6-alkylguanine-DNA alkyltransferase. Bioorg. Med. Chem. 2016, 24, 2097–2107. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.H.; Fan, T.J.; Zhang, N.; Ren, T.; Zhao, L.J.; Zhong, R.G. Identification of the structural features of guanine derivatives as MGMT inhibitors using 3D-QSAR modeling combined with molecular docking. Molecules 2016, 21, 823. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.M.; Ren, T.; Lai, X.X.; Sun, G.H.; Zhao, L.J.; Zhang, N.; Zhong, R.G. Synthesis and antitumor activity evaluation of a novel combinitrosourea prodrug: BGCNU. ACS Med. Chem. Lett. 2017, 8, 174–178. [Google Scholar] [CrossRef]
- Sun, G.H.; Zhao, L.J.; Fan, T.J.; Ren, T.; Zhong, R.G. Measurement of O6-alkylguanine-DNA alkyltransferase activity in tumour cells using stable isotope dilution HPLC-ESI-MS/MS. J. Chromatogr. B 2016, 1033, 138–146. [Google Scholar] [CrossRef]
- Baumann, R.P.; Penketh, P.G.; Seow, H.A.; Shyam, K.; Sartorelli, A.C. Generation of oxygen deficiency in cell culture using a two-enzyme system to evaluate agents targeting hypoxic tumor cells. Radiat. Res. 2008, 170, 651–660. [Google Scholar] [CrossRef]
- Lee, C.T.; Yang, W.T.; Parr, R.G. Development of the Colic-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B 1987, 37, 785–789. [Google Scholar] [CrossRef]
- Becke, A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1992, 98, 5648–5652. [Google Scholar] [CrossRef]
- Zhao, Y.; Truhlar, D.G. The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: Two new functionals and systematic testing of four M06-class functionals and 12 other functionals. Theor. Chem. Acc. 2008, 120, 215–241. [Google Scholar]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09, Gaussian, Inc.: Wallingford, CT, USA, 2013.
- Wibley, J.E.A.; Pegg, A.E.; Moody, P.C.E. Crystal structure of the human O6-alkylguanine-DNA alkyltransferase. Nucleic Acids Res. 2000, 28, 393–401. [Google Scholar] [CrossRef]
- Driggers, C.M.; Dayal, P.V.; Ellis, H.R.; Karplus, P.A. Crystal structure of escherichia coli ssuE: Defining a general catalytic cycle for FMN reductases of the flavodoxin-like superfamily. Biochemistry 2014, 53, 3509–3519. [Google Scholar] [CrossRef] [PubMed]
- Race, P.R.; Lovering, A.L.; Green, R.M.; Ossor, A.; White, S.A.; Searle, P.F.; Wrighton, C.J.; Hyde, E.I. Structural and mechanistic studies of Escherichia coli nitroreductase with the antibiotic nitrofurazone. J. Biol. Chem. 2005, 280, 13256–13264. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Babin, V.; Berryman, J.T.; Betz, R.M.; Cai, Q.; Cerutti, D.S.; Cheatham, T.E., III; Darden, T.A.; Duke, R.E.; Gohlke, H.; et al. Amber 14; University of California: San Francisco, CA, USA, 2014. [Google Scholar]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Wu, C.; Chowdhury, S.; Lee, M.C.; Xiong, G.; Zhang, W.; Yang, R.; Cieplak, P.; Luo, R.; Lee, T.; et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 2003, 24, 1999–2012. [Google Scholar] [CrossRef]
- Ryckaert, J.-P.; Ciccotti, G.; Berendsen, H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints:molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23, 321–341. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Kollman, P.A.; Massova, I.; Reyes, C.; Kuhn, B.; Huo, S.; Chong, L.; Lee, M.; Lee, T.; Duan, Y.; Wang, W.; et al. Calculating structures and free energies of complex molecules: Combining molecular mechanics and continuum models. Acc. Chem. Res. 2000, 33, 889–897. [Google Scholar] [CrossRef]
- Massova, I.; Kollman, P.A. Combined molecular mechanical and continuum solvent approach (MM-PBSA/GBSA) to predict ligand binding. Perspect. Drug Discov. Des. 2000, 18, 113–135. [Google Scholar] [CrossRef]
- Sitkoff, D.; Sharp, K.A.; Honig, B. Accurate Calculation of Hydration Free Energies Using Macroscopic Solvent Models. J. Phys. Chem. 1994, 98, 1978–1988. [Google Scholar] [CrossRef]



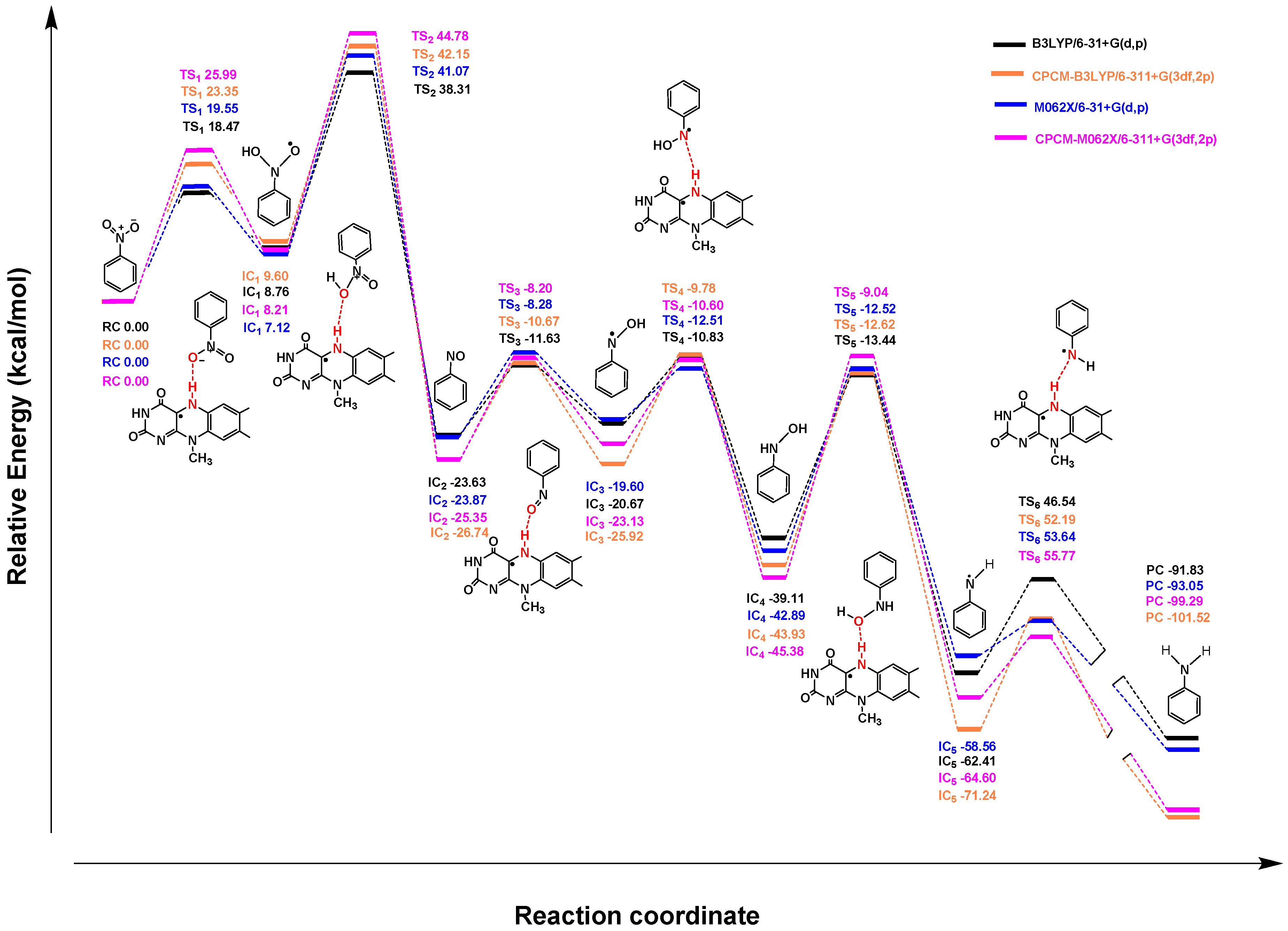
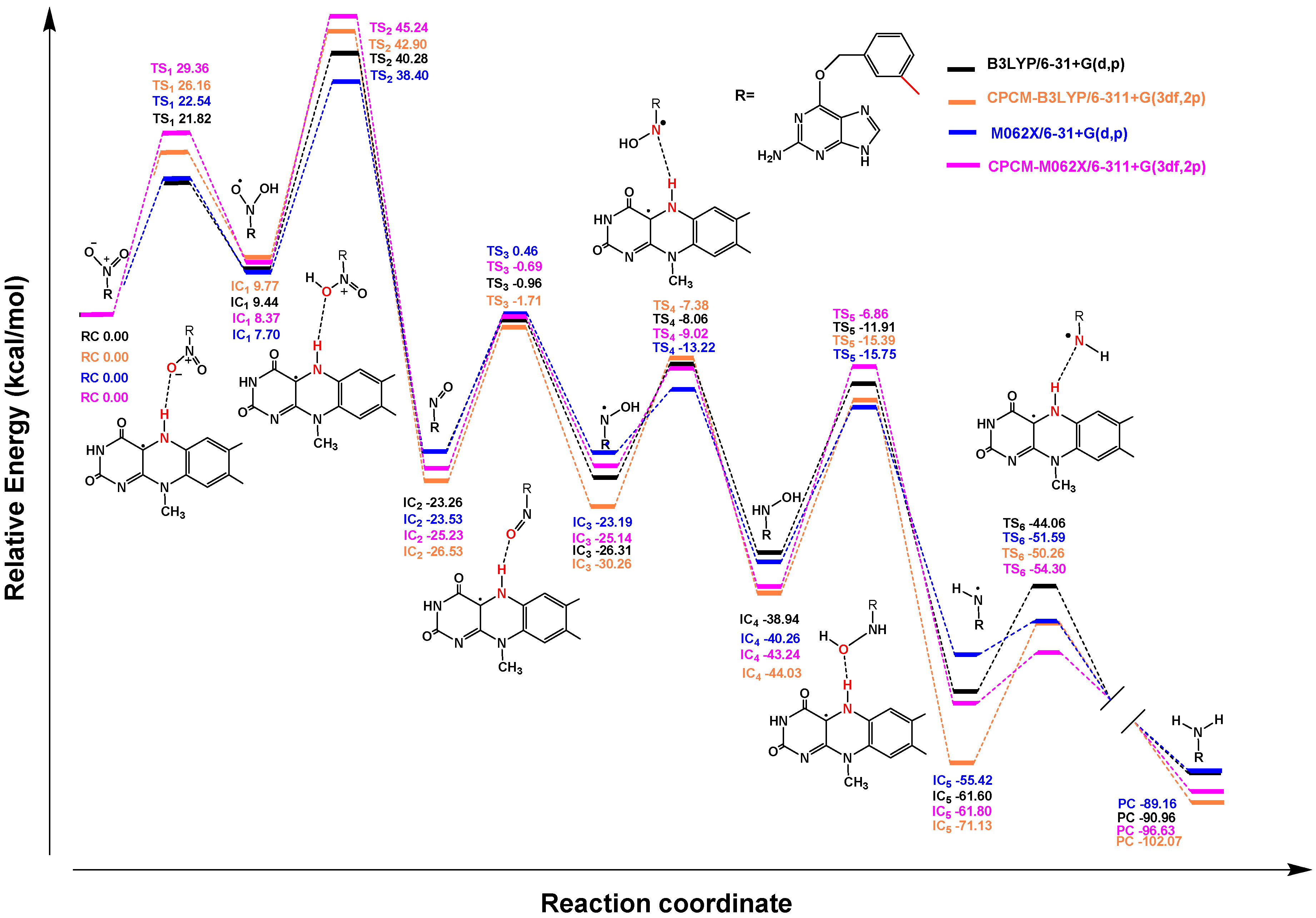
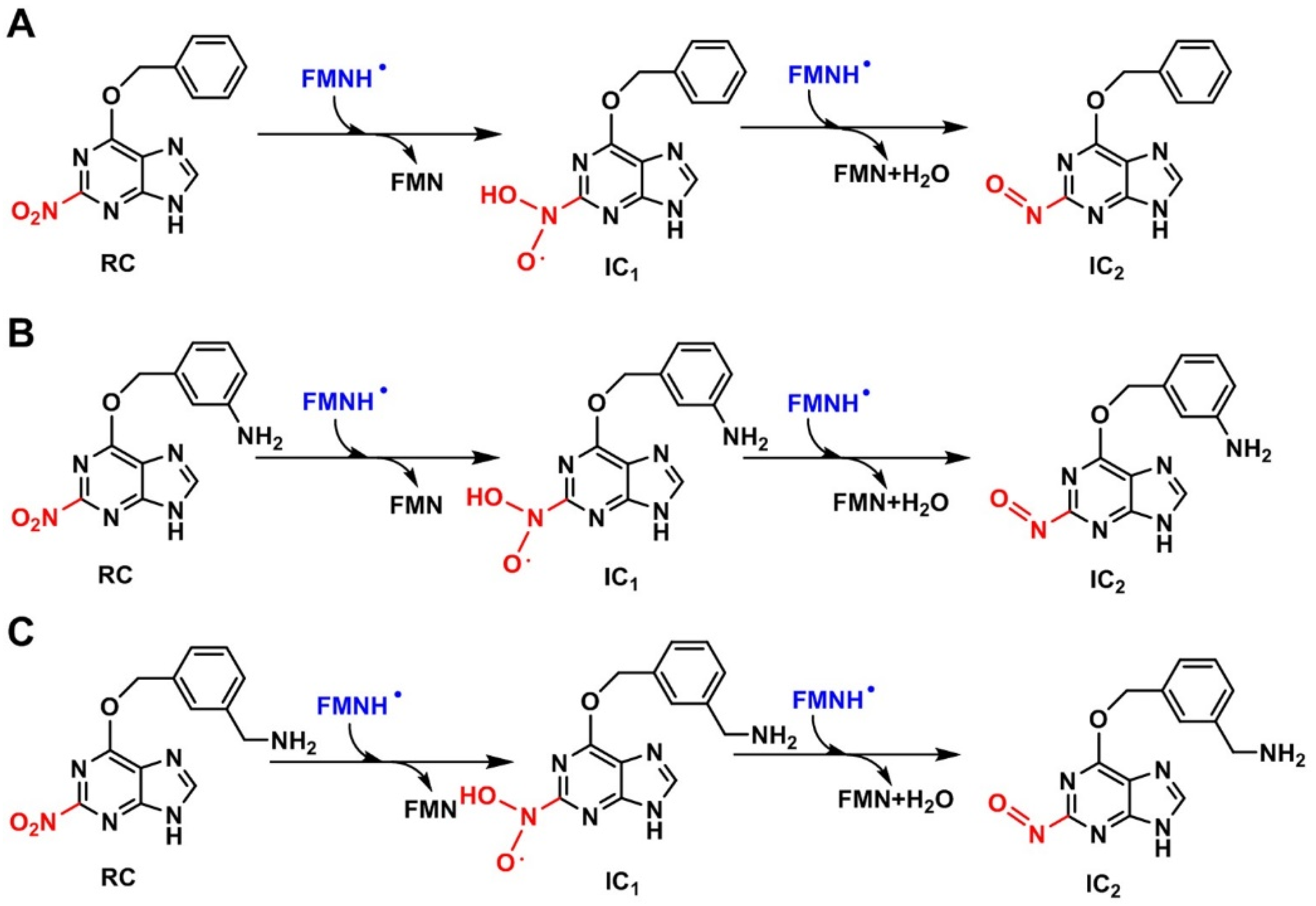
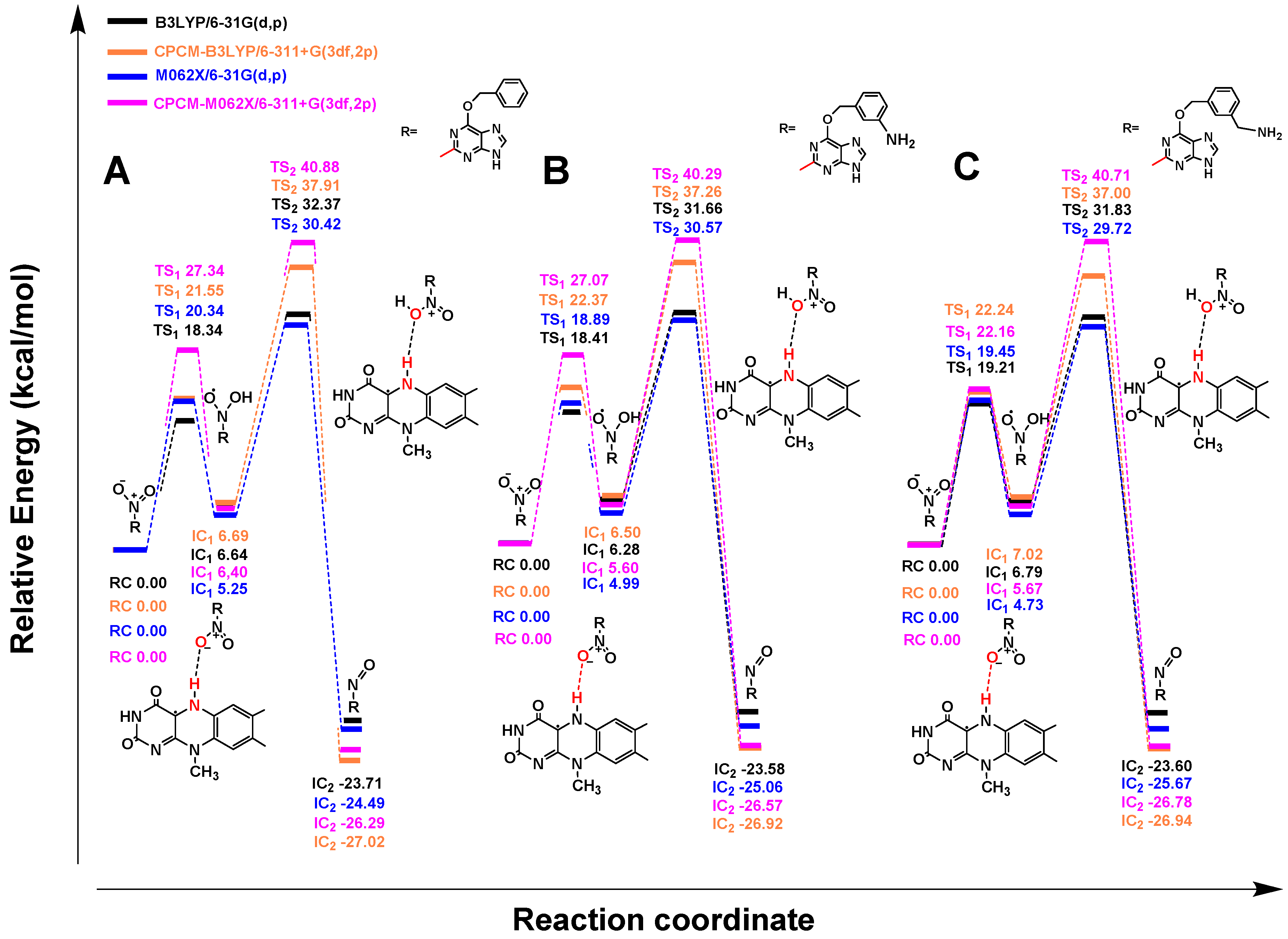
| Theoretical Levels | 3-NBG | 2-NBP | ANBP | AMNBP |
|---|---|---|---|---|
| B3LYP/6-31+G(d,p) | 30.83 | 25.73 | 25.37 | 25.04 |
| CPCM-B3LYP/6-311+G(3df,2p) | 33.13 | 30.95 | 30.76 | 29.98 |
| M062X/6-31+G(d,p) | 30.71 | 25.18 | 25.57 | 24.99 |
| CPCM-M062X/6-311+G(3df,2p) | 36.88 | 34.49 | 34.69 | 35.04 |
| Molecule | Fitness Score | Hydrogen Bonds | ||
|---|---|---|---|---|
| Amount | Length/Å | |||
| 3-NBG | 49.5377 | 2 | O6-Glu268 | 2.4 |
| N7-Glu268 | 2.9 | |||
| 2-NBP | 49.9341 | 2 | O6-Glu268 | 2.1 |
| N7-Glu268 | 2.8 | |||
| ANBP | 53.4563 | 5 | O6-Glu268 | 2.3 |
| N7-Glu268 | 2.8 | |||
| N-Leu266 | 2.6 | |||
| N-Asp264 | 2.4 | |||
| N-Lys78 | 3.1 | |||
| AMNBP | 53.2185 | 3 | O-Glu268 | 2.9 |
| N-Glu268 | 2.8 | |||
| N-Leu266 | 2.8 | |||
| Compounds | Fitness Score | Orientation 1 | Hydrogen Bonds | ||
|---|---|---|---|---|---|
| Amount | Length/Å | ||||
| 3-NBG | 47.0434 | No | / | / | |
| 2-NBP | 49.7631 | No | / | / | |
| ANBP | 49.4182 | No | / | / | |
| AMNBP | 50.8505 | No | / | / | |
| O6-BG | 54.8831 | Yes | 4 | O6–Ser159 | 3.2 |
| N2–Val148 | 3.0 | ||||
| N2–Cys145 | 2.4 | ||||
| N4–Tyr114 | 2.9 | ||||
| ABG | 53.3393 | Yes | 5 | N2–Lys165 | 3.0 |
| O6–Ser159 | 2.9 | ||||
| N2–Val148 | 3.0 | ||||
| N2–Cys145 | 2.5 | ||||
| N–Arg135 | 3.0 | ||||
| AMBG | 53.9127 | Yes | 5 | O6–Ser159 | 2.0 |
| N2–Val148 | 2.4 | ||||
| N2–Cys145 | 1.7 | ||||
| N–Asn137 | 1.7 | ||||
| N3–Tyr114 | 2.1 | ||||
| Free Energy (Kcal/mol) | O6-BG | ABG | AMBG |
|---|---|---|---|
| ΔEele | −20.72 ± 1.35 | −37.54 ± 1.74 | −34.92 ± 1.74 |
| ΔEvdw | −38.57 ± 1.35 | −40.55 ± 1.89 | −39.19 ± 2.67 |
| ΔEgas a | −59.29 ± 1.60 | −78.09 ± 3.62 | −74.12 ± 2.64 |
| ΔGnonpolar | −4.35 ± 0.06 | −4.43 ± 0.04 | −5.05 ± 0.20 |
| ΔGpolar | 30.68 ± 1.16 | 46.90 ± 2.53 | 43.71 ± 1.74 |
| ΔGsol b | 26.33 ± 1.14 | 42.47 ± 2.49 | 38.66 ± 1.74 |
| ΔGele c | 9.96 ± 1.97 | 9.36 ± 0.79 | 8.79 ± 1.76 |
| ΔGbinding d | −32.96 ± 1.75 | −35.63 ± 1.13 | −35.45 ± 3.30 |
| ΔΔGbinding d | 0 | −2.67 | −2.49 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, W.; Sun, G.; Fan, T.; Liu, J.; Zhang, N.; Zhao, L.; Zhong, R. Reductive Activity and Mechanism of Hypoxia- Targeted AGT Inhibitors: An Experimental and Theoretical Investigation. Int. J. Mol. Sci. 2019, 20, 6308. https://doi.org/10.3390/ijms20246308
Xiao W, Sun G, Fan T, Liu J, Zhang N, Zhao L, Zhong R. Reductive Activity and Mechanism of Hypoxia- Targeted AGT Inhibitors: An Experimental and Theoretical Investigation. International Journal of Molecular Sciences. 2019; 20(24):6308. https://doi.org/10.3390/ijms20246308
Chicago/Turabian StyleXiao, Weinan, Guohui Sun, Tengjiao Fan, Junjun Liu, Na Zhang, Lijiao Zhao, and Rugang Zhong. 2019. "Reductive Activity and Mechanism of Hypoxia- Targeted AGT Inhibitors: An Experimental and Theoretical Investigation" International Journal of Molecular Sciences 20, no. 24: 6308. https://doi.org/10.3390/ijms20246308
APA StyleXiao, W., Sun, G., Fan, T., Liu, J., Zhang, N., Zhao, L., & Zhong, R. (2019). Reductive Activity and Mechanism of Hypoxia- Targeted AGT Inhibitors: An Experimental and Theoretical Investigation. International Journal of Molecular Sciences, 20(24), 6308. https://doi.org/10.3390/ijms20246308