Silencing of lncRNA AK045490 Promotes Osteoblast Differentiation and Bone Formation via β-Catenin/TCF1/Runx2 Signaling Axis
Abstract
1. Introduction
2. Results
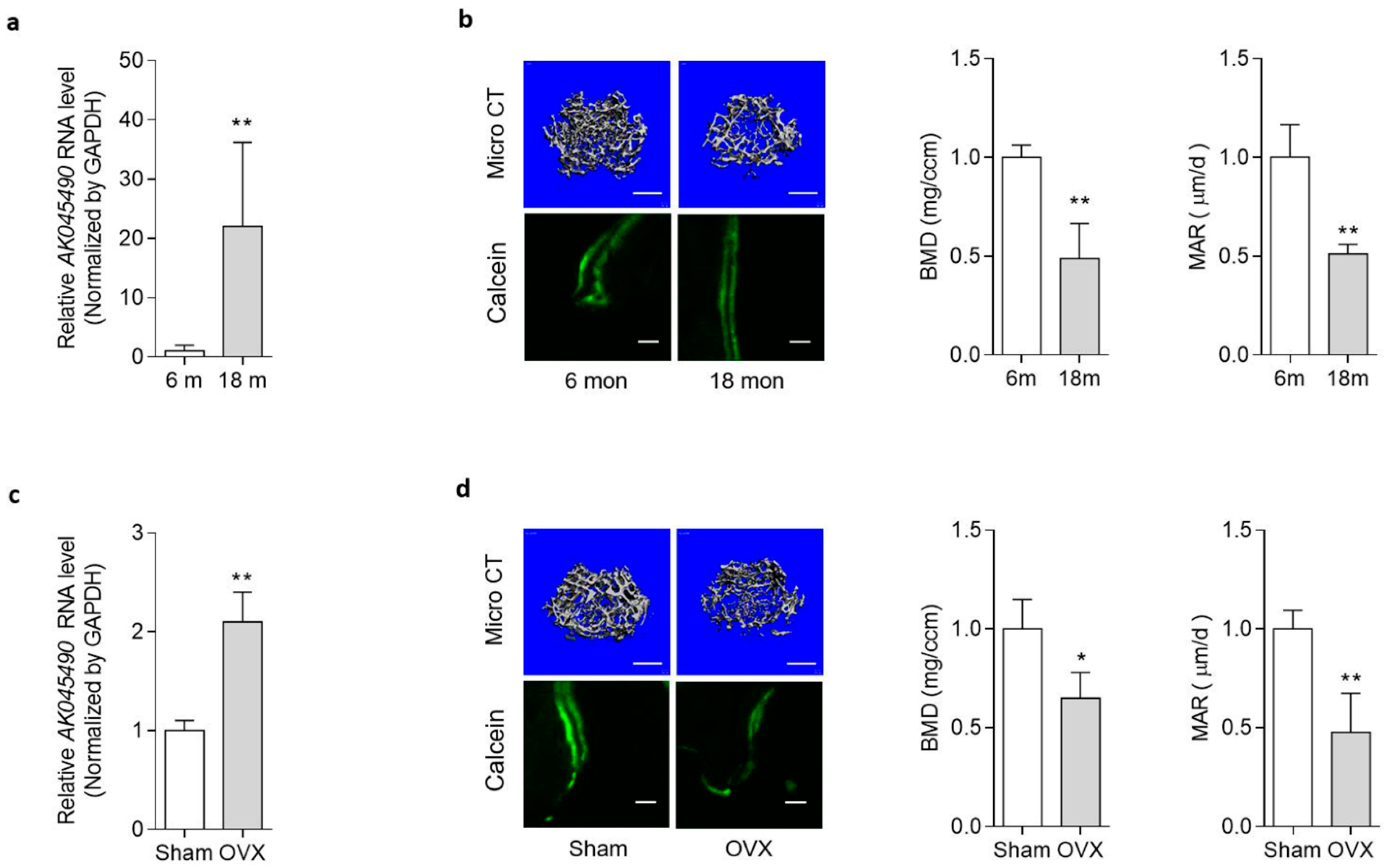
2.1. Elevated AK045490 Expression in Bone Was Accompanied by Deteriorated Bone Microstructure and Decreased Bone Formation in Osteoporotic Mice
2.2. AK045490 Inhibited Osteoblast Differentiation
2.3. Knockdown of AK045490 Promoted β-Catenin Nuclear Translocation and Up-Regulates the Expression of TCF1, LEF1 and Runx2
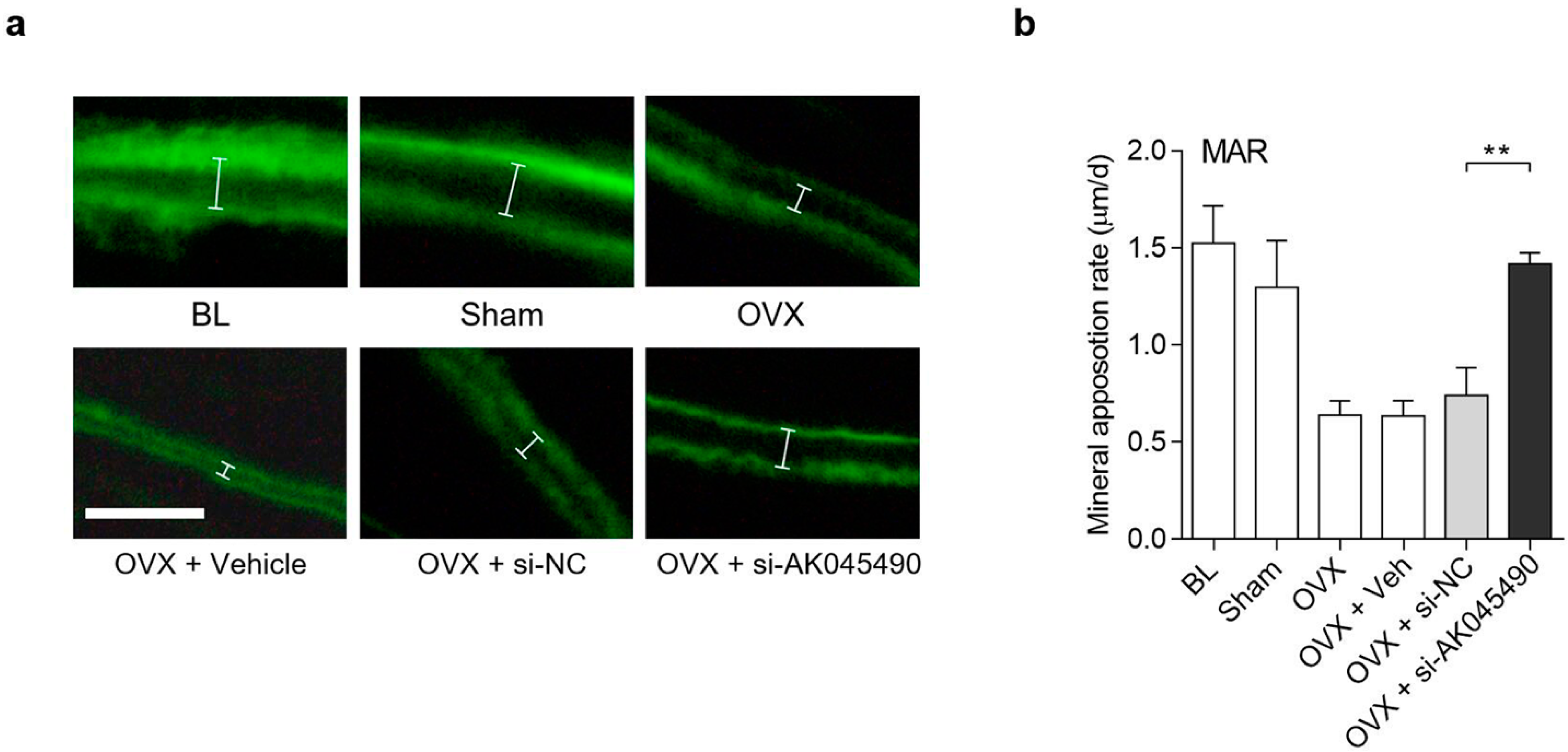
2.4. Promoting Effect of AK045490 siRNA on Calvaria Bone Formation in OVX Mice
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Mice Model
4.3. RNA Isolation and Real-Time PCR (RT-PCR) Analysis
4.4. MicroCT Analysis
4.5. Bone Histology and Histomorphometry
4.6. siRNA Transfection In Vitro
4.7. Western Blot
4.8. ALP Staining
4.9. Alizarin Red Staining
4.10. Immunohistofluorescence (IHF) Staining
4.11. siRNA Transfection In Vivo
4.12. Analysis of lncRNA–microRNA–mRNA Interaction
4.13. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ALP | alkaline phosphatase |
| BFR | bone formation rate |
| BL | baseline |
| BMD | bone mineral density |
| BMSC | bone-derived mesenchymal stem cell |
| ceRNA | competing endogenous RNA |
| Col Iα1 | collagen type I |
| ECL | electro-chemi-luminescence |
| IHF | immunohistofluorescence |
| LEF1 | lymphoid enhancer-binding factor 1 |
| lncRNA | long noncoding RNA |
| Linc-ROR | large intergenic noncoding RNA regulator of reprogramming |
| MAR | mineralizing apposition rate |
| Micro CT | micro computed tomography |
| MSC | mesenchymal stem cell |
| Sham | sham-operated |
| Ocn | osteocalcin |
| OVX | ovariectomized |
| PFA | paraformaldehyde |
| Runx2 | runt-related transcription factor 2 |
| TCF1 | transcription factor T cell factor 1 |
| WB | western blot |
References
- Hendrickx, G.; Boudin, E.; Van Hul, W. A look behind the scenes: The risk and pathogenesis of primary osteoporosis. Nat. Rev. Rheumatol. 2015, 11, 462–474. [Google Scholar] [CrossRef] [PubMed]
- Eastell, R.; O’Neill, T.W.; Hofbauer, L.C.; Langdahl, B.; Reid, I.R.; Gold, D.T.; Cummings, S.R. Postmenopausal osteoporosis. Nat. Rev. Dis. Primers 2016, 2. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.M.; Moura, S.R.; Teixeira, J.H.; Barbosa, M.A.; Santos, S.G.; Almeida, M.I. Long noncoding RNAs: A missing link in osteoporosis. Bone Res. 2019, 7. [Google Scholar] [CrossRef] [PubMed]
- Kour, S.; Rath, P.C. Long noncoding RNAs in aging and age-related diseases. Ageing Res. Rev. 2016, 26, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.L.; Jia, L.F.; Li, X.B.; Guo, R.Z.; Huang, Y.P.; Zheng, Y.F.; Li, W.R. Long Noncoding RNAs: New Players in the Osteogenic Differentiation of Bone Marrow- and Adipose-Derived Mesenchymal Stem Cells. Stem Cell Rev. Rep. 2018, 14, 297–308. [Google Scholar] [CrossRef]
- Huynh, N.P.T.; Anderson, B.A.; Guilak, F.; McAlinden, A. Emerging roles for long noncoding RNAs in skeletal biology and disease. Connect. Tissue Res. 2017, 58, 116–141. [Google Scholar] [CrossRef]
- Yin, C.; Tian, Y.; Yu, Y.; Wang, H.Y.; Wu, Z.X.; Huang, Z.Z.; Zhang, Y.; Li, D.J.; Yang, C.F.; Wang, X.; et al. A novel long noncoding RNA AK016739 inhibits osteoblast differentiation and bone formation. J. Cell. Physiol. 2019, 234, 11524–11536. [Google Scholar] [CrossRef]
- Feng, L.; Shi, L.; Lu, Y.F.; Wang, B.; Tang, T.; Fu, W.M.; He, W.; Li, G.; Zhang, J.F. Linc-ROR Promotes Osteogenic Differentiation of Mesenchymal Stem Cells by Functioning as a Competing Endogenous RNA for miR-138 and miR-145. Mol. Ther.-Nucl. Acids 2018, 11, 345–353. [Google Scholar] [CrossRef]
- Gaur, T.; Lengner, C.J.; Hovhannisyan, H.; Bhat, R.A.; Bodine, P.V.N.; Komm, B.S.; Javed, A.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; et al. Canonical WNT signaling promotes osteogenesis by directly stimulating Runx2 gene expression. J. Biol. Chem. 2005, 280, 33132–33140. [Google Scholar] [CrossRef]
- Compston, J.E.; McClung, M.R.; Leslie, W.D. Osteoporosis. Lancet 2019, 393, 364–376. [Google Scholar] [CrossRef]
- Li, D.; Yang, C.; Yin, C.; Zhao, F.; Chen, Z.; Tian, Y.; Dang, K.; Jiang, S.; Zhang, W.; Zhang, G.; et al. LncRNA, Important Players in Bone Development and Disease. Endocr. Metab. Immune Disord. Drug Targets 2019. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.Y.; Dong, R.; Diao, S.; Du, J.; Fan, Z.P.; Wang, F. Differential long noncoding RNA/mRNA expression profiling and functional network analysis during osteogenic differentiation of human bone marrow mesenchymal stem cells. Stem Cell Res. Ther. 2017, 8, 30. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.F.; Su, P.H.; Li, R.Z.; Yan, K.; Chen, Z.H.; Shang, P.; Qian, A.R. Knockdown of microtubule actin crosslinking factor 1 inhibits cell proliferation in MC3T3-E1 osteoblastic cells. BMB Rep. 2015, 48, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.P.; Zheng, Y.F.; Jia, L.F.; Li, W.R. Long Noncoding RNA H19 Promotes Osteoblast Differentiation Via TGF-beta 1/Smad3/HDAC Signaling Pathway by Deriving miR-675. Stem Cells 2015, 33, 3481–3492. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.F.; Liu, Y.; Fu, W.M.; Xu, J.; Wang, B.; Sun, Y.X.; Wu, T.Y.; Xu, L.L.; Chan, K.M.; Zhang, J.F.; et al. Long noncoding RNA H19 accelerates tenogenic differentiation and promotes tendon healing through targeting miR-29b-3p and activating TGF-beta 1 signaling. FASEB J. 2017, 31, 954–964. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wang, S.L.; Tang, C.L.; Chen, W.J. Upregulation of long non-coding RNA HIF 1 alpha-anti-sense 1 induced by transforming growth factor-beta-mediated targeting of sirtuin 1 promotes osteoblastic differentiation of human bone marrow stromal cells. Mol. Med. Rep. 2015, 12, 7233–7238. [Google Scholar] [CrossRef]
- Zhang, J.L.; Tao, Z.W.; Wang, Y.L. Long non-coding RNA DANCR regulates the proliferation and osteogenic differentiation of human bone-derived marrow mesenchymal stem cells via the p38 MAPK pathway. Int. J. Mol. Med. 2018, 41, 213–219. [Google Scholar] [CrossRef]
- Li, C.L.; Wang, S.Y.; Xing, Z.; Lin, A.F.; Liang, K.; Song, J.; Hui, Q.S.; Yao, J.; Chen, Z.Y.; Park, P.K.; et al. A ROR1 HER3 IncRNA signalling axis modulates the Hippo YAP pathway to regulate bone metastasis. Nat. Cell Biol. 2017, 19, 106–119. [Google Scholar] [CrossRef]
- Liao, J.Y.; Yu, X.Y.; Hu, X.; Fan, J.M.; Wang, J.; Zhang, Z.C.; Zhao, C.; Zeng, Z.Y.; Shu, Y.; Zhang, R.Y.; et al. lncRNA H19 mediates BMP9-induced osteogenic differentiation of mesenchymal stem cells (MSCs) through Notch signaling. Oncotarget 2017, 8, 53581–53601. [Google Scholar] [CrossRef]
- Zhang, Y.; Yin, C.; Hu, L.F.; Chen, Z.H.; Zhao, F.; Li, D.J.; Ma, J.H.; Ma, X.L.; Su, P.H.; Qiu, W.X.; et al. MACF1 Overexpression by Transfecting the 21 kbp Large Plasmid PEGFP-C1A-ACF7 Promotes Osteoblast Differentiation and Bone Formation. Hum. Gene Ther. 2018, 29, 259–270. [Google Scholar] [CrossRef]
- Hu, L.; Su, P.; Yin, C.; Zhang, Y.; Li, R.; Yan, K.; Chen, Z.; Li, D.; Zhang, G.; Wang, L.; et al. Microtubule actin crosslinking factor 1 promotes osteoblast differentiation by promoting beta-catenin/TCF1/Runx2 signaling axis. J. Cell. Physiol. 2018, 233, 1574–1584. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.P.; Li, Y.P.; Paulson, C.; Shao, J.Z.; Zhang, X.L.; Wu, M.R.; Chen, W. Wnt and the Wnt signaling pathway in bone development and disease. Front. Biosci. 2014, 19, 379–407. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.Q.; Xu, Z.M.; Duan, C.C.; Liu, W.W.; Sun, J.C.; Han, B. Role of TCF/LEF Transcription Factors in Bone Development and Osteogenesis. Int. J. Med. Sci. 2018, 15, 1415–1422. [Google Scholar] [CrossRef] [PubMed]
- Issa, Y.A.; Kamal, L.; Abu Rayyan, A.; Dweik, D.; Pierce, S.; Lee, M.K.; King, M.C.; Walsh, T.; Kanaan, M. Mutation of KREMEN1, a modulator of Wnt signaling, is responsible for ectodermal dysplasia including oligodontia in Palestinian families. Eur. J. Hum. Genet. 2016, 24, 1430–1435. [Google Scholar] [CrossRef]
- Nakamura, T.; Nakamura, T.; Matsumoto, K. The functions and possible significance of Kremen as the gatekeeper of Wnt signalling in development and pathology. J. Cell. Mol. Med. 2008, 12, 391–408. [Google Scholar] [CrossRef]
- Wu, M.R.; Chen, W.; Lu, Y.; Zhu, G.C.; Hao, L.; Li, Y.P. G alpha 13 negatively controls osteoclastogenesis through inhibition of the Akt-GSK3 beta-NFATc1 signalling pathway. Nat. Commun. 2017, 8, 13700. [Google Scholar] [CrossRef]
- Gillespie, J.R.; Bush, J.R.; Bell, G.I.; Aubrey, L.A.; Dupuis, H.; Ferron, M.; Kream, B.; DiMattia, G.; Patel, S.; Woodgett, J.R.; et al. GSK-3 beta Function in Bone Regulates Skeletal Development, Whole-Body Metabolism, and Male Life Span. Endocrinology 2013, 154, 3702–3718. [Google Scholar] [CrossRef]
- Li, C.J.; Xiao, Y.; Yang, M.; Su, T.; Sun, X.; Guo, Q.; Huang, Y.; Luo, X.H. Long noncoding RNA Bmncr regulates mesenchymal stem cell fate during skeletal aging. J. Clin. Investig. 2018, 128, 5251–5266. [Google Scholar] [CrossRef]
- Li, B.Z.; Han, H.Y.; Song, S.; Fan, G.; Xu, H.X.; Zhou, W.Q.; Qiu, Y.C.; Qian, C.A.; Wang, Y.J.; Yuan, Z.H.; et al. HOXC10 Regulates Osteogenesis of Mesenchymal Stromal Cells Through Interaction with Its Natural Antisense Transcript lncHOXC-AS3. Stem Cells 2019, 37, 247–256. [Google Scholar] [CrossRef]
- Liu, Y.B.; Lin, L.P.; Zou, R.; Zhao, Q.H.; Lin, F.Q. Silencing long non-coding RNA MEG3 accelerates tibia fraction healing by regulating the Wnt/beta-catenin signalling pathway. J. Cell. Mol. Med. 2019, 23, 3855–3866. [Google Scholar] [CrossRef]
- Mulati, M.; Kobayashi, Y.; Takahashi, A.; Numata, H.; Saito, M.; Hiraoka, Y.; Ochi, H.; Sato, S.; Ezura, Y.; Yuasa, M.; et al. The long noncoding RNA Crnde regulates osteoblast proliferation through the Wnt/beta-catenin signaling pathway in mice. Bone 2019, 130, 115076. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]


| Target gene | Primer Direction | Sequence (5′–3′) |
|---|---|---|
| AK045490 | Forward: | GCATTGTATCTCGCTCCACA |
| Reverse: | TGTTGCCTACCTGCTTACTGC | |
| Alp | Forward: | GTTGCCAAGCTGGGAAGAACAC |
| Reverse: | CCCACCCCGCTATTCCAAAC | |
| Ocn | Forward: | GAAGGCAACAGTCGATTCACC |
| Reverse: | GACTGTCTTGCCCCAAGTTCC | |
| Col Ⅰα1 | Forward: | GAAGGCAACAGTCGATTCACC |
| Reverse: | GACTGTCTTGCCCCAAGTTCC | |
| Tcf1 | Forward: | CAGAATCCACAGATACAGCA |
| Reverse: | CAGCCTTTGAAATCTTCATC | |
| Let1 | Forward: | GATCCCCTTCAAGGACGAAG |
| Reverse: | GGCTTGTCTGACCACCTCAT | |
| Runx2 | Forward: | CGCCCCTCCCTGAACTCT |
| Reverse: | TGCCTGCCTGGGATCTGTA | |
| Gapdh | Forward: | TGCACCACCAACTGCTTAG |
| Reverse: | GGATGCAGGGATGATGTTC |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, D.; Tian, Y.; Yin, C.; Huai, Y.; Zhao, Y.; Su, P.; Wang, X.; Pei, J.; Zhang, K.; Yang, C.; et al. Silencing of lncRNA AK045490 Promotes Osteoblast Differentiation and Bone Formation via β-Catenin/TCF1/Runx2 Signaling Axis. Int. J. Mol. Sci. 2019, 20, 6229. https://doi.org/10.3390/ijms20246229
Li D, Tian Y, Yin C, Huai Y, Zhao Y, Su P, Wang X, Pei J, Zhang K, Yang C, et al. Silencing of lncRNA AK045490 Promotes Osteoblast Differentiation and Bone Formation via β-Catenin/TCF1/Runx2 Signaling Axis. International Journal of Molecular Sciences. 2019; 20(24):6229. https://doi.org/10.3390/ijms20246229
Chicago/Turabian StyleLi, Dijie, Ye Tian, Chong Yin, Ying Huai, Yipu Zhao, Peihong Su, Xue Wang, Jiawei Pei, Kewen Zhang, Chaofei Yang, and et al. 2019. "Silencing of lncRNA AK045490 Promotes Osteoblast Differentiation and Bone Formation via β-Catenin/TCF1/Runx2 Signaling Axis" International Journal of Molecular Sciences 20, no. 24: 6229. https://doi.org/10.3390/ijms20246229
APA StyleLi, D., Tian, Y., Yin, C., Huai, Y., Zhao, Y., Su, P., Wang, X., Pei, J., Zhang, K., Yang, C., Dang, K., Jiang, S., Miao, Z., Li, M., Hao, Q., Zhang, G., & Qian, A. (2019). Silencing of lncRNA AK045490 Promotes Osteoblast Differentiation and Bone Formation via β-Catenin/TCF1/Runx2 Signaling Axis. International Journal of Molecular Sciences, 20(24), 6229. https://doi.org/10.3390/ijms20246229





