Mutation of Aspartate 238 in FAD Synthase Isoform 6 Increases the Specific Activity by Weakening the FAD Binding
Abstract
1. Introduction
2. Results
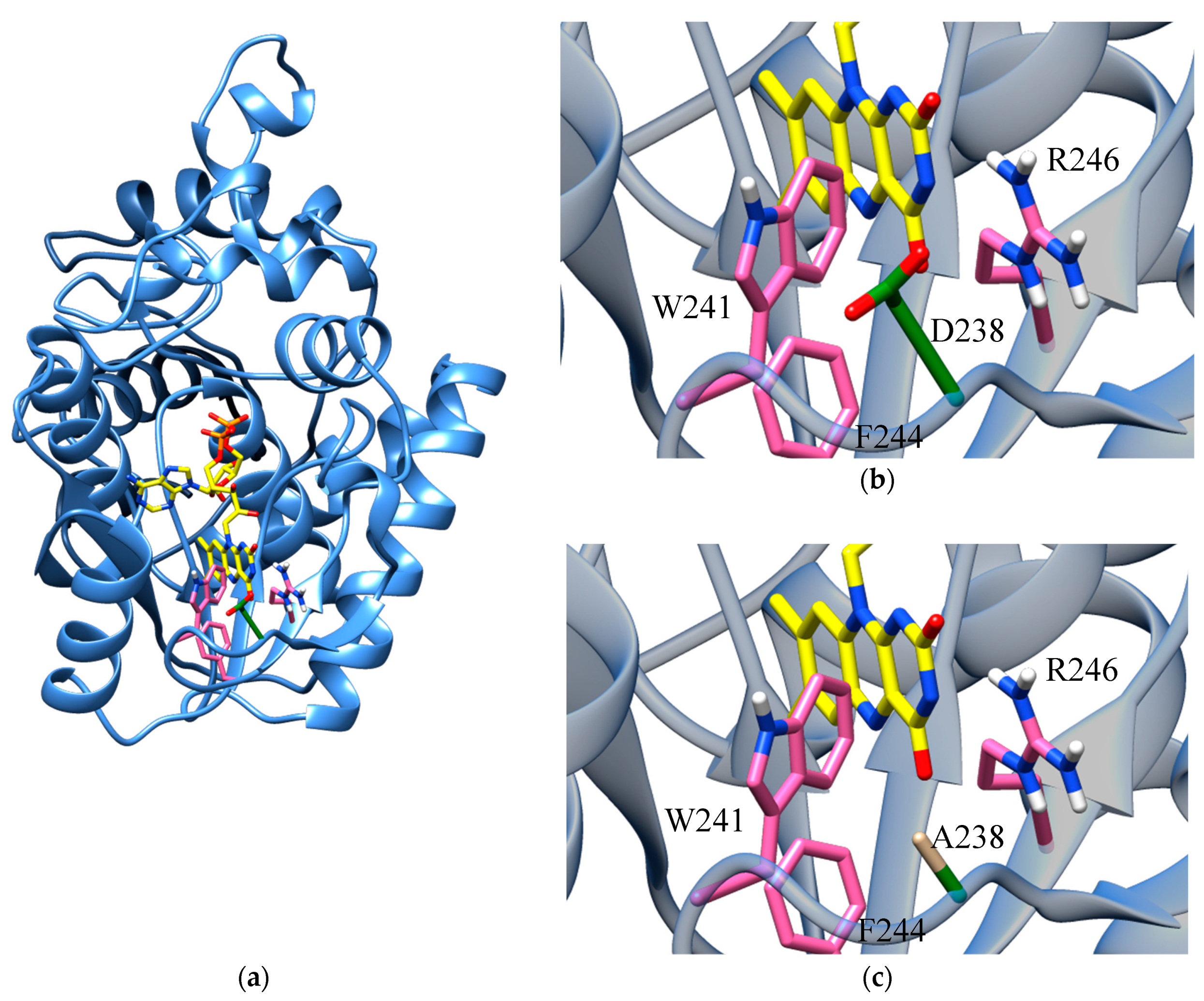
2.1. Homology Model of D238A-hFADS6
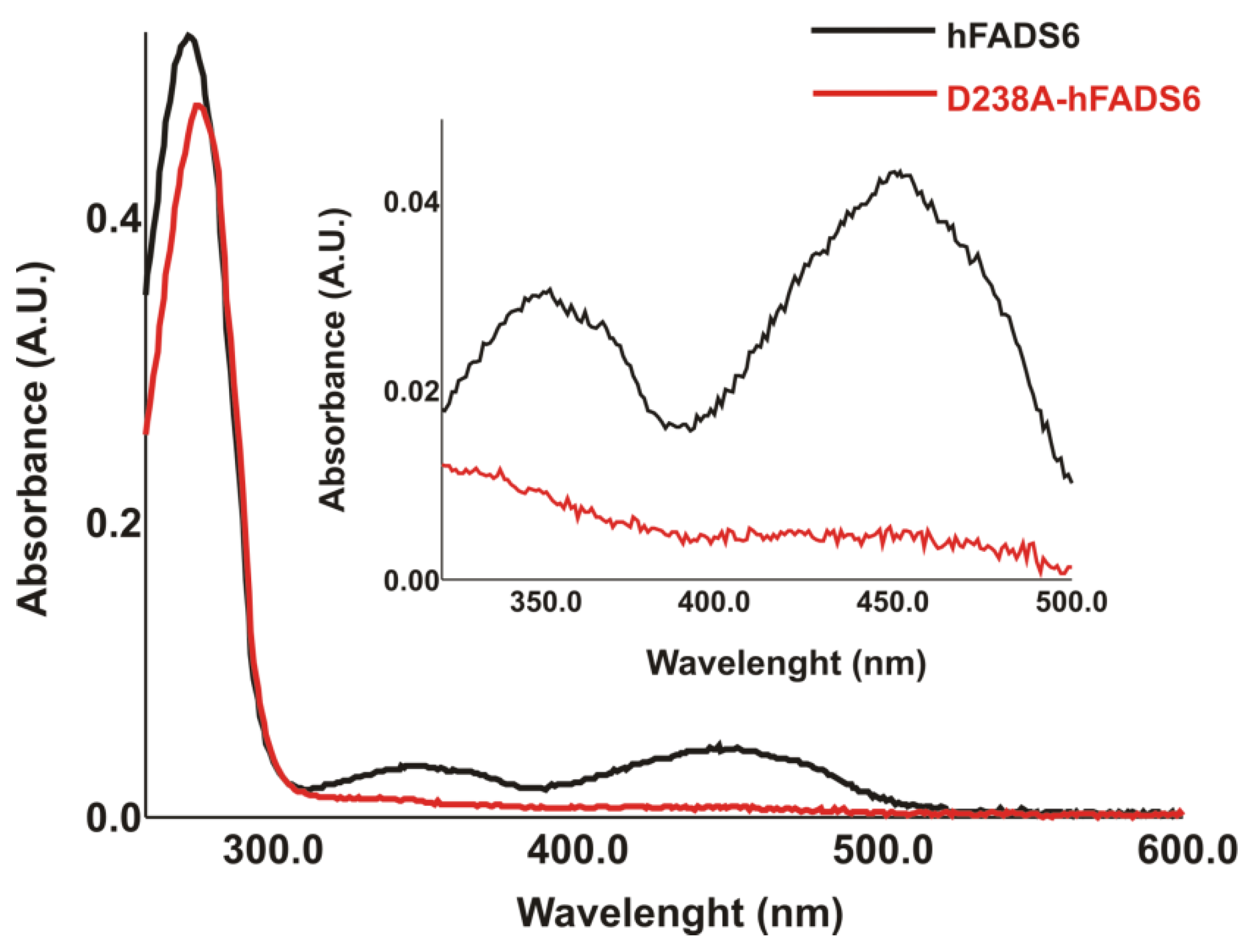
2.2. Cloning, Expression, and Purification of the D238A-hFADS6 Isoform
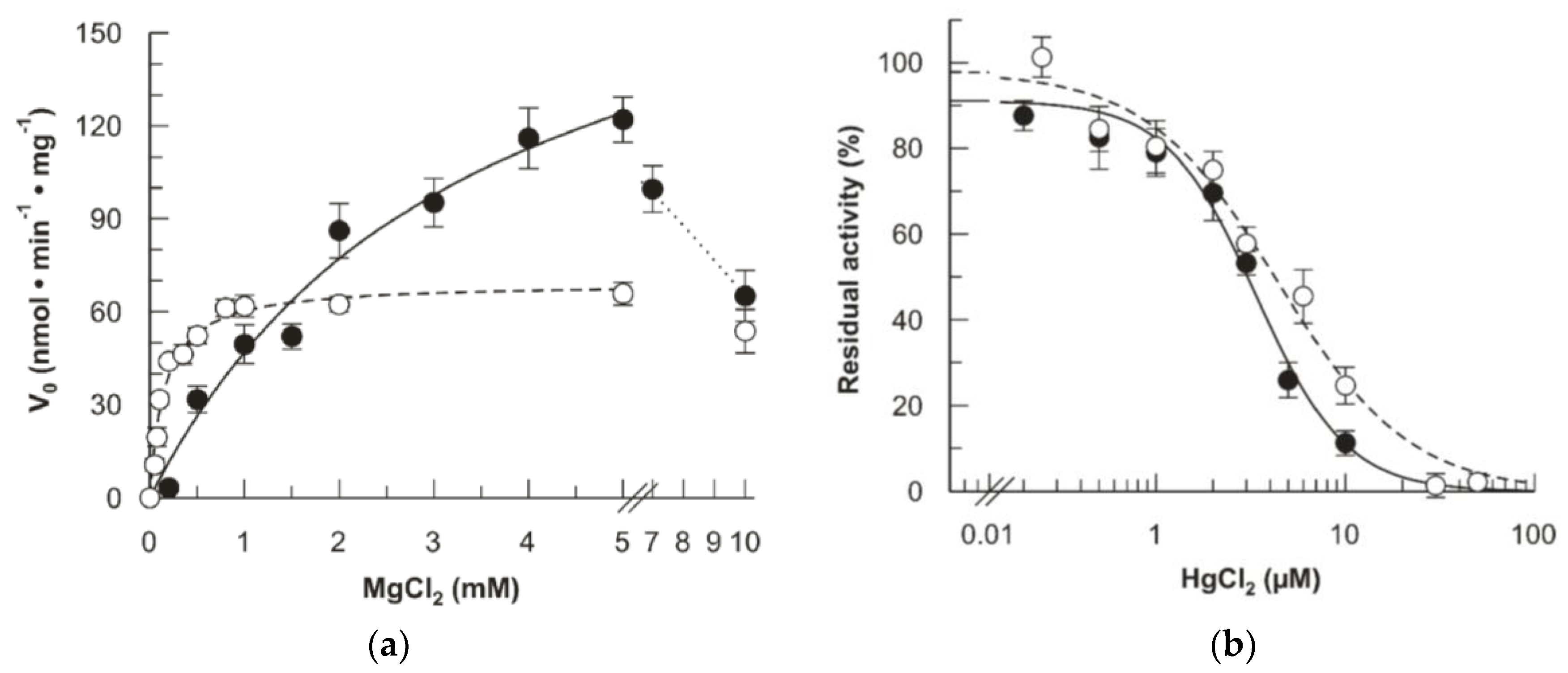
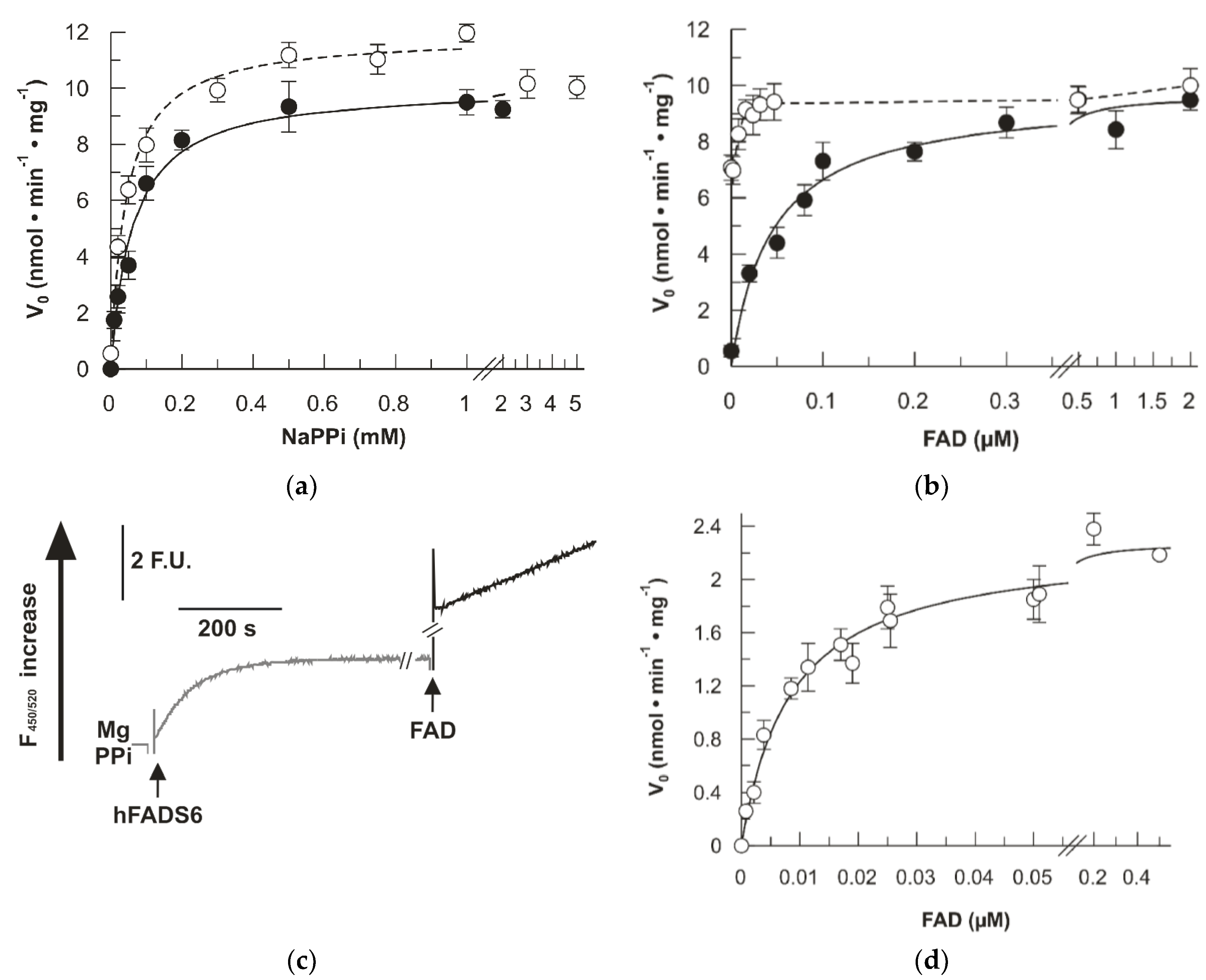
2.3. Kinetics of the D238A hFADS6
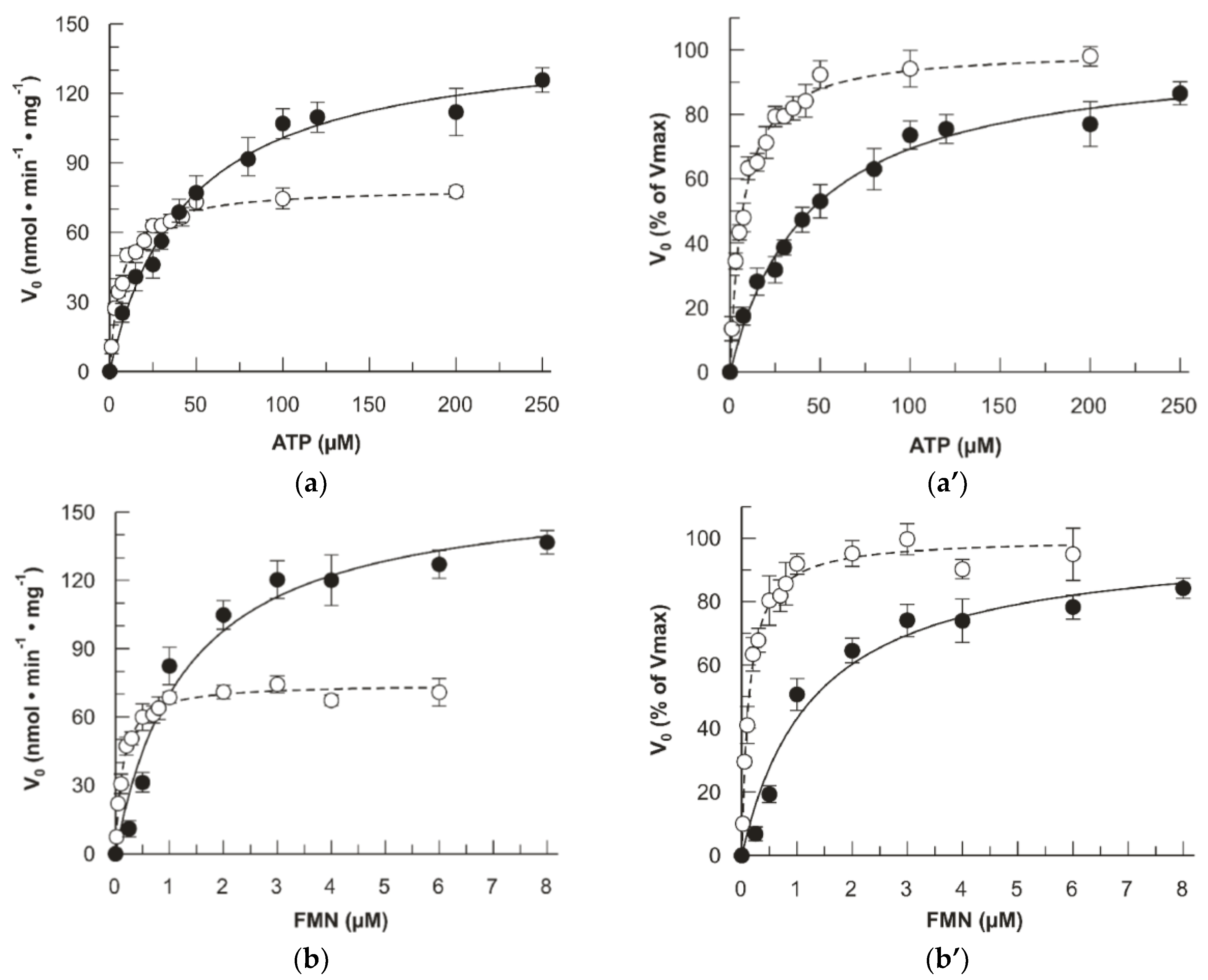
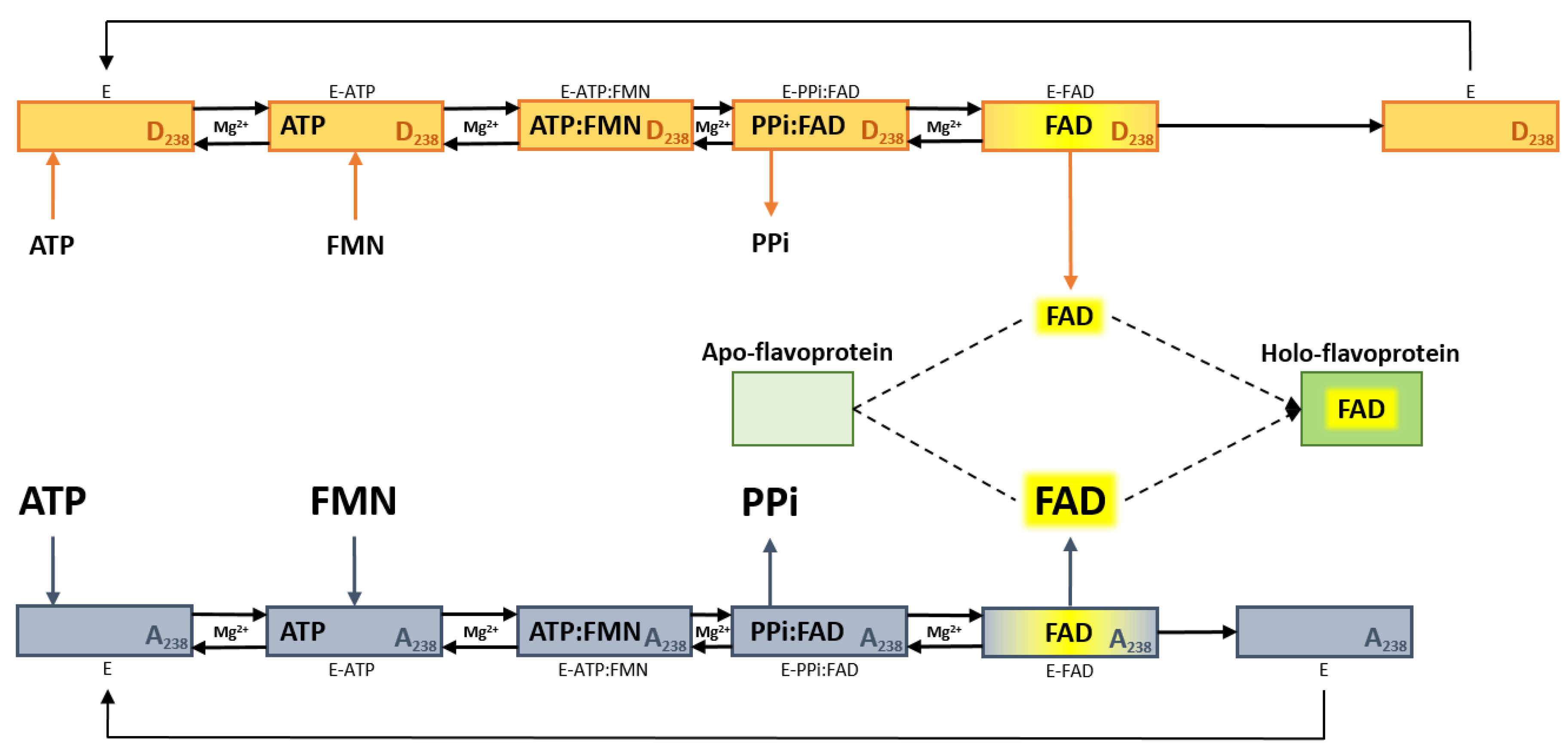
2.4. Impact of the D238A Mutation in the Binding Kinetics of Flavinic Substrates
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Site-Directed Mutagenesis of the hFADS6
4.3. Expression of the WT hFADS6 and hFADS6-D238A Proteins in E. coli
4.4. Purification of Recombinant hFADS6-D238A
4.5. 3D Modeling and Docking of the WT hFADS6 and hFADS6-D238A Proteins
4.6. Measurements of Enzyme Catalyzed Rates for FAD Synthesis and FAD Pyrophosphorolysis
4.7. Kinetics for the Binding of Flavinic Ligands to WT hFADS6 and hFADS6-D238A Proteins
4.8. Isothermal Titration Calorimetry (ITC)
4.9. Electrophoretic Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| FADS | FAD synthase |
| FMNAT | FMN adenylyl transferase |
| hFADS6 | human FAD synthase isoform 6 |
| Rf | riboflavin |
| FMN | flavin mono nucleotide |
| FAD | flavin adenine dinucleotide |
| PAPS | phosphoadenosine 5-phosphosulfate |
| FADSy | FAD synthase domain |
| MPTb | molybdopterin binding |
| FADHy | FAD hydrolase domain |
| BVVLS | Brown–Vialetto-van Laere syndrome |
| RR-MADD PMSF | riboflavin responsive multiple acyl-CoA dehydrogenase deficiency phenylmethyl sulfonyl fluoride |
References
- Barile, M.; Giancaspero, T.A.; Leone, P.; Galluccio, M.; Indiveri, C. Riboflavin transport and metabolism in humans. J. Inherit. Metab. Dis. 2016, 39, 545–557. [Google Scholar] [CrossRef] [PubMed]
- Lienhart, W.D.; Gudipati, V.; Macheroux, P. The human flavoproteome. Arch. Biochem. Biophys. 2013, 535, 150–162. [Google Scholar] [CrossRef] [PubMed]
- Green, P.; Wiseman, M.; Crow, Y.J.; Houlden, H.; Riphagen, S.; Lin, J.P.; Raymond, F.L.; Childs, A.M.; Sheridan, E.; Edwards, S.; et al. Brown-Vialetto-Van Laere syndrome, a ponto-bulbar palsy with deafness, is caused by mutations in c20orf54. Am. J. Hum. Genet. 2010, 86, 485–489. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, B.; Bosch, A.M. Clinical presentation and outcome of riboflavin transporter deficiency: mini review after five years of experience. J. Inherit. Metab. Dis. 2016, 39, 559–564. [Google Scholar] [CrossRef] [PubMed]
- Ryder, B.; Tolomeo, M.; Nochi, Z.; Colella, M.; Barile, M.; Olsen, R.K.; Inbar-Feigenberg, M. A Novel Truncating FLAD1 Variant, Causing Multiple Acyl-CoA Dehydrogenase Deficiency (MADD) in an 8-Year-Old Boy. JIMD Rep. 2019, 45, 37–44. [Google Scholar] [PubMed]
- Balasubramaniam, S.; Christodoulou, J.; Rahman, S. Disorders of riboflavin metabolism. J. Inherit. Metab. Dis. 2019, 42, 608–619. [Google Scholar] [CrossRef]
- Olsen, R.K.J.; Konarikova, E.; Giancaspero, T.A.; Mosegaard, S.; Boczonadi, V.; Matakovic, L.; Veauville-Merllie, A.; Terrile, C.; Schwarzmayr, T.; Haack, T.B.; et al. Riboflavin-Responsive and -Non-responsive Mutations in FAD Synthase Cause Multiple Acyl-CoA Dehydrogenase and Combined Respiratory-Chain Deficiency. Am. J. Hum. Genet. 2016, 98, 1130–1145. [Google Scholar] [CrossRef]
- Yonezawa, A.; Inui, K. Novel riboflavin transporter family RFVT/SLC52: identification, nomenclature, functional characterization and genetic diseases of RFVT/SLC52. Mol. Asp. Med. 2013, 34, 693–701. [Google Scholar] [CrossRef]
- Console, L.; Tolomeo, M.; Colella, M.; Barile, M.; Indiveri, C. Reconstitution in Proteoliposomes of the Recombinant Human Riboflavin Transporter 2 (SLC52A2) Overexpressed in E. coli. Int. J. Mol. Sci. 2019, 20, 4416. [Google Scholar] [CrossRef]
- Brizio, C.; Galluccio, M.; Wait, R.; Torchetti, E.M.; Bafunno, V.; Accardi, R.; Gianazza, E.; Indiveri, C.; Barile, M. Over-expression in Escherichia coli and characterization of two recombinant isoforms of human FAD synthetase. Biochem. Biophys. Res. Commun. 2006, 344, 1008–1016. [Google Scholar] [CrossRef]
- Liuzzi, V.C.; Giancaspero, T.A.; Gianazza, E.; Banfi, C.; Barile, M.; De Giorgi, C. Silencing of FAD synthase gene in Caenorhabditis elegans upsets protein homeostasis and impacts on complex behavioral patterns. Biochim. Biophys. Acta 2012, 1820, 521–531. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Repetto, B.; Glerum, D.M.; Tzagoloff, A. Cloning and characterization of FAD1, the structural gene for flavin adenine dinucleotide synthetase of Saccharomyces cerevisiae. Mol. Cell Biol. 1995, 15, 264–271. [Google Scholar] [CrossRef] [PubMed]
- Huerta, C.; Borek, D.; Machius, M.; Grishin, N.V.; Zhang, H. Structure and mechanism of a eukaryotic FMN adenylyltransferase. J. Mol. Biol. 2009, 389, 388–400. [Google Scholar] [CrossRef] [PubMed]
- Leulliot, N.; Blondeau, K.; Keller, J.; Ulryck, N.; Quevillon-Cheruel, S.; van Tilbeurgh, H. Crystal structure of yeast FAD synthetase (Fad1) in complex with FAD. J. Mol. Biol. 2010, 398, 641–646. [Google Scholar] [CrossRef] [PubMed]
- Torchetti, E.M.; Brizio, C.; Colella, M.; Galluccio, M.; Giancaspero, T.A.; Indiveri, C.; Roberti, M.; Barile, M. Mitochondrial localization of human FAD synthetase isoform 1. Mitochondrion 2010, 10, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Barile, M.; Giancaspero, T.A.; Brizio, C.; Panebianco, C.; Indiveri, C.; Galluccio, M.; Vergani, L.; Eberini, I.; Gianazza, E. Biosynthesis of flavin cofactors in man: implications in health and disease. Curr. Pharm. Des. 2013, 19, 2649–2675. [Google Scholar] [CrossRef] [PubMed]
- Giancaspero, T.A.; Colella, M.; Brizio, C.; Difonzo, G.; Fiorino, G.M.; Leone, P.; Brandsch, R.; Bonomi, F.; Iametti, S.; Barile, M. Remaining challenges in cellular flavin cofactor homeostasis and flavoprotein biogenesis. Front. Chem. 2015, 3, 30. [Google Scholar] [CrossRef]
- Miccolis, A.; Galluccio, M.; Nitride, C.; Giancaspero, T.A.; Ferranti, P.; Iametti, S.; Indiveri, C.; Bonomi, F.; Barile, M. Significance of redox-active cysteines in human FAD synthase isoform 2. Biochim. Biophys. Acta 2014, 1844, 2086–2095. [Google Scholar] [CrossRef]
- Giancaspero, T.A.; Galluccio, M.; Miccolis, A.; Leone, P.; Eberini, I.; Iametti, S.; Indiveri, C.; Barile, M. Human FAD synthase is a bi-functional enzyme with a FAD hydrolase activity in the molybdopterin binding domain. Biochem. Biophys. Res. Commun. 2015, 465, 443–449. [Google Scholar] [CrossRef]
- Leone, P.; Galluccio, M.; Brizio, C.; Barbiroli, A.; Iametti, S.; Indiveri, C.; Barile, M. The hidden side of the human FAD synthase 2. Int. J. Biol. Macromol. 2019, 138, 986–995. [Google Scholar] [CrossRef]
- Leone, P.; Galluccio, M.; Barbiroli, A.; Eberini, I.; Tolomeo, M.; Vrenna, F.; Gianazza, E.; Iametti, S.; Bonomi, F.; Indiveri, C.; et al. Bacterial Production, Characterization and Protein Modeling of a Novel Monofuctional Isoform of FAD Synthase in Humans: An Emergency Protein? Molecules 2018, 23, 116. [Google Scholar] [CrossRef]
- Miccolis, A.; Galluccio, M.; Giancaspero, T.A.; Indiveri, C.; Barile, M. Bacterial over-expression and purification of the 3’phosphoadenosine 5’phosphosulfate (PAPS) reductase domain of human FAD synthase: functional characterization and homology modeling. Int. J. Mol. Sci. 2012, 13, 16880–16898. [Google Scholar] [CrossRef]
- Torchetti, E.M.; Bonomi, F.; Galluccio, M.; Gianazza, E.; Giancaspero, T.A.; Iametti, S.; Indiveri, C.; Barile, M. Human FAD synthase (isoform 2): A component of the machinery that delivers FAD to apo-flavoproteins. FEBS J. 2011, 278, 4434–4449. [Google Scholar] [CrossRef]
- Huerta, C.; Grishin, N.V.; Zhang, H. The “super mutant” of yeast FMN adenylyltransferase enhances the enzyme turnover rate by attenuating product inhibition. Biochemistry 2013, 52, 3615–3617. [Google Scholar] [CrossRef]
- Weber, G. Fluorescence of riboflavin and flavin-adenine dinucleotide. Biochem. J. 1950, 47, 114–121. [Google Scholar] [CrossRef]
- Sebastián, M.; Velázquez-Campoy, A.; Medina, M. The RFK catalytic cycle of the pathogen Streptococcus pneumoniae shows species-specific features in prokaryotic FMN synthesis. J. Enzyme Inhib. Med. Chem. 2018, 33, 842–849. [Google Scholar] [CrossRef]
- Pebay-Peyroula, E.; Dahout-Gonzalez, C.; Kahn, R.; Trezeguet, V.; Lauquin, G.J.; Brandolin, G. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature 2003, 426, 39–44. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Shapovalov, M.V.; Dunbrack, R.L., Jr. A smoothed backbone-dependent rotamer library for proteins derived from adaptive kernel density estimates and regressions. Structure 2011, 19, 844–858. [Google Scholar] [CrossRef]
- Barile, M.; Passarella, S.; Bertoldi, A.; Quagliariello, E. Flavin adenine dinucleotide synthesis in isolated rat liver mitochondria caused by imported flavin mononucleotide. Arch. Biochem Biophys 1993, 305, 442–447. [Google Scholar] [CrossRef]
- Barile, M.; Brizio, C.; De Virgilio, C.; Delfine, S.; Quagliariello, E.; Passarella, S. Flavin adenine dinucleotide and flavin mononucleotide metabolism in rat liver—The occurrence of FAD pyrophosphatase and FMN phosphohydrolase in isolated mitochondria. Eur. J. Biochem. 1997, 249, 777–785. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Brizio, C.; Brandsch, R.; Bufano, D.; Pochini, L.; Indiveri, C.; Barile, M. Over-expression in Escherichia coli, functional characterization and refolding of rat dimethylglycine dehydrogenase. Protein Expr. Purif. 2004, 37, 434–442. [Google Scholar] [CrossRef]



| 6His-hFADS6 | 6His-D238A-hFADS6 | |
|---|---|---|
| Forward reaction | ||
| Km FMN (µM) | 0.13 ± 0.01 | 1.3 ± 0.3 |
| Km ATP (µM) | 6.9 ± 0.5 | 44 ± 4 |
| Ac50 Mg2+ (mM) | 0.15 ± 0.02 | 3.5 ± 0.9 |
| Reverse reaction | ||
| Km PPi (mM) | 0.042 ± 0.006 | 0.060 ± 0.008 |
| Km FAD (µM) | 0.0079 ± 0.0017 * | 0.045 ± 0.008 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leone, P.; Galluccio, M.; Quarta, S.; Anoz-Carbonell, E.; Medina, M.; Indiveri, C.; Barile, M. Mutation of Aspartate 238 in FAD Synthase Isoform 6 Increases the Specific Activity by Weakening the FAD Binding. Int. J. Mol. Sci. 2019, 20, 6203. https://doi.org/10.3390/ijms20246203
Leone P, Galluccio M, Quarta S, Anoz-Carbonell E, Medina M, Indiveri C, Barile M. Mutation of Aspartate 238 in FAD Synthase Isoform 6 Increases the Specific Activity by Weakening the FAD Binding. International Journal of Molecular Sciences. 2019; 20(24):6203. https://doi.org/10.3390/ijms20246203
Chicago/Turabian StyleLeone, Piero, Michele Galluccio, Stefano Quarta, Ernesto Anoz-Carbonell, Milagros Medina, Cesare Indiveri, and Maria Barile. 2019. "Mutation of Aspartate 238 in FAD Synthase Isoform 6 Increases the Specific Activity by Weakening the FAD Binding" International Journal of Molecular Sciences 20, no. 24: 6203. https://doi.org/10.3390/ijms20246203
APA StyleLeone, P., Galluccio, M., Quarta, S., Anoz-Carbonell, E., Medina, M., Indiveri, C., & Barile, M. (2019). Mutation of Aspartate 238 in FAD Synthase Isoform 6 Increases the Specific Activity by Weakening the FAD Binding. International Journal of Molecular Sciences, 20(24), 6203. https://doi.org/10.3390/ijms20246203









