MicroRNA-137 Inhibits Cancer Progression by Targeting Del-1 in Triple-Negative Breast Cancer Cells
Abstract
1. Introduction
2. Results
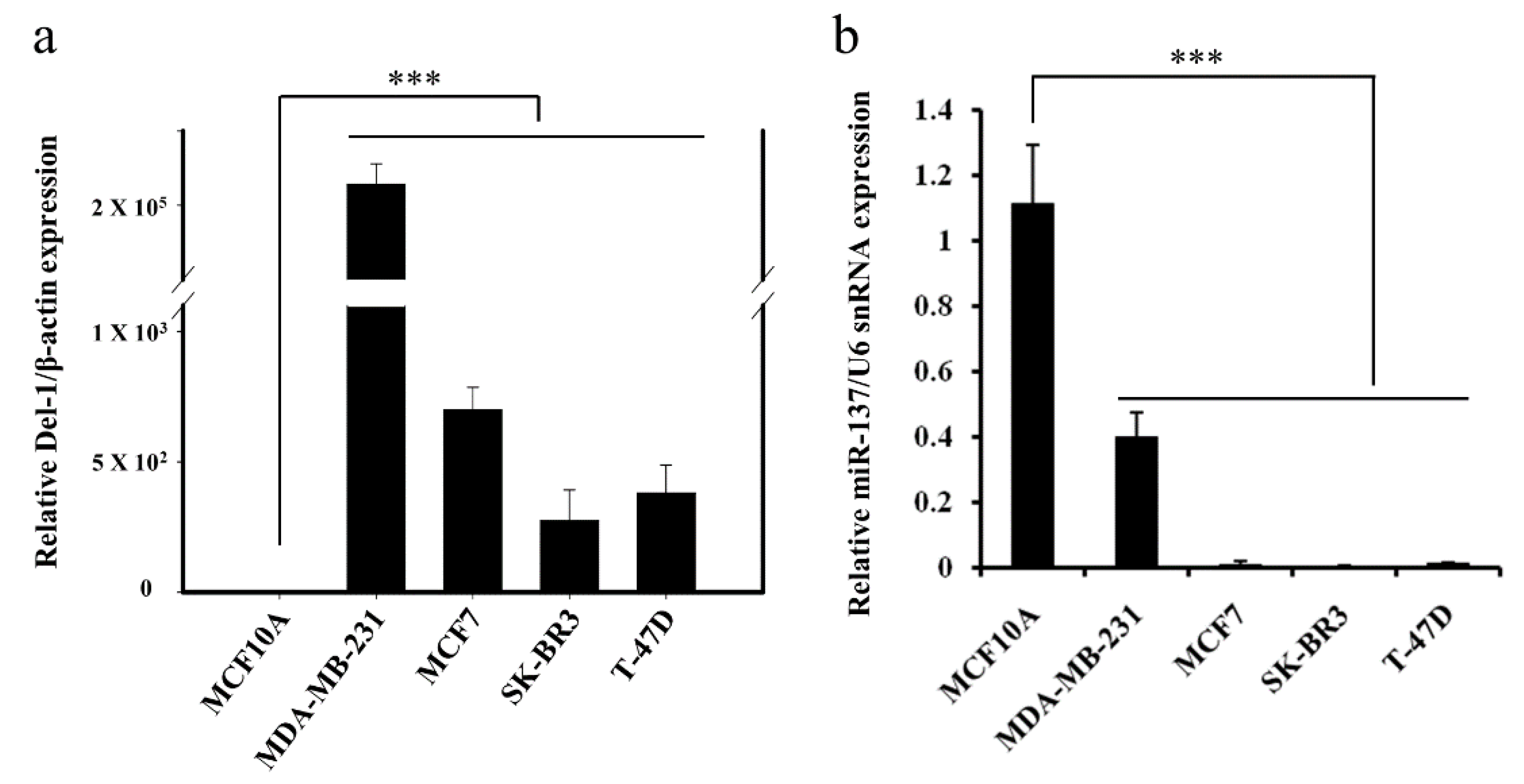
2.1. Negative Correlation between miR-137 and Del-1 Expression in TNBC Cell Lines
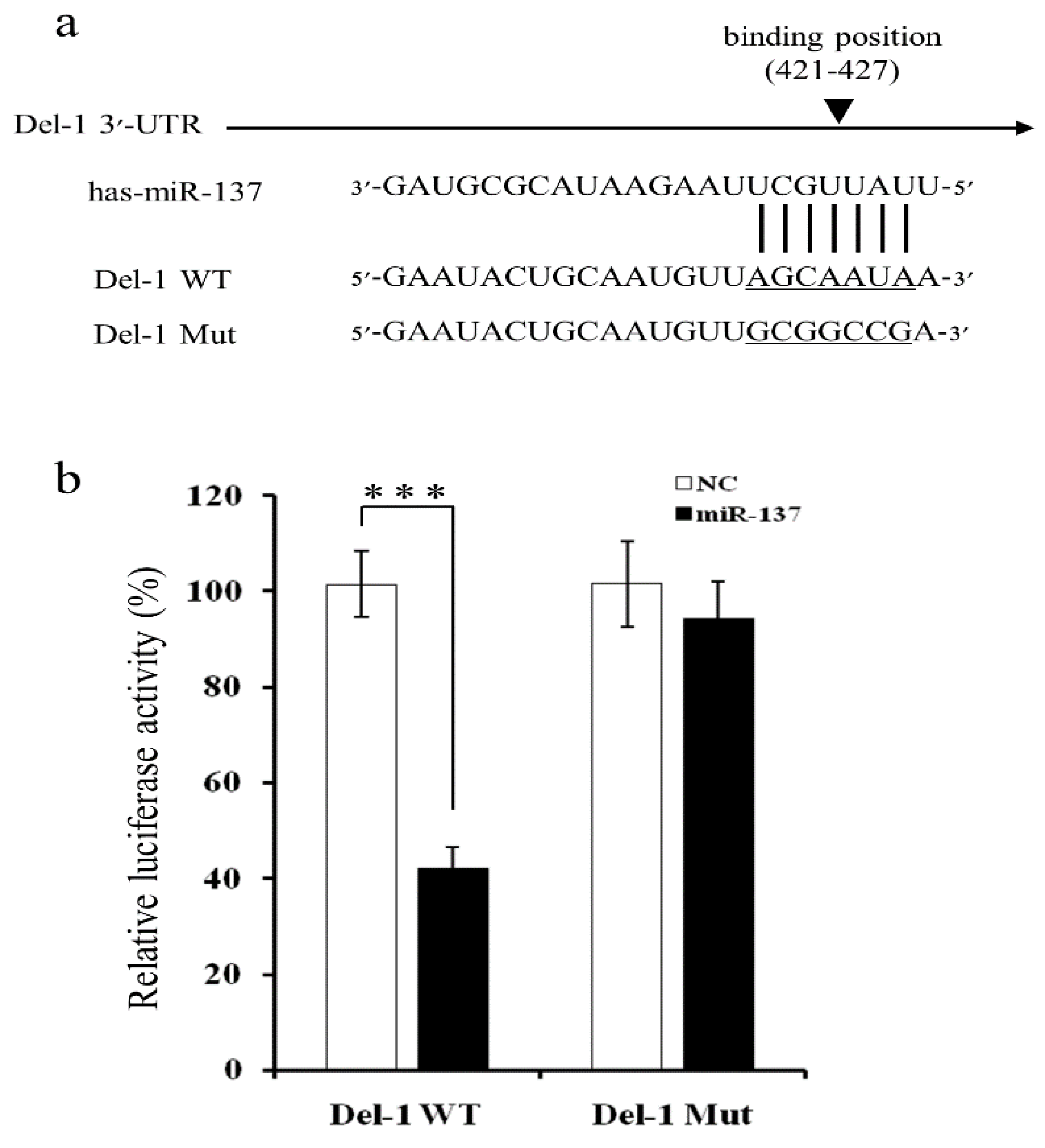
2.2. MiR-137 Directly Targets the 3′-UTR of Del-1 to Inhibit Del-1 Expression
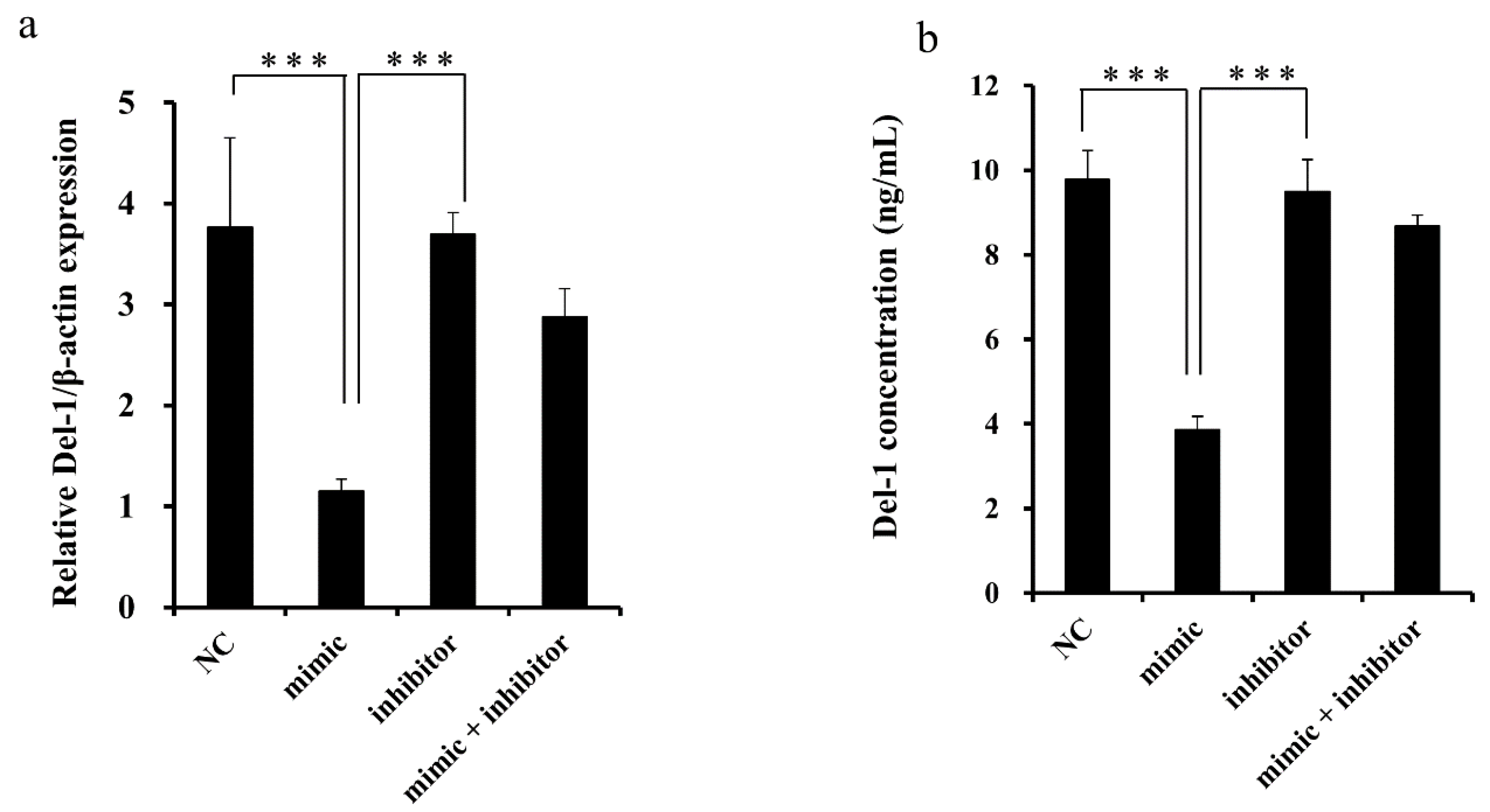
2.3. miR-137 Regulation of Endogenous Del-1 Expression
2.4. miR-137 Inhibits MDA-MB-231 Cell Proliferation, Migration, and Invasion
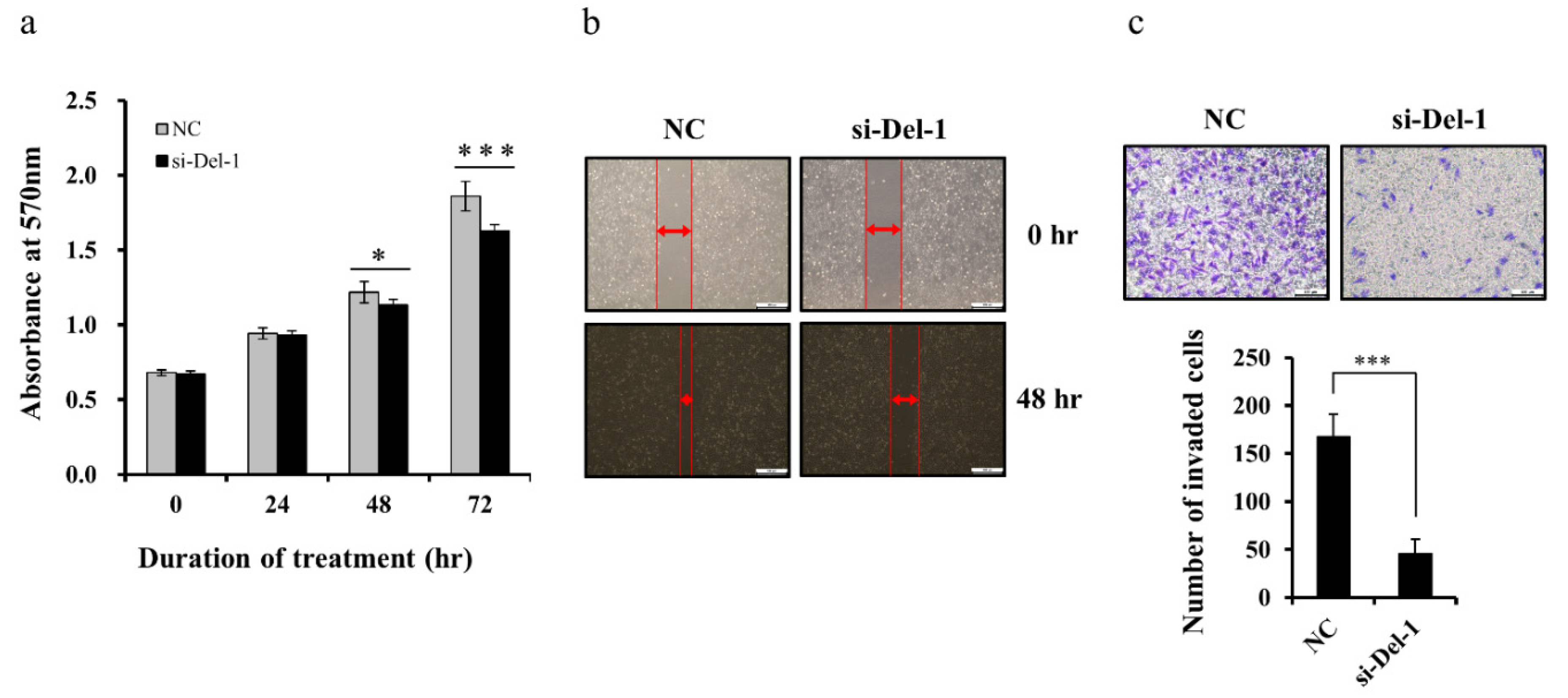
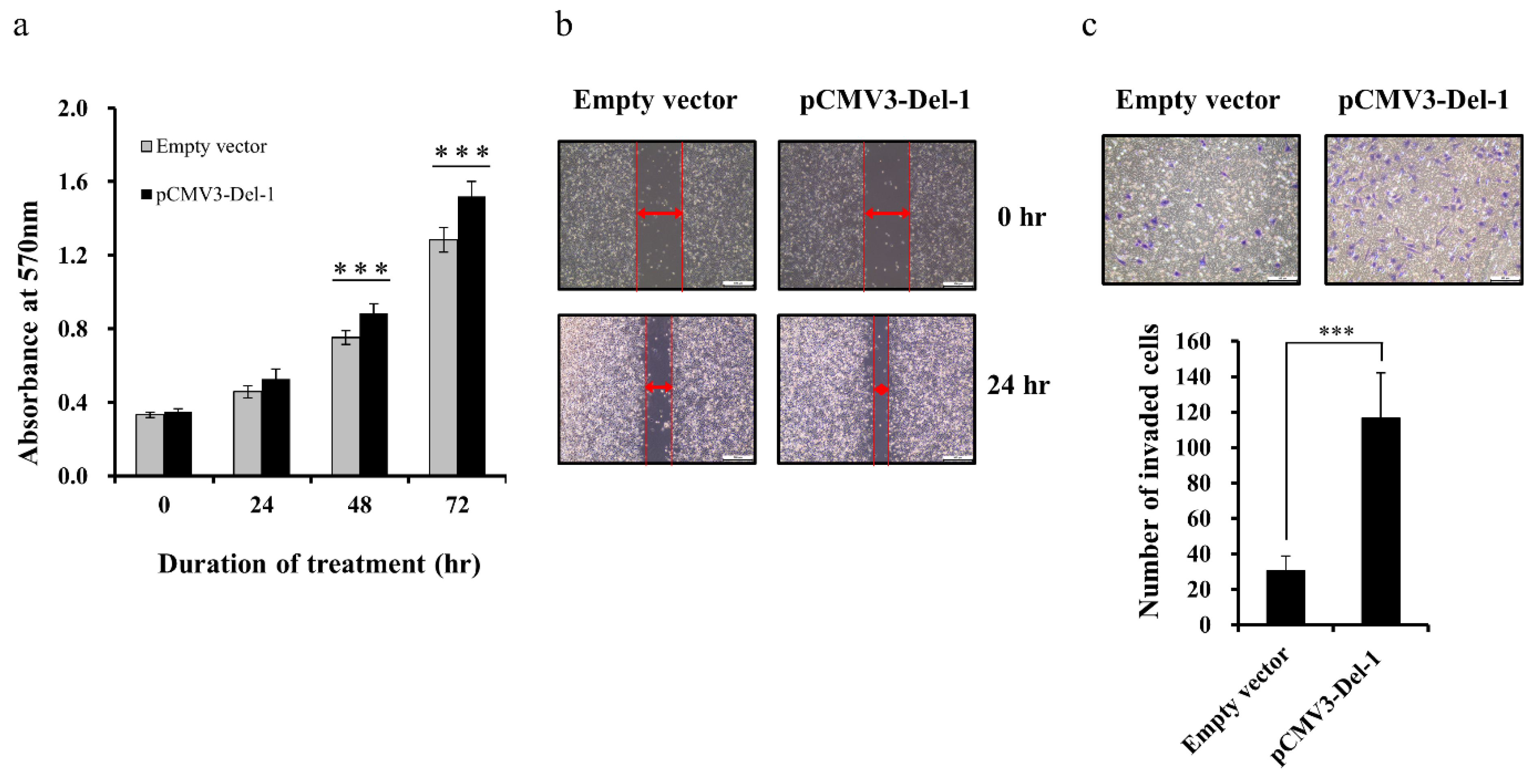
2.5. Del-1 Expression Regulates Proliferation, Migration, and Invasion of MDA-MB-231 Cells
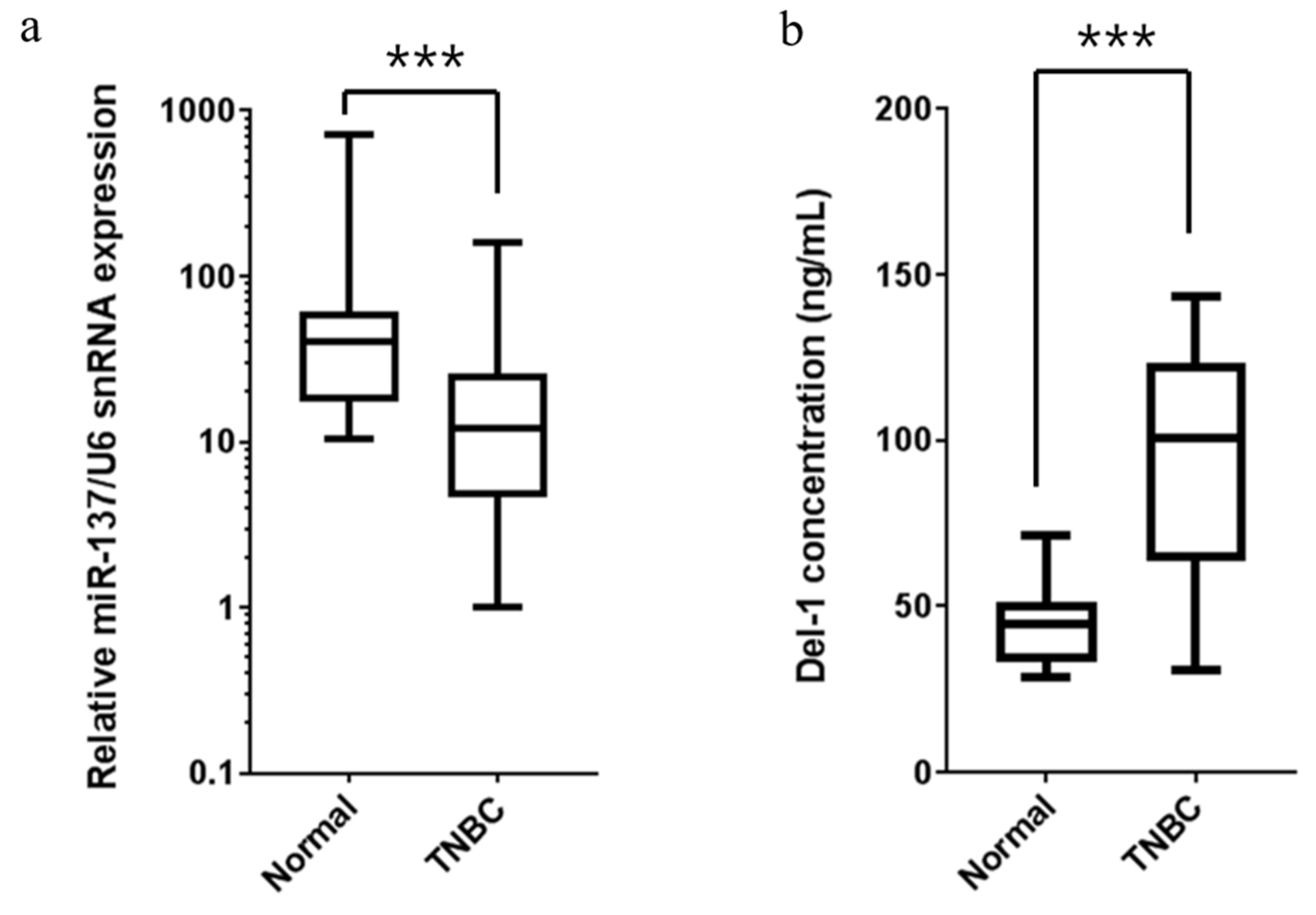
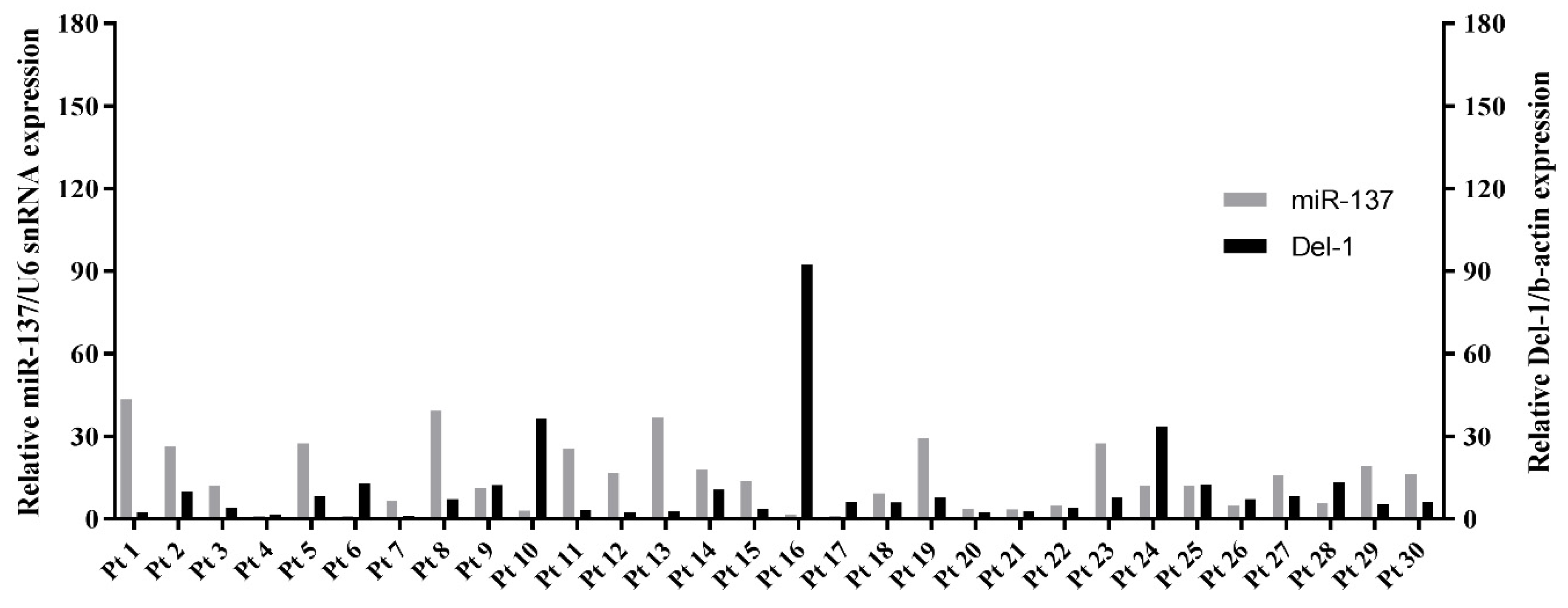
2.6. Expression of miR-137 and Del-1 in TNBC Patients
3. Discussion
4. Materials and Methods
4.1. Candidate miRNA Selection
4.2. Cell Lines and Cell Culture
4.3. RNA Extraction and Quantitative RT-PCR
4.4. miRNA Transfection
4.5. Plasmid Vector Construction for Luciferase Reporter Assays and Mutagenesis
4.6. Dual-Luciferase Reporter Assay
4.7. ELISA
4.8. MTT Assay for Cell Proliferation
4.9. Wound Scratch Assay for Cell Migration
4.10. Transwell Assay for Cell Invasion
4.11. Expression of miR-137 and Del-1 in Tissues
4.12. Knockdown and Overexpression of Del-1 in MDA-MB-231 Cells
4.13. Statistical Analyses
4.14. Ethical Considerations
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| HER2 | human epidermal growth factor receptor 2 |
| Del-1 | developmental endothelial locus-1 |
| Edil-3 | epidermal growth factor-like repeats and discoidin domains-3 |
| WT | wild type |
| Mut | mutant type |
| ELISA | enzyme-linked immunosorbent assay |
| NC | control group |
| siRNA | small-interfering RNA |
| TNBC | triple negative breast cancer |
| SD | standard deviation |
References
- Moon, P.G.; Lee, J.E.; Cho, Y.E.; Lee, S.J.; Jung, J.H.; Chae, Y.S.; Bae, H.I.; Kim, Y.B.; Kim, I.S.; Park, H.Y.; et al. Identification of developmental endothelial locus-1 on circulating extracellular vesicles as a novel biomarker for early breast cancer detection. Clin. Cancer Res. 2016, 22, 1757–1766. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Lee, S.H.; Lee, M.N.; Oh, G.T.; Choi, K.C.; Choi, E.Y. p53 regulates the transcription of the anti-inflammatory molecule developmental endothelial locus-1 (Del-1). Oncotarget 2013, 4, 1976–1985. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lee, S.H.; Kim, D.Y.; Jing, F.; Kim, H.; Yun, C.O.; Han, D.J.; Choi, E.Y. Del-1 overexpression potentiates lung cancer cell proliferation and invasion. Biochem. Biophys. Res. Commun. 2015, 468, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Lim, L.P.; Lau, N.C.; Garrett-Engele, P.; Grimson, A.; Schelter, J.M.; Castle, J.; Bartel, D.P.; Linsley, P.S.; Johnson, J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005, 433, 769–773. [Google Scholar] [CrossRef] [PubMed]
- Ambs, S.; Prueitt, R.L.; Yi, M.; Hudson, R.S.; Howe, T.M.; Petrocca, F.; Wallace, T.A.; Liu, C.G.; Volinia, S.; Calin, G.A. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res. 2008, 68, 6162–6170. [Google Scholar] [CrossRef]
- Hobert, O. Gene regulation by transcription factors and microRNAs. Science 2008, 319, 1785–1786. [Google Scholar] [CrossRef]
- Iorio, M.V.; Croce, C.M. MicroRNAs in cancer: Small molecules with a huge impact. J. Clin. Oncol. 2009, 27, 5848–5856. [Google Scholar] [CrossRef]
- Lee, J.E.; Moon, P.G.; Cho, Y.E.; Kim, Y.B.; Kim, I.S.; Park, H.; Baek, M.C. Identification of EDIL3 on extracellular vesicles involved in breast cancer cell invasion. J. Proteom. 2016, 131, 17–28. [Google Scholar] [CrossRef]
- Sun, J.C.; Liang, X.T.; Pan, K.; Wang, H.; Zhao, J.J.; Li, J.J.; Ma, H.Q.; Chen, Y.B.; Xia, J.C. High expression level of EDIL3 in HCC predicts poor prognosis of HCC patients. World J. Gastroenterol. 2010, 16, 4611–4615. [Google Scholar] [CrossRef]
- Beckham, C.J.; Olsen, J.; Yin, P.N.; Wu, C.H.; Ting, H.J.; Hagen, F.K.; Scosyrev, E.; Messing, E.M.; Lee, Y.F. Bladder cancer exosomes contain EDIL-3/Del1 and facilitate cancer progression. J. Urol. 2014, 192, 583–592. [Google Scholar] [CrossRef]
- Goldhirsch, A.; Ingle, J.N.; Gelber, R.D.; Coates, A.S.; Thürlimann, B.; Senn, H.J. Panel members. Thresholds for therapies: Highlights of the St Gallen International Expert Consensus on the primary therapy of early breast cancer 2009. Ann. Oncol. 2009, 20, 1319–1329. [Google Scholar] [CrossRef] [PubMed]
- Goldhirsch, A. Personalized adjuvant therapies: Lessons from the past: The opening address by the St. Gallen 2013 award recipient. Breast 2013, 22, S3–S7. [Google Scholar] [CrossRef] [PubMed]
- Eskan, M.A.; Jotwani, R.; Abe, T.; Chmelar, J.; Lim, J.H.; Liang, S.; Ciero, P.A.; Krauss, J.L.; Li, F. The leukocyte integrin antagonist Del-1 inhibits IL-17-mediated inflammatory bone loss. Nat. Immunol. 2012, 13, 465–473. [Google Scholar] [CrossRef] [PubMed]
- Choi, E.Y.; Chavakis, E.; Czabanka, M.A.; Langer, H.F.; Fraemohs, L.; Economopoulou, M.; Kundu, R.K.; Orlandi, A.; Zheng, Y.Y. Del-1, an endogenous leukocyte-endothelial adhesion inhibitor, limits inflammatory cell recruitment. Science 2008, 322, 1101–1104. [Google Scholar] [CrossRef] [PubMed]
- Mitroulis, I.; Kang, Y.Y.; Gahmberg, C.G.; Siegert, G.; Hajishengallis, G.; Chavakis, T.; Choi, E.Y. Developmental endothelial locus-1 attenuates complement-dependent phagocytosis through inhibition of Mac-1-integrin. Thromb. Haemost. 2014, 111, 1004–1006. [Google Scholar] [CrossRef] [PubMed]
- Choi, E.Y.; Lim, J.H.; Neuwirth, A.; Economopoulou, M.; Chatzigeorgiou, A.; Chung, K.J.; Bittner, S.; Lee, S.H.; Langer, H.; Samus, M. Developmental endothelial locus-1 is a homeostatic factor in the central nervous system limiting neuroinflammation and demyelination. Mol. Psychiatry 2015, 20, 880–888. [Google Scholar] [CrossRef]
- Dong, S.; Jin, M.; Li, Y.; Ren, P.; Liu, J. MiR-137 acts as a tumor suppressor in papillary thyroid carcinoma by targeting CXCL12. Oncol. Rep. 2016, 35, 2151–2158. [Google Scholar] [CrossRef]
- Du, Y.; Chen, Y.; Wang, F.; Gu, L. miR-137 plays tumor suppressor roles in gastric cancer cell lines by targeting KLF12 and MYO1C. Tumor Biol. 2016, 37, 13557–13569. [Google Scholar] [CrossRef]
- Han, Y.; Bi, Y.; Bi, H.; Diao, C.; Zhang, G.; Cheng, K.; Yang, Z. miR-137 suppresses the invasion and procedure of EMT of human breast cancer cell line MCF-7 through targeting CtBP1. Hum. Cell 2016, 29, 30–36. [Google Scholar] [CrossRef]
- Neault, M.; Mallette, F.A.; Richard, S. miR-137 modulates a tumor suppressor network-inducing senescence in pancreatic cancer cells. Cell Rep. 2016, 14, 1966–1978. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, Y.; Lou, G.; Zhao, L.; Xu, Z.; Zhang, Y.; He, F. MiR-137 targets estrogen-related receptor alpha and impairs the proliferative and migratory capacity of breast cancer cells. PLoS ONE 2012, 7, e39102. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Liang, J.; Wang, Q.; Li, Z.; Du, Y.; Xu, X. MicroRNA-137 suppresses tongue squamous carcinoma cell proliferation, migration and invasion. Cell Prolif. 2016, 49, 628–635. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Li, K.; Huang, W.; Zhang, X. MiR-137 inhibited cell proliferation and migration of vascular smooth muscle cells via targeting IGFBP-5 and modulating the mTOR/STAT3 signaling. PLoS ONE 2017, 10, e0186245. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zou, Y.; Zheng, R.; Ma, X. MiR-137 inhibits cell proliferation in acute lymphoblastic leukemia by targeting JARID1B. Eur. J. Haematol. 2019, 103, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Nuzzo, S.; Catuogno, S.; Capuozzo, M.; Fiorelli, A.; Swiderski, P.; Boccella, S.; de Nigris, F.; Esposito, C.L. Axl-Targeted Delivery of the Oncosuppressor miR-137 in Non-small-Cell Lung Cancer. Mol. Ther. Nucleic Acids 2019, 6, 256–263. [Google Scholar] [CrossRef]

| No. | miRNA Name | Predicted Database |
|---|---|---|
| 1 | miR-137 | Miranda, TargetScan, miRDB |
| 2 | miR-496 | Miranda, TargetScan, miRDB |
| 3 | miR-361-5P | TargetScan, miRDB |
| 4 | miR-425 | TargetScan, miRDB |
| 5 | miR-9500 | TargetScan, miRDB |
| 6 | miR-2113 | TargetScan, miRDB |
| 7 | miR-3685 | TargetScan, miRDB |
| 8 | miR-3163 | TargetScan, miRDB |
| 9 | miR-4672 | TargetScan, miRDB |
| 10 | miR-3662 | TargetScan, miRDB |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.J.; Jeong, J.-H.; Kang, S.H.; Kang, J.; Kim, E.A.; Lee, J.; Jung, J.H.; Park, H.Y.; Chae, Y.S. MicroRNA-137 Inhibits Cancer Progression by Targeting Del-1 in Triple-Negative Breast Cancer Cells. Int. J. Mol. Sci. 2019, 20, 6162. https://doi.org/10.3390/ijms20246162
Lee SJ, Jeong J-H, Kang SH, Kang J, Kim EA, Lee J, Jung JH, Park HY, Chae YS. MicroRNA-137 Inhibits Cancer Progression by Targeting Del-1 in Triple-Negative Breast Cancer Cells. International Journal of Molecular Sciences. 2019; 20(24):6162. https://doi.org/10.3390/ijms20246162
Chicago/Turabian StyleLee, Soo Jung, Jae-Hwan Jeong, Seung Hee Kang, Jieun Kang, Eun Ae Kim, Jeeyeon Lee, Jin Hyang Jung, Ho Yong Park, and Yee Soo Chae. 2019. "MicroRNA-137 Inhibits Cancer Progression by Targeting Del-1 in Triple-Negative Breast Cancer Cells" International Journal of Molecular Sciences 20, no. 24: 6162. https://doi.org/10.3390/ijms20246162
APA StyleLee, S. J., Jeong, J.-H., Kang, S. H., Kang, J., Kim, E. A., Lee, J., Jung, J. H., Park, H. Y., & Chae, Y. S. (2019). MicroRNA-137 Inhibits Cancer Progression by Targeting Del-1 in Triple-Negative Breast Cancer Cells. International Journal of Molecular Sciences, 20(24), 6162. https://doi.org/10.3390/ijms20246162





