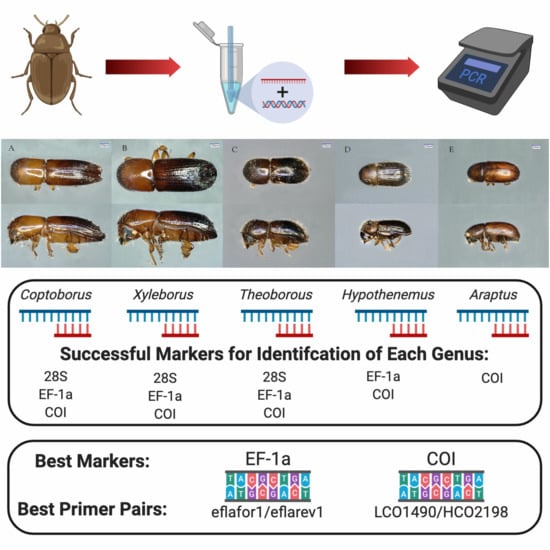
Rapid Molecular Identification of Scolytinae (Coleoptera: Curculionidae)
Abstract
1. Introduction
2. Results
2.1. Quantity and Quality of Extracted DNA
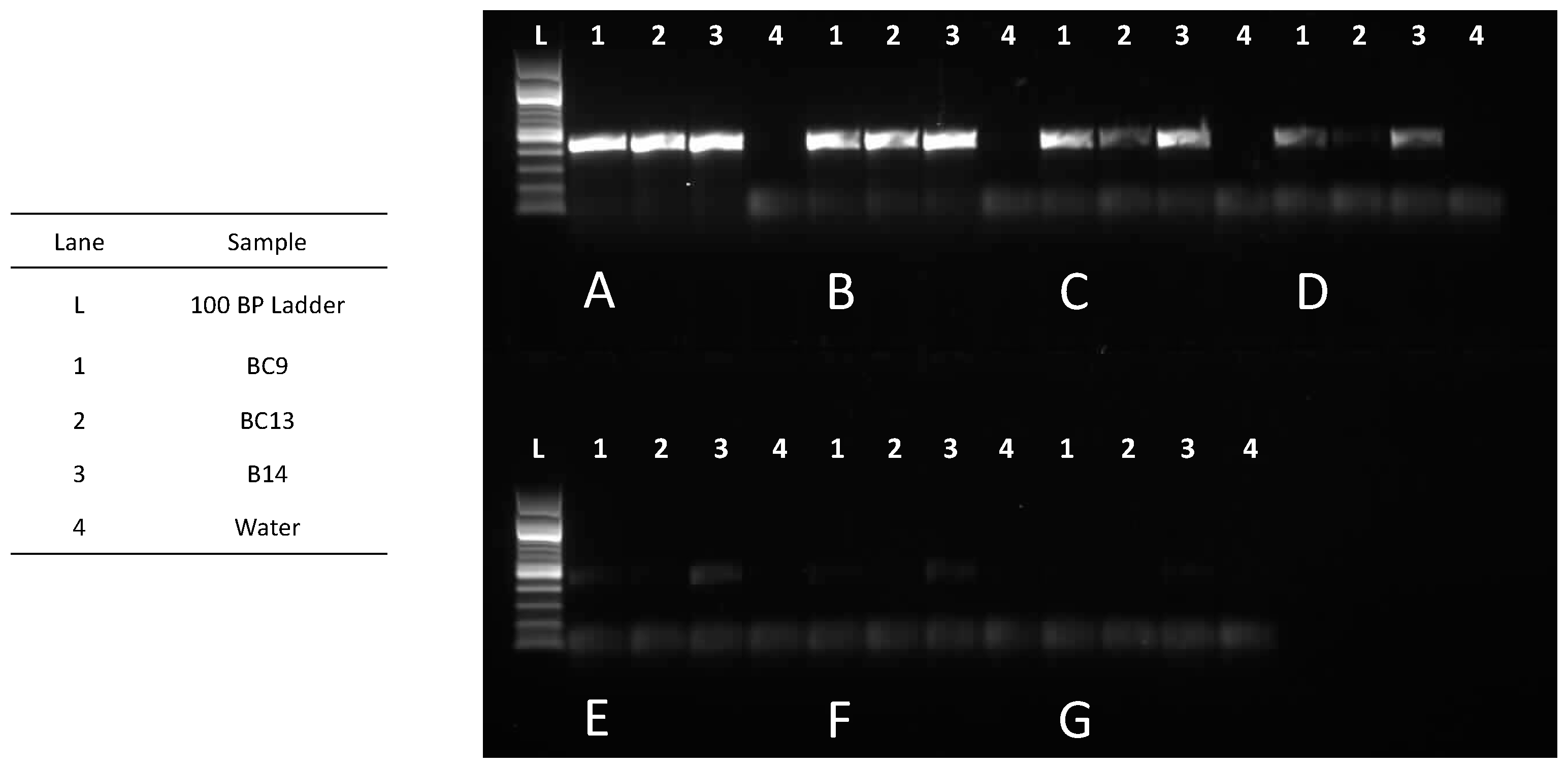
2.2. DNA Threshold for Molecular Identification Using D2F1/D3R2
2.3. Primer Pairs for Molecular Identification of Diverse Genera
3. Discussion
3.1. Quantity and Quality of Extracted DNA
3.2. DNA Threshold for Molecular Identification Using D2F1/D3R2
3.3. Primer Pairs for Molecular Identification of Diverse Genera
4. Materials and Methods
4.1. Insect Specimens
4.2. DNA Extraction
4.3. DNA Quality
4.4. DNA Threshold for Molecular Identification Using D2F1/D3R2
4.5. Primer Pairs for Molecular Identification of Diverse Genera
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hulcr, J.; Dunn, R.R. The sudden emergence of pathogenicity in insect-fungus symbioses threatens naive forest ecosystems. Proc. R. Soc. 2011, 278, 2866–2873. [Google Scholar] [CrossRef] [PubMed]
- Ploetz, R.C.; Hulcr, J.; Wingfield, M.J.; de Beer, Z.W. Destructive Tree Diseases Associated with Ambrosia and Bark Beetles: Black Swan Events in Tree Pathology? Plant Dis. 2013, 97, 856–872. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.C.; Haack, R.A.; Negron, J.F.; Witcosky, J.J.; Seybold, S.J. Invasive Bark Beetles. Available online: http://purl.access.gpo.gov/GPO/LPS90197 (accessed on 25 November 2019).
- Aukema, J.E.; McCullough, D.G.; Von Holle, B.; Liebhold, A.M.; Britton, K.; Frankel, S.J. Historical Accumulation of Nonindigenous Forest Pests in the Continental United States. Bioscience 2010, 60, 886–897. [Google Scholar] [CrossRef]
- Webber, J. A Natural Biological-Control of Dutch Elm Disease. Nature 1981, 292, 449–451. [Google Scholar] [CrossRef]
- Hanula, J.L.; Mayfield, A.E.; Fraedrich, S.W.; Rabaglia, R.J. Biology and host associations of redbay ambrosia beetle (Coleoptera: Curculionidae: Scolytinae), exotic vector of laurel wilt killing redbay trees in the southeastern United States. J. Econ. Entomol. 2008, 101, 1276–1286. [Google Scholar] [CrossRef]
- Kendra, P.E.; Montgomery, W.S.; Niogret, J.; Pruett, G.E.; Mayfield, A.E., III; MacKenzie, M.; Deyrup, M.A.; Bauchan, G.R.; Ploetz, R.C.; Epsky, N.D. North American Lauraceae: Terpenoid emissions, relative attraction and boring preferences of redbay ambrosia beetle, Xyleborus glabratus (Coleoptera: Curculionidae: Scolytinae). PLoS ONE 2014, 9, e102086. [Google Scholar] [CrossRef]
- Tisserat, N.; Cranshaw, W.; Leatherman, D.; Utley, C.; Alexander, K. Black walnut mortality in Colorado caused by the walnut twig beetle and thousand cankers disease. Phytopathology 2009, 10, 10. [Google Scholar] [CrossRef]
- Eskalen, A.; Gonzalez, A.; Wang, D.H.; Twizeyimana, M.; Mayorquin, J.S.; Lynch, S.C. First Report of a Fusarium sp. and Its Vector Tea Shot Hole Borer (Euwallacea fornicatus) Causing Fusarium Dieback on Avocado in California. Plant Dis. 2012, 96, 1070. [Google Scholar] [CrossRef]
- Kendra, P.E.; Owens, D.; Montgomery, W.S.; Narvaez, T.I.; Bauchan, G.R.; Schnell, E.Q.; Tabanca, N.; Carrillo, D. α-Copaene is an attractant, synergistic with quercivorol, for improved detection of Euwallacea nr. fornicatus (Coleoptera: Curculionidae: Scolytinae). PLoS ONE 2017, 12, e0179416. [Google Scholar] [CrossRef]
- Haack, R.A. Exotic bark- and wood-boring Coleoptera in the United States: Recent establishments and interceptions. Can. J. For. Res. 2016, 36, 269–288. [Google Scholar] [CrossRef]
- Haack, R.A.; Cavey, J.F. Insects Intercepted on Wood Articles at ports-of-Entry in the United States: 1985-1996. Newsl. Mich. Entomol. Soc. 1997, 42, 1–6. [Google Scholar]
- Haack, R.A.; Cavey, J.F. Insects Intercepted on Solid Wood Packing Materials at United States Ports-of-Entry: 1985–1998. Available online: https://www.nrs.fs.fed.us/pubs/2116 (accessed on 25 November 2019).
- Haack, R.A. Intercepted Scolytidae (Coleoptera) at U.S. ports of entry: 1985–2000. Integr. Pest Manag. Rev. 2001, 6, 253–282. [Google Scholar] [CrossRef]
- Cognato, A.I.; Sperling, F.A.H. Phylogeny of Ips DeGeer species (Coleoptera: Scolytidae) inferred from mitochondrial cytochrome oxidase I DNA sequence. Mol. Phylog. Evol. 2000, 14, 445–460. [Google Scholar] [CrossRef] [PubMed]
- Jordal, B.H.; Sequeira, A.S.; Cognato, A.I. The age and phylogeny of wood boring weevils and the origin of subsociality. Mol. Phylog. Evol. 2011, 59, 708–724. [Google Scholar] [CrossRef]
- O’Donnell, K.; Libeskind-HadasJiri, R. Invasive Asian Fusarium – Euwallacea ambrosia beetle mutualists pose a serious threat to forests, urban landscapes and the avocado industry. Phytoparasitica 2016, 44, 435–442. [Google Scholar] [CrossRef]
- Jordal, B.H.; Gillespie, J.J.; Cognato, A.I. Secondary structure alignment and direct optimization of 28S rDNA sequences provide limited phylogenetic resolution in bark and ambrosia beetles (Curculionidae: Scolytinae). Zool. Scr. 2008, 37, 43–56. [Google Scholar] [CrossRef]
- Hebert, P.D.; Cywinska, A.; Ball, S.L.; deWaard, J.R. Biological identifications through DNA barcodes. Proc. Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef]
- Dole, S.A.; Jordal, B.H.; Cognato, A.I. Polyphyly of Xylosandrus reitter inferred from nuclear and mitochondrial genes (Coleoptera: Curculionidae: Scolytinae). Mol. Phylog. Evol. 2010, 54, 773–782. [Google Scholar] [CrossRef]
- Normark, B.B.; Jordal, B.H.; Farrell, B.D. Origin of a haplodiploid beetle lineage. Proc. Biol. Soc. 1999, 266, 2253–2259. [Google Scholar] [CrossRef]
- Andreev, D.; Breilid, H.; Kirkendall, L.; Brun, L.O.; Ffrench-Constant, R.H. Lack of nucleotide variability in a beetle pest with extreme inbreeding. Insect Mol. Biol. 1998, 7, 197–200. [Google Scholar] [CrossRef] [PubMed]
- Georgiou, C.D.; Papapostolou, I. Assay for the quantification of intact/fragmented genomic DNA. Anal. Biochem. 2006, 358, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Sim, S.B.; Yoneishi, N.M.; Brill, E.; Geib, S.M.; Follett, P.A. Molecular Markers Detect Cryptic Predation on Coffee Berry Borer (Coleoptera: Curculionidae) by Silvanid and Laemophloeid Flat Bark Beetles (Coleoptera: Silvanidae, Laemophloeidae) in Coffee Beans. J. Econom. Entomol. 2016, 109, 100–105. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, A.; Maddox, C. Bark beetles (Coleoptera: Curculionidae: Scolytinae) of importance to the Australian macadamia industry: An integrative taxonomic approach to species diagnostics. Aust. J. Entomol. 2010, 49, 104–113. [Google Scholar] [CrossRef]
- Masood, A.; Stoeckle, B.C.; Kuehn, R.; Saeed, S. Cross Species Transfer of Microsatellite Loci in Scolytidae Species Mostly Associated with Mango (Mangifera indica L.; Anacardiaceae) Quick Decline Disease. Pak. J. Zool. 2011, 43, 411–414. [Google Scholar]
- Kendra, P.E.; Sanchez, J.S.; Montgomery, W.S.; Okins, K.E.; Niogret, J.; Pena, J.E.; Epsky, N.D.; Heath, R.R. Diversity of Scolytinae (Coleoptera: Curculionidae) Attracted to Avocado, Lychee, and Essential Oil Lures. Fla. Entomol. 2011, 94, 123–130. [Google Scholar] [CrossRef]
- Cognato, A.I.; Sun, J.H. DNA based cladograms augment the discovery of a new Ips species from China (Coleoptera: Curculionidae: Scolytinae). Cladistics 2007, 23, 539–551. [Google Scholar] [CrossRef]
- Marchal, L.; Cazeres, S.; Kergoat, G.J.; Letellier, K.; Mitchell, A.; Nattier, R.; Mille, C. A new pest of lychees in New Caledonia. N. Z. J. Zool. 2017, 44, 49–64. [Google Scholar] [CrossRef][Green Version]
- Campbell, P.R.; Geering, A.D.W. Biosecurity Capacity Building for the Australian Avocado Industry: Laurel Wilt. Available online: http://www.avocadosource.com/wac7/Section_03/CampbellPaul2011.pdf (accessed on 25 November 2019).
- Simon, C.; Frati, F.; Beckenbach, A.; Crespi, B.; Liu, H.; Flook, P. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann. Entomol. Soc. Am. 1994, 87, 651–701. [Google Scholar] [CrossRef]



| Gene | Primer | Sequence (5′-3′) | Amplicon Size (bp) | Reference |
|---|---|---|---|---|
| 28S | D2F1 | ACTGTTGGCGACGATGTTCT | 500–570 | [18] |
| D3R2 | TCTTCGCCCCTATACCC | |||
| COI | LCO1490 | GGTCAACAAATCATAAAGATATTGG | 630–650 | [19] |
| HCO2198 | TAAACTTCAGGGTGACCAAAAAATCA | |||
| EF-1a | eflafor1 | TACGTAACCATCATTGATGCTYCC | 500 | [20] |
| eflarev1 | CTTCTTTACGTTCAATGGACCATCC | |||
| ets149 | ATCGAGAAGTICGAGAAGGAGGCYCARGAAATGGG | 585 | [21] | |
| efa754 | CCACCAATTTTGTAGACATC | |||
| ITS | ITS2F | GTGGATCCTGTGAACTGCAGGACACATG | 460 | [22] |
| ITS2R | GTGAATTCATGCTTAAATTTAGGGGGTA |
| Primer Pair | Variable Assessed | Xyleborus | Coptoborus | Theoborus | Hypothenemus | Araptus |
|---|---|---|---|---|---|---|
| D2F1 D3R2 | Amplification | 1/1 | 5/5 | 4/4 | 3/4 | 3/3 |
| Quality | 1/1 | 5/5 | 4/4 | 3/4 | 3/3 | |
| Identification | 1/1 | 5/5 | 4/4 | 0/4 | 0/3 | |
| eflafor1 eflarev1 | Amplification | 1/1 | 4/5 | 4/4 | 3/4 | 2/3 |
| Quality | 1/1 | 4/5 | 4/4 | 2/4 | 2/3 | |
| Identification | 1/1 | 4/5 | 4/4 | 3/4 | 0/3 | |
| ets149 efa754 | Amplification | 1/1 | 4/5 | 4/4 | 3/4 | 3/3 |
| Quality | 1/1 | 3/5 | 3/4 | 2/4 | 0/3 | |
| Identification | 1/1 | 4/5 | 3/4 | 2/4 | 0/3 | |
| ITS2F ITS2R | Amplification | 1/1 | 3/5 | 0/4 | 0/4 | 0/3 |
| Quality | 1/1 | 3/5 | 0/4 | 0/4 | 0/3 | |
| Identification | 0/1 | 0/5 | 0/4 | 0/4 | 0/3 | |
| LCO1490 HCO2198 | Amplification | 1/1 | 5/5 | 2/4 | 4/4 | 3/3 |
| Quality | 1/1 | 2/5 | 1/4 | 0/4 | 2/3 | |
| Identification | 1/1 | 2/5 | 2/4 | 1/4 | 2/3 |
| 28S (D2F1/D3R2) | |||||
|---|---|---|---|---|---|
| Sample | Genus | PCR Concentration (ng/μL) | GenBank Match | Accession Number | % Identical |
| B2 | Coptoborus pseudotenuis | 44.6 | Coptoborus pseudotenuis | HM099689 | 100.0% |
| B4 | C. pseudotenuis | 45 | Coptoborus pseudotenuis | HM099689 | 100.0% |
| B6 | C. pseudotenuis | 16.6 | Coptoborus pseudotenuis | HM099689 | 100.0% |
| B7a | C. pseudotenuis | 46.5 | Coptoborus pseudotenuis | HM099689 | 100.0% |
| B7b | C. pseudotenuis | 38.2 | Coptoborus pseudotenuis | HM099689 | 100.0% |
| B3 | Xyleborus ferrugineus | 42.4 | Xyleborus volvulus | HM099763 | 100.0% |
| X. perforans | HM099747 | 100.0% | |||
| X. bispinatus | HM099741 | 100.0% | |||
| X. affinis | GU808581 | 100.0% | |||
| B14 | Theoborus sp. | 39.2 | Theoborus sp. | HM099718 | 99.7% |
| BC1 | Theoborus sp. | 23 | Theoborus sp. | HM099718 | 99.7% |
| BC9 | Theoborus sp. | 42.8 | Theoborus sp. | HM099718 | 99.7% |
| BC13 | Theoborus sp. | 37.7 | Theoborus sp. | HM099718 | 100.0% |
| B16 | Hypothenemus sp. | 2.31 | Theoborus sp. | HM099718 | 99.7% |
| B17 | Hypothenemus sp. | 6.07 | Theoborus sp. | HM099718 | 99.7% |
| B19 | Hypothenemus sp. | 4.74 | Theoborus sp. | HM099718 | 99.7% |
| B20 | Hypothenemus sp. | No amplification | |||
| J1 | Araptus sp. | 4.91 | Ips duplicatus | JX263733 | 93.2% |
| Araptus sp. | AF375297 | 87.9% | |||
| J2 | Araptus sp. | 9.32 | Ips duplicatus | JX263733 | 91.4% |
| Araptus sp. | AF375297 | 87.0% | |||
| J4 | Araptus sp. | 8.08 | Ips duplicatus. | JX263733 | 91.5% |
| Araptus sp. | AF375297 | 87.0% | |||
| EF-1a (eflafor1/eflarev1) | |||||
|---|---|---|---|---|---|
| Sample | Genus | PCR Concentration (ng/μL) | GenBank Match | Accession Number | % Identical |
| B2 | Coptoborus pseudotenuis | 4.37 | Coptoborus pseudotenuis | AF508880 | 99.7% |
| B4 | C. pseudotenuis | 4.36 | Coptoborus pseudotenuis | AF508880 | 99.7% |
| B6 | C. pseudotenuis | No amplification | |||
| B7a | C. pseudotenuis | 11 | Coptoborus pseudotenuis | AF508880 | 100.0% |
| B7b | C. pseudotenuis | 5.1 | Coptoborus pseudotenuis | AF508880 | 100.0% |
| B3 | Xyleborus ferrugineus | 2.92 | Xyleborus ferrugineus | KP941383 | 100.0% |
| B14 | Theoborus sp. Theoborus sp. | 15.8 | Theoborus sp. | HM064194 | 99.6% |
| T. theobromae | AF259881 | 99.6% | |||
| BC1 | Theoborus sp. | 10.2 | Theoborus sp. | HM064194 | 99.7% |
| BC9 | Theoborus sp. | 14.1 | Theoborus sp. | HM064194 | 99.6% |
| BC13 | Theoborus sp. | 19 | Theoborus sp. | HM064194 | 99.7% |
| B16 | Hypothenemus sp. | 10.3 | Hypothenemus nr. eruditus | JX264092 | 97.0% |
| B17 | Hypothenemus sp. | No amplification | |||
| B19 | Hypothenemus sp. | 16.1 | H. nr. eruditus | JX264092 | 97.7% |
| B20 | Hypothenemus sp. | 5 | Hypothenemus sp. | AF186658 | 96.0% |
| H. nr. eruditus | JX264092 | 96.0% | |||
| J1 | Araptus sp. | No amplification | |||
| J2 | Araptus sp. | 11.2 | Phelloterus sp. | KY805860 | 91.6% |
| J4 | Araptus sp. | 7.36 | Phelloterus sp. | KY805860 | 91.6% |
| Araptus attenuatus | FJ347565 | 89.6% | |||
| EF-1a (ets149/efa754) | |||||
|---|---|---|---|---|---|
| Sample | Genus | PCR Concentration (ng/μL) | GenBank Match | Accession Number | % Identical |
| B2 | Coptoborus pseudotenuis | 12.9 | Coptoborus pseudotenuis | AF508880 | 99.7% |
| B4 | C. pseudotenuis | 12.4 | Coptoborus pseudotenuis | AF508880 | 99.0% |
| B6 | C. pseudotenuis | No amplification | |||
| B7a | C. pseudotenuis | 16.3 | Coptoborus pseudotenuis | AF508880 | 100.0% |
| B7b | C. pseudotenuis | 5.1 | Coptoborus pseudotenuis | AF508880 | 100.0% |
| B3 | Xyleborus ferrugineus | 38.9 | Xyleborus ferrugineus | KP941383 | 94.7% |
| B14 | Theoborus sp. | 9.53 | Theoborus theobromae | AF259881 | 99.6% |
| BC1 | Theoborus sp. | 10.2 | Theoborus sp. | HM064194 | 99.7% |
| BC9 | Theoborus sp. | 5.56 | Poor sequence | ||
| BC13 | Theoborus sp. | 9.05 | Theoborus theobromae | AF259881 | 100.0% |
| B16 | Hypothenemus sp. | 25.9 | Hypothenemus nr. eruditus | JX264092 | 98.1% |
| B17 | Hypothenemus sp. | No amplification | |||
| B19 | Hypothenemus sp. | 25.9 | Hypothenemus nr. eruditus | JX264092 | 99.3% |
| B20 | Hypothenemus sp. | 20.4 | Poor sequence | ||
| J1 | Araptus sp. | 7.62 | Poor sequence | ||
| J2 | Araptus sp. | 38.2 | Poor sequence | ||
| J4 | Araptus sp. | 14.9 | Poor sequence | ||
| ITS2 (ITS2F/ITS2R) | |||||
|---|---|---|---|---|---|
| Sample | Genus | PCR Concentration (ng/μL) | GenBank Match | Accession Number | % Identical |
| B2 | Coptoborus pseudotenuis | 12.70 | No match | ||
| B4 | C. pseudotenuis | No amplification | |||
| B6 | C. pseudotenuis | No amplification | |||
| B7a | C. pseudotenuis | 25.80 | No match | ||
| B7b | C. pseudotenuis | 12.10 | Pityogenes chalcographus | JQ066311 | 100.0% |
| Galapaganus spp. | EU748796 | 100.0% | |||
| B3 | Xyleborus ferrugineus | 21.00 | Pityogenes chalcographus | JQ066311 | 100.0% |
| Galapaganus spp. | EU748796 | 100.0% | |||
| B14 | Theoborus sp. | No amplification | |||
| BC1 | Theoborus sp. | No amplification | |||
| BC9 | Theoborus sp. | No amplification | |||
| BC13 | Theoborus sp. | No amplification | |||
| B16 | Hypothenemus sp. | No amplification | |||
| B17 | Hypothenemus sp. | No amplification | |||
| B19 | Hypothenemus sp. | No amplification | |||
| B20 | Hypothenemus sp. | No amplification | |||
| J1 | Araptus sp. | No amplification | |||
| J2 | Araptus sp. | No amplification | |||
| J4 | Araptus sp. | No amplification | |||
| COI (LCO1490/HCO2198) | |||||
|---|---|---|---|---|---|
| Sample | Genus | PCR Concentration (ng/μL) | GenBank Match | Accession Number | % Identical |
| B2 | Coptoborus pseudotenuis | 23.2 | Poor sequence | ||
| B4 | C. pseudotenuis | 12 | Poor sequence | ||
| B6 | C. pseudotenuis | 7.75 | Poor sequence | ||
| B7a | C. pseudotenuis | 12.7 | Coptodyras sp. | HM064072 | 86.4% |
| C. pseudotenuis | HM064071 | 85.5% | |||
| B7b | C. pseudotenuis | 21.8 | Coptodyras sp. | HM064072 | 84.7% |
| C. pseudotenuis | HM064071 | 84.3% | |||
| B3 | Xyleborus ferrugineus | 60 | Xyleborus ferrugineus | KP941251 | 98.0% |
| B14 | Theoborus sp. | No amplification | |||
| BC1 | Theoborus sp. | No amplification | |||
| BC9 | Theoborus sp. | 5.87 | Theoborus sp. | HM064100 | 94.7% |
| BC13 | Theoborus sp. | 38.9 | Theoborus sp. | HM064100 | 91.2% |
| B16 | Hypothenemus sp. | 51 | Poor sequence | ||
| B17 | Hypothenemus sp. | 6.26 | Poor sequence | ||
| B19 | Hypothenemus sp. | 38.5 | Poor sequence | ||
| B20 | Hypothenemus sp. | 21.3 | Hypothenemus sp. | MK768187 | 97.8% |
| Hypothenemus sp. | KX035186 | 97.8% | |||
| J1 | Araptus sp. | 6.45 | Poor sequence | ||
| J2 | Araptus sp. | 8.38 | Araptus carinifrons | MK768387 | 84.1% |
| J4 | Araptus sp. | 7.25 | Araptus carinifrons | MK767301 | 84.4% |
| Gene | Primer | Annealing Temp (°C) | Annealing Time (s) | Touch Down Temp (°C)/cycle | Number of Cycles |
|---|---|---|---|---|---|
| 28S | D2F1 | 58 | 45 | −0.1 | 33 |
| D3R2 | |||||
| COI | LCO1490 | 51 | 45 | −0.2 | 40 |
| HCO2198 | |||||
| EF-1a | eflafor1 | 56 | 45 | −0.1 | 38 |
| eflarev1 | |||||
| ets149 | 51 | 45 | −0.1 | 40 | |
| efa754 | |||||
| ITS | ITS2F | 56 | 30 | −0.1 | 40 |
| ITS2R |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Albo, J.E.; Marelli, J.-P.; Puig, A.S. Rapid Molecular Identification of Scolytinae (Coleoptera: Curculionidae). Int. J. Mol. Sci. 2019, 20, 5944. https://doi.org/10.3390/ijms20235944
Albo JE, Marelli J-P, Puig AS. Rapid Molecular Identification of Scolytinae (Coleoptera: Curculionidae). International Journal of Molecular Sciences. 2019; 20(23):5944. https://doi.org/10.3390/ijms20235944
Chicago/Turabian StyleAlbo, Jonathan Eric, Jean-Philippe Marelli, and Alina Sandra Puig. 2019. "Rapid Molecular Identification of Scolytinae (Coleoptera: Curculionidae)" International Journal of Molecular Sciences 20, no. 23: 5944. https://doi.org/10.3390/ijms20235944
APA StyleAlbo, J. E., Marelli, J.-P., & Puig, A. S. (2019). Rapid Molecular Identification of Scolytinae (Coleoptera: Curculionidae). International Journal of Molecular Sciences, 20(23), 5944. https://doi.org/10.3390/ijms20235944