1. Introduction
Alzheimer’s Disease (AD) is a progressive and irreversible complex neurodegenerative disease which accounts for above 75% of all dementia cases in the world [
1]. According to statistics, about 35% of people in the world over 80 years of age suffer from AD [
2]. Typical neuropathological features of AD include an increase in the number of senile plaques (SPs) and neurofibrillary tangles (NFTs) [
3]. Post-mortem studies have indicated that these neuropathological features originate in the medial temporal lobe limbic system, including the hippocampus and amygdala, and spread to other brain structures as the disease progresses [
4]. Therefore, it is necessary to investigate the molecular mechanism of AD from the perspective of multiple brain structures.
AD is a genetically complex neurologic disease; many susceptible genetic loci have been identified by several studies, including those that have used the genetic linkage approach and genome-wide association study (GWAS) [
5]. These loci include apolipoprotein E (
APOE) [
6] and other less significant loci such as
BIN1,
CLU,
ABCA7,
CR1,
PICALM,
MS4A6A,
CD33,
MS4A4E, and
CD2AP [
7,
8]. Single nucleotide polymorphism (SNP) interaction is also of importance in AD research. The effect of a single SNP is small and only a minority of SNPs contribute to the development AD due to “missing heritability” [
9]. Recent work on AD has focused on epistasis, which is the interaction between SNPs. Studying these interactions has been reported to have strong potential for revealing the underlying mechanism of AD [
10]. One meta-analysis examined the interactions between known AD susceptibility loci and reported a significant interaction between SNP in
APOE and SNP in
PICALM [
11]. In a whole genome-level interaction analysis, Gusareva et al. found an interaction between rs6455128 (
KHDRBS2) and rs7989332 (
CRYL1) [
12].
GWAS studies of AD have typically studied qualitative traits. However, studying quantitative traits offers several advantages over qualitative traits, including higher statistical power and smaller required sample size [
13]. Quantitative trait-based GWAS is also more objective because the interpretation of results relies on the relationship between identified causative SNPs and hypothetical mechanisms of a particular trait [
14]. Quantitative traits can include metabolite concentrations, brain structure volumes, or atrophy rates. For example,
APOE and 14 other novel genes show significant associations with the cerebrospinal fluid (CSF) amyloid β42 (Aβ42)/total tau (T-tau) concentration ratio [
15], and gene
GRIN2B shows a significant association with temporal lobe volume [
16]. Subcortical structures play a critical functional role in basic and higher cognitive ability and are potentially valuable biomarkers of AD [
17,
18]. Features of subcortical structures, such as their volume, have been frequently used as quantitative traits in AD studies. Most previous quantitative trait-based genetic studies have focused on the implication of genetic loci or loci interactions on subcortical structures. Potkin et al. found five new loci which reached a significant level in the hippocampus atrophy-based GWAS [
19]. Moloch et al. performed a gene-set enrichment analysis using statistics from a large-scale genome-wide association study of hippocampal volume [
20]. Hibar et al. performed a genome-wide interaction study using two datasets and identified a significant interaction between rs1345203 (
ELF3) and rs1213205, and this interaction was associated with temporal lobe volume variation [
21]. Shashwath et al. tested all SNP interactions within the 212 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and identified 125 interactions that may be associated with the right hippocampus atrophy [
22].
Multiple subcortical structures work together as modules when performing a specific complex task. A phenotypic modular classification analysis found that structural and functional modules exist in the brain [
23]. For example, Chen et al. [
24] discovered six modules based on the correlation of cortical thickness; these modules are related to closely overlapping known functional domains, which indicates that functional specialization and integration coexist in the human brain [
25]. Thus, analysis of the relationship between subcortical structure modules and genetic factors may reflect the underlying biological mechanism of neurodegenerative disorders such as AD.
The purpose of this study was to investigate the effect of genetic polymorphisms and their interactions on AD-related quantitative traits. Subcortical structure module volumes were used as quantitative traits for the GWAS and SNP interaction analysis. Our study provides a new perspective and goes some way to reveal the relationship between brain structures and genetic factors.
3. Discussion
AD is a progressive, irreversible and genetically complex neurodegenerative disease. Several genetic loci have been identified for AD risk. In this experiment, we performed a quantitative trait module-based genetic study of AD. First, all selected subcortical structures were clustered into five specialized brain area modules according to Pearson’s correlation coefficient. Second, module volumes were used as quantitative traits in the genome-wide association studies for screening the significant SNPs for each module. GO and KEGG pathway enrichment analyses showed few overlaps between different modules, especially for KEGG annotation. Finally, we made detailed analyses of the top five main effect SNPs and three interactive effect SNPs, as well as their relationships with each subcortical structure module and AD.
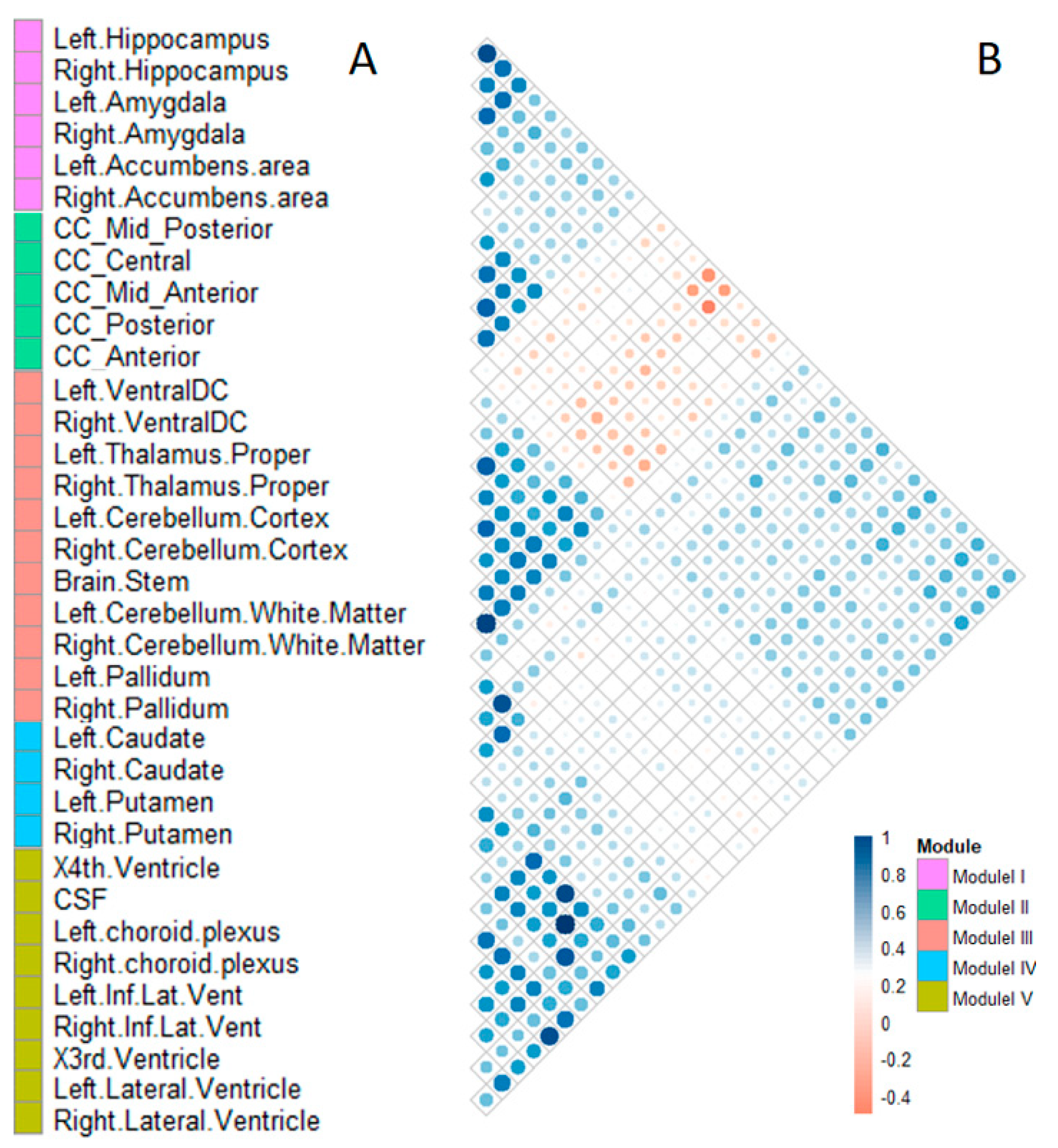
We clustered 35 selected subcortical structures into five modules. The structures in module I are parts of the limbic system, which is closely related to emotion, memory, and cognition [
26], and located in the medial temporal lobe of the cerebrum. The pathological features of the AD first appear in the medial temporal lobe, especially in the hippocampus [
27]. The hippocampus–amygdala complex is the most important part of the memory system and plays an important role in human memory function [
4]. In addition, the accumbens areas accept large amounts of aggregated fibers from the hippocampus and amygdala and play an important role in several cognitive processes. Module II included five structures belonging to the corpus callosum. The corpus callosum is the largest connective white matter fiber tract in the human brain and contains 200–250 million axonal projections that connect the left and right hemispheres. Diffusion tensor imaging (DTI) findings of patients with AD had a lower fractional anisotropy (FA) value of white matter fibers in the corpus callosum than normal control samples indicate that this structure causes the bilateral spread of AD within the brain [
28]. The atrophy of the corpus callosum is a useful biomarker for the diagnosis of AD [
29]. Module III comprised 11 structures, including the ventral-dorsal cord (Ventral DC), thalamus proper, cerebellum cortex, cerebellum white matter, pallidum, and brainstem. The pallidum connects the cerebral cortex and thalamus, and the thalamus is the major component of the ventral diencephalons, which plays an important role in consciousness levels. The brainstem, thalamus, and cerebellum are associated with basic life processes, including respiration, heart rate, arousal, movement, balance, and sensation. Module IV comprised four phenotypes, including the caudate and putamen, in both hemispheres. The subcortical structures in this module are involved in independent memory systems belonging to the basal ganglia neostriatum. The neostriatum is an important structure that mainly produces dopamine. There are many dopamine receptors in the dopaminergic system, which are mainly produced in the basal ganglia neostriatum. Module V comprised nine structures, including the choroid plexus, temporal horn of lateral ventricle, lateral ventricle, third ventricle, fourth ventricle, and CSF. These ventricular system structures are related to the storage and transportation of CSF. CSF is an important diagnostic marker of early AD, and the choroid plexus is a plexus of cells that produce CSF in the ventricles of the brain. Several studies have found that biochemical changes in CSF could reflect brain tissue damage, and CSF is an important diagnostic biomarker of AD [
30]. According to our results, some subcortical structures have similar patterns of volume changes although they are in different brain areas. For example, caudate, putamen, nucleus accumbens, and pallidum all belong to basal ganglia anatomically, but nucleus accumbens is more similar to hippocampus/amygdala (module I) on volume change patterns, while pallidum is more similar to thalamus and others (module III) on volume change patterns.
We proposed a modular analysis for the human brain subcortical structures in the Alzheimer’s Disease Neuroimaging Initiative (ADNI) cohort. Prior studies have focused on modular analysis on cortical structures, and several modular organizations were discovered [
24,
31]. To our knowledge, this is the first research on the whole brain subcortical structure module using volumetric measurements. Thirty five subcortical structures were clustered into five modules, each corresponding to a particular brain structure/area. At present, the hippocampus and amygdala in the module I have been widely studied in genetic studies of AD. For example, both volumes of hippocampus and amygdala are associated with
APOE ε4 in AD patients [
32]. Hibar et al. identified four novel loci associated with hippocampal volume [
33]. However, there have been few genetic studies on the subcortical structures in modules II, III, IV, and V. In this study, we highlight the need to consider other subcortical structures in AD, including those identified in the other four modules. For this purpose, subcortical structure module volumes were used as quantitative traits for GWAS and interaction analysis.
GO and KEGG pathway enrichment analyses revealed few overlaps between the different modules, especially for the KEGG enrichments, which indicates that the modular clustering was reasonable. KEGG categorizes genes into meaningful biological pathways, and results were more straightforward [
34], thus, we only discuss the KEGG results in detail.
Adherens junction proteins are related to maintaining the blood-brain barrier [
35] while high plasma concentrations of aldosterone may cause hippocampus dysfunction through the blood-brain barrier [
36]. Thus, the two KEGG pathways in module I have both been associated with the blood-brain barrier. Blood-brain barrier disruption has been identified as a key mechanism in the early stages of AD [
37]. For module III, degradation of taste and cancer are often considered as complications of AD [
38]. For module IV, the activation of phospholipase D (PLD) may be regulated by dopamine receptor D5 [
39], which is mainly generated in the neostriatum. For module V, axon guidance might play a role in AD [
40] and CSF protein has been reported to participate in the axon guidance signaling pathway [
41]. Serotonergic synapses have been associated with memory conditions, and serotonin is abundant in CSF [
42]. Kim et al. [
43] found that the MAPK signaling pathway is implicated in the development of AD through the regulation of phosphorylation of Amyloid Beta Precursor Protein (APP) and Tau, the main contents in CSF. The Ras and MAPK signaling pathways are activated by the same activity factors such as Grb2 [
44]. Module II shares two KEGG pathways, ALS pathway and Rap1 signaling pathway, with module V. ALS is another common neurodegeneration disease as well as AD and Parkinson’s disease (PD) and may involve the same risk pathway as AD and PD [
38]. Module III shares one pathway, CAMs pathway, with module V. Multiple studies have found that the CAMs pathway is strongly associated with AD and plays an integral role in the interaction between immune cells and peripheral nerve cells [
45].
We selected the top five significant SNPs as the main effect SNPs for each module for further analysis, and the top three interactive effect SNPs were selected from the interaction analysis. In each module, the interactive effect SNPs all interacted with the same subset of the main effect SNPs. The potential pathogenic mechanisms of module-associated genes are listed in
Table 12.
For module I, the main effect loci were associated with the volume change of the limbic system. Indeed,
APOE has been found to be associated with the atrophies of the hippocampus and amygdala [
32].
NECTIN2,
APOC1, and
TOMM40 are neighbor genes of
APOE on chromosome 19 [
46,
47]. In biology, these loci were involved in the deposition of β-amyloid protein and the abnormal phosphorylation of Tau protein [
48].
TNFRSF8 (Tumor Necrosis Factor Receptor Superfamily Member 8) is associated with neuroinflammation and is a down-regulated gene in the hippocampus from the AD brain but not the normal brain [
49]. Neuroinflammation plays a critical role in AD progression and accelerates the development of amyloid-β and tau pathology [
50,
51]. We speculated that the interactions between
TNFRSF8 and the main effect loci in module I may be involved in the neuroinflammation-induced developments of AD pathological features, which may cause cell death and lead to atrophy of the limbic system in the pathogenesis of AD.
For module II, the main effect loci were associated with the volume change of the corpus callosum and it could be inferred that the abnormal expressions of these loci may cause the atrophy of the corpus callosum. Lines of evidence support our inference.
ALDH1L1 encodes aldehyde dehydrogenase-1 protein, and a lower expression level of
ALDH1L1 may cause white matter damage in AD [
52].
CDH18 encodes calcium-dependent cadherin, and the expression of cadherin plays a key role in the development of neural fiber tracts [
53].
DENND5A encodes differentially expressed in neoplastic vs normal cells (DENN) domain-containing 5A protein, while its highest level has been found during the development of neuronal development [
54].
FGD4 (FYVE, RhoGEF and PH Domain Containing 4 protein) contains an actin filament-binding domain and is related to cytoskeleton and cell shape. The cytoskeleton is associated with the maintenance of cell shape, and the breakdown of cytoskeletal protein may cause damage to corpus callosum [
55]. With these observations, the interactive effect loci
FGD4 may be related to the integrity of corpus callosum, and the underlying regulatory mechanisms need further investigations.
For module III,
RAB13 (Ras-related protein) is a member of small G-proteins. Small G-proteins are regulators of transmembrane transport.
NRXN3 (neurexin 3) is a receptor and cell-adhesion molecule in the central nervous system.
KIAA1217, also known as sickle tail protein homolog (SKT), is involved in actin binding [
56].
CYTH1 encodes cytohesin-1, which is involved in protein transportation.
RCHY1 encodes E3 ubiquitin ligase and plays an important role in cell proliferation, differentiation, and apoptosis [
57].
CARHSP1 (Calcium Regulated Heat Stable Protein 1) is related to nucleic acid binding and mRNA 3′-UTR binding.
UNC5D is a netrin receptor and plays a role in cell–cell adhesion and cell guidance. Currently, studies about the genetic factors of AD are more concentrated in the cerebrum, with less attention paid to brainstem, thalamus, and cerebellum. Our result indicated that
RAB13,
NRXN3,
KIAA1217,
CYTH1, and
RCHY1 were associated with the volume changes of module III, including thalamus, cerebellum, pallidum, and brainstem, and interactive effect loci
CARHSP1 and
UNC5D may be likely to be associated with basic biological processes.
For module IV, the main effect loci
CSNK1E and
ROR1 in module IV are associated with the synthesis of dopamine, and the dysfunctions of
CSNK1E and
ROR1 may cause the atrophy of the basal ganglia neostriatum. Indeed, a decreased level of dopamine in putamen has been proved to be related to the degeneration of dopaminergic neurons, which may lead to the atrophy of the caudate nucleus [
58].
ADORA3 encodes an adenosine A3 receptor protein, a family member of adenosine receptors, which are G-protein-coupled receptors that are involved in a variety of intracellular signaling pathways and physiological functions. Adenosine receptors play a fundamental role in the modulation of dopaminergic neurotransmission [
59]. We speculated that the interaction between
ADORA3 and
CSNK1E has an effect on the reduction of dopaminergic neurons and the atrophy of the basal ganglia neostriatum.
For module V,
NEBL (nebulette, also called
LASP2) encodes an actin-binding protein associated with cell attachment, migration, and cellular communication [
60].
NGL1 (netrin G1 protein) is a presynaptic adhesion molecule [
61].
ST3GAL4 (ST3 beta-galactoside alpha-2,3-sialyltransferase-4 protein) is involved in protein glycosylation [
62].
NNT (nicotinamide nucleotide transhydrogenase) is associated with insulin secretion [
63].
RFX5 encodes a regulatory factor X5, and the homologous gene
RFX6 regulates the production of insulin in the islet [
64]. Our results indicated that the interactive effect loci
RFX5 was associated with the secretion of insulin. Higher insulin levels in the brain are correlated with lower rates of whole-brain atrophy in AD [
65]. Whole-brain atrophy is related to ventricular enlargement [
66]. We speculated that
NNT and
RFX5 may be involved in the secretion of insulin and the abnormity of these loci may be related to the whole brain atrophy, leading to the enlargements of ventricular system structures in AD. The underly mechanisms of interactions between
RFX5 and
NGL1, and
ST3GAL4 need further investigations.