Inhibition of the Low Molecular Weight Protein Tyrosine Phosphatase (LMPTP) as a Potential Therapeutic Strategy for Hepatic Progenitor Cells Lipotoxicity—Short Communication
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Isolation and Culture
2.2. In Vitro Study
2.3. RNA Isolation and Gene Expression Profiling Analysis
2.4. Statistical Analysis
3. Results
3.1. HPC Genotype and Morphology
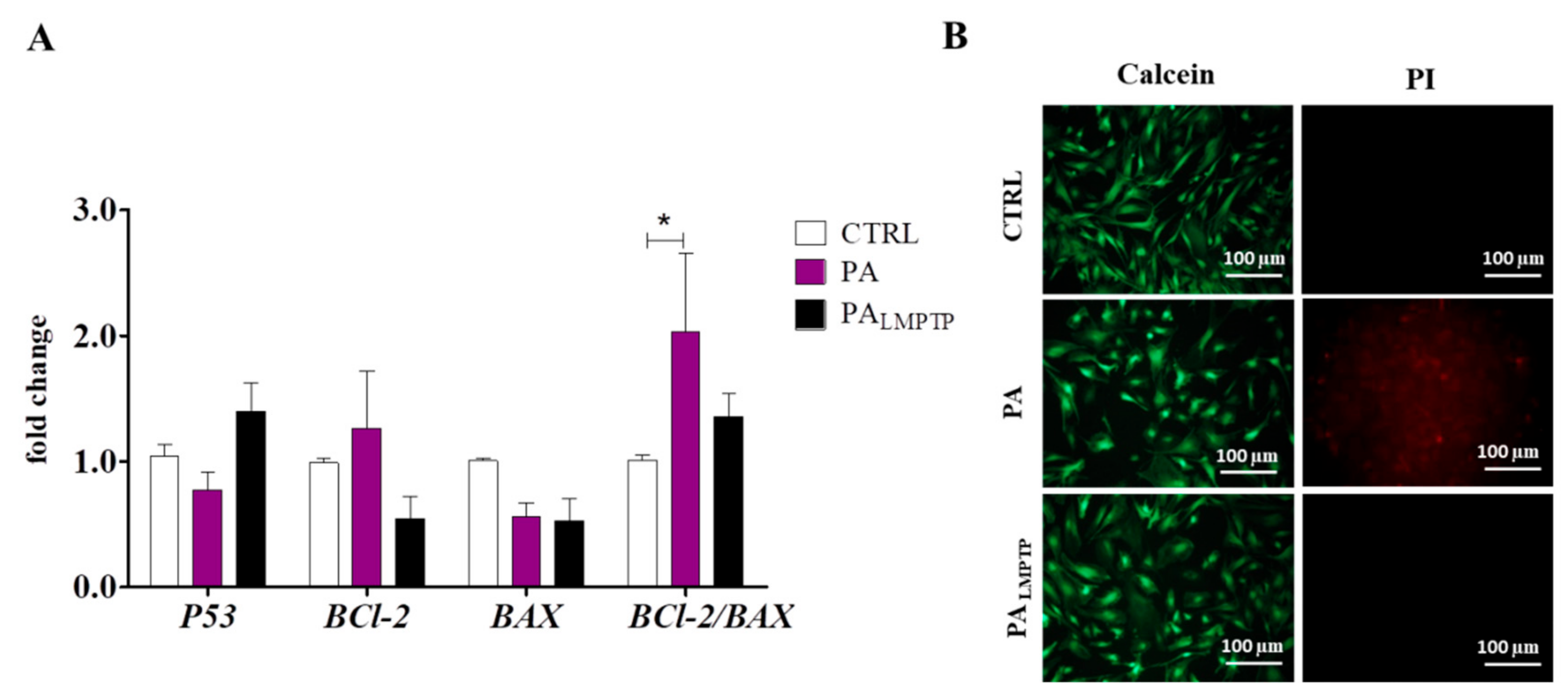
3.2. Expression of Apoptosis-Related Genes
3.3. LMPTP Inhibition Alters Expression of Genes Involved in Autophagy, Mitochondria Dynamics, and Unfolded Protein Response (UPR) in IR-Induced HPCs
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Frank, N.; Geor, R.J.; Bailey, S.R.; Durham, A.E.; Johnson, P.J. Equine Metabolic Syndrome. J. Vet. Intern. Med. 2010, 24, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Johnson, T.O.; Ermolieff, J.; Jirousek, M.R. Protein tyrosine phosphatase 1B inhibitors for diabetes. Nat. Rev. Drug Discov. 2002, 1, 696–709. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, S.H.; Kohli, R.; Gores, G.J. Mechanisms of Lipotoxicity in NAFLD and Clinical Implications. J. Pediatr. Gastroenterol. Nutr. 2011, 53, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Alicka, M.; Marycz, K. The Effect of Chronic Inflammation and Oxidative and Endoplasmic Reticulum Stress in the Course of Metabolic Syndrome and Its Therapy. Stem Cells Int. 2018, 2018, 4274361. [Google Scholar] [CrossRef]
- Kaczmarek, K.; Janicki, B.; Głowska, M. Insulin resistance in the horse: A review. J. Appl. Anim. Res. 2016, 44, 424–430. [Google Scholar] [CrossRef]
- Marycz, K.; Kornicka, K.; Szlapka-Kosarzewska, J.; Weiss, C. Excessive Endoplasmic Reticulum Stress Correlates with Impaired Mitochondrial Dynamics, Mitophagy and Apoptosis, in Liver and Adipose Tissue, but Not in Muscles in EMS Horses. Int. J. Mol. Sci. 2018, 19. [Google Scholar] [CrossRef]
- Boucher, J.; Kleinridders, A.; Kahn, C.R. Insulin Receptor Signaling in Normal and Insulin-Resistant States. Cold Spring Harb. Perspect. Biol. 2014, 6, a009191. [Google Scholar] [CrossRef]
- Stanford, S.M.; Aleshin, A.E.; Zhang, V.; Ardecky, R.J.; Hedrick, M.P.; Zou, J.; Ganji, S.R.; Bliss, M.R.; Yamamoto, F.; Bobkov, A.A.; et al. Diabetes reversal by inhibition of the low-molecular-weight tyrosine phosphatase. Nat. Chem. Biol. 2017, 13, 624–632. [Google Scholar] [CrossRef]
- Saltiel, A.R.; Kahn, C.R. Insulin signalling and the regulation of glucose and lipid metabolism. Nature 2001, 414, 799–806. [Google Scholar] [CrossRef]
- Abdelsalam, S.S.; Korashy, H.M.; Zeidan, A.; Agouni, A. The Role of Protein Tyrosine Phosphatase (PTP)-1B in Cardiovascular Disease and Its Interplay with Insulin Resistance. Biomolecules 2019, 9, 286. [Google Scholar] [CrossRef]
- Ardecky, R.J.; Hedrick, M.P.; Stanford, S.M.; Bliss, M.R.; Zou, J.; Gosalia, P.; Yamamoto, F.; Milewski, M.; Barron, N.; Sun, Q.; et al. Allosteric Small Molecule Inhibitors of LMPTP. In Probe Reports from the NIH Molecular Libraries Program; National Center for Biotechnology Information: Bethesda, MD, USA, 2010. [Google Scholar]
- Bottini, N.; MacMurray, J.; Peters, W.; Rostamkhani, M.; Comings, D.E. Association of the acid phosphatase (ACP1) gene with triglyceride levels in obese women. Mol. Genet. Metab. 2002, 77, 226–229. [Google Scholar] [CrossRef]
- Iannaccone, U.; Bergamaschi, A.; Magrini, A.; Marino, G.; Bottini, N.; Lucarelli, P.; Bottini, E.; Gloria-Bottini, F. Serum glucose concentration and ACP1 genotype in healthy adult subjects. Metab. Clin. Exp. 2005, 54, 891–894. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.K.; Yu, X.X.; Watts, L.M.; Michael, M.D.; Sloop, K.W.; Rivard, A.R.; Leedom, T.A.; Manchem, V.P.; Samadzadeh, L.; McKay, R.A.; et al. Reduction of Low Molecular Weight Protein-tyrosine Phosphatase Expression Improves Hyperglycemia and Insulin Sensitivity in Obese Mice. J. Biol. Chem. 2007, 282, 14291–14299. [Google Scholar] [CrossRef] [PubMed]
- Stanford, S.; Zhang, V.; Bottini, N. The Tyrosine Phosphatase LMPTP Is a Key Promoter of Adipogenesis. Diabetes 2018, 67, 2421. [Google Scholar] [CrossRef]
- Hao, P.-P.; Lee, M.-J.; Yu, G.-R.; Kim, I.-H.; Cho, Y.-G.; Kim, D.-G. Isolation of EpCAM+/CD133− hepatic progenitor cells. Mol. Cells 2013, 36, 424–431. [Google Scholar] [CrossRef]
- Yang, M.; Wei, D.; Mo, C.; Zhang, J.; Wang, X.; Han, X.; Wang, Z.; Xiao, H. Saturated fatty acid palmitate-induced insulin resistance is accompanied with myotube loss and the impaired expression of health benefit myokine genes in C2C12 myotubes. Lipids Health Dis. 2013, 12, 104. [Google Scholar] [CrossRef]
- Chomczynski, P.; Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef]
- Cassimeris, L.; Engiles, J.B.; Galantino-Homer, H. Detection of endoplasmic reticulum stress and the unfolded protein response in naturally-occurring endocrinopathic equine laminitis. Bmc Vet. Res. 2019, 15. [Google Scholar] [CrossRef]
- Lo, R.C.; Ng, I.O. Hepatic Progenitor Cells: Their Role and Functional Significance in the New Classification of Primary Liver Cancers. Liver Cancer 2013, 2, 84–92. [Google Scholar]
- Baig, M.T.; Ali, G.; Awan, S.J.; Shehzad, U.; Mehmood, A.; Mohsin, S.; Khan, S.N.; Riazuddin, S. Serum from CCl4-induced acute rat injury model induces differentiation of ADSCs towards hepatic cells and reduces liver fibrosis. Growth Factors 2017, 35, 144–160. [Google Scholar] [CrossRef]
- Giancotti, A.; Monti, M.; Nevi, L.; Safarikia, S.; D’Ambrosio, V.; Brunelli, R.; Pajno, C.; Corno, S.; Di Donato, V.; Musella, A.; et al. Functions and the Emerging Role of the Foetal Liver into Regenerative Medicine. Cells 2019, 8, 914. [Google Scholar] [CrossRef] [PubMed]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.; Horwitz, E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef]
- Liu, L.; Shi, G.-P. CD31: Beyond a marker for endothelial cells. Cardiovasc. Res. 2012, 94, 3–5. [Google Scholar] [CrossRef] [PubMed]
- Listenberger, L.L.; Ory, D.S.; Schaffer, J.E. Palmitate-induced apoptosis can occur through a ceramide-independent pathway. J. Biol. Chem. 2001, 276, 14890–14895. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chang, F.; Li, F.; Fu, H.; Wang, J.; Zhang, S.; Zhao, J.; Yin, D. Palmitate promotes autophagy and apoptosis through ROS-dependent JNK and p38 MAPK. Biochem. Biophys. Res. Commun. 2015, 463, 262–267. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; McStay, G.P. Regulation of Mitochondrial Dynamics by Proteolytic Processing and Protein Turnover. Antioxidants (Basel) 2018, 7, 15. [Google Scholar]
- Guo, T.; Liu, T.; Sun, Y.; Liu, X.; Xiong, R.; Li, H.; Li, Z.; Zhang, Z.; Tian, Z.; Tian, Y. Sonodynamic therapy inhibits palmitate-induced beta cell dysfunction via PINK1/Parkin-dependent mitophagy. Cell Death Dis. 2019, 10, 457. [Google Scholar] [CrossRef]
- Tabas, I.; Ron, D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat. Cell Biol. 2011, 13, 184–190. [Google Scholar] [CrossRef]
- Kaur, S.; Siddiqui, H.; Bhat, M.H. Hepatic Progenitor Cells in Action: Liver Regeneration or Fibrosis? Am. J. Pathol. 2015, 185, 2342–2350. [Google Scholar] [CrossRef]
- Shin, S.; Walton, G.; Aoki, R.; Brondell, K.; Schug, J.; Fox, A.; Smirnova, O.; Dorrell, C.; Erker, L.; Chu, A.S.; et al. Foxl1-Cre-marked adult hepatic progenitors have clonogenic and bilineage differentiation potential. Genes Dev. 2011, 25, 1185–1192. [Google Scholar] [CrossRef]
- Dorrell, C.; Erker, L.; Schug, J.; Kopp, J.L.; Canaday, P.S.; Fox, A.J.; Smirnova, O.; Duncan, A.W.; Finegold, M.J.; Sander, M.; et al. Prospective isolation of a bipotential clonogenic liver progenitor cell in adult mice. Genes Dev. 2011, 25, 1193–1203. [Google Scholar] [CrossRef] [PubMed]
- Gloria-Bottini, F.; Gerlini, G.; Lucarini, N.; Borgiani, P.; Amante, A.; La Torre, M.; Antonacci, E.; Bottini, E. Phosphotyrosine protein phosphatases and diabetic pregnancy: An association between low molecular weight acid phosphatase and degree of glycemic control. Experientia 1996, 52, 340–343. [Google Scholar] [CrossRef] [PubMed]
- Carpino, G.; Renzi, A.; Onori, P.; Gaudio, E. Role of Hepatic Progenitor Cells in Nonalcoholic Fatty Liver Disease Development: Cellular Cross-Talks and Molecular Networks. Int. J. Mol. Sci. 2013, 14, 20112–20130. [Google Scholar] [CrossRef] [PubMed]
- Follis, A.V.; Llambi, F.; Merritt, P.; Chipuk, J.E.; Green, D.R.; Kriwacki, R.W. Pin1-Induced Proline Isomerization in Cytosolic p53 Mediates BAX Activation and Apoptosis. Mol. Cell 2015, 59, 677–684. [Google Scholar] [CrossRef]
- Goldstein, I.; Ezra, O.; Rivlin, N.; Molchadsky, A.; Madar, S.; Goldfinger, N.; Rotter, V. p53, a novel regulator of lipid metabolism pathways. J. Hepatol. 2012, 56, 656–662. [Google Scholar] [CrossRef]
- Wei, Y.; Wang, D.; Topczewski, F.; Pagliassotti, M.J. Saturated fatty acids induce endoplasmic reticulum stress and apoptosis independently of ceramide in liver cells. Am. J. Physiol. -Endocrinol. Metab. 2006, 291, E275–E281. [Google Scholar] [CrossRef]
- Ly, L.D.; Xu, S.; Choi, S.-K.; Ha, C.-M.; Thoudam, T.; Cha, S.-K.; Wiederkehr, A.; Wollheim, C.B.; Lee, I.-K.; Park, K.-S. Oxidative stress and calcium dysregulation by palmitate in type 2 diabetes. Exp. Mol. Med. 2017, 49, e291–e291. [Google Scholar] [CrossRef]
- Zhang, T.; Liu, J.; Shen, S.; Tong, Q.; Ma, X.; Lin, L. SIRT3 promotes lipophagy and chaperon-mediated autophagy to protect hepatocytes against lipotoxicity. Cell Death Differ. 2019, 1–16. [Google Scholar] [CrossRef]
- Singh, R.; Kaushik, S.; Wang, Y.; Xiang, Y.; Novak, I.; Komatsu, M.; Tanaka, K.; Cuervo, A.M.; Czaja, M.J. Autophagy regulates lipid metabolism. Nature 2009, 458, 1131–1135. [Google Scholar] [CrossRef]
- Schneider, J.L.; Suh, Y.; Cuervo, A.M. Deficient chaperone-mediated autophagy in liver leads to metabolic dysregulation. Cell Metab. 2014, 20, 417–432. [Google Scholar] [CrossRef]
- Tasset, I.; Cuervo, A.M. Role of chaperone-mediated autophagy in metabolism. Febs J. 2016, 283, 2403–2413. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-S.; Jeong, S.-K.; Kim, H.-R.; Kim, D.-S.; Chae, S.-W.; Chae, H.-J. Metformin regulates palmitate-induced apoptosis and ER stress response in HepG2 liver cells. Immunopharmacol. Immunotoxicol. 2010, 32, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Maiers, J.L.; Malhi, H. Endoplasmic Reticulum Stress in Metabolic Liver Diseases and Hepatic Fibrosis. Semin Liver Dis. 2019, 39, 235–248. [Google Scholar] [CrossRef] [PubMed]
- Sano, R.; Reed, J.C. ER stress-induced cell death mechanisms. Biochim. Et Biophys. Acta (Bba) - Mol. Cell Res. 2013, 1833, 3460–3470. [Google Scholar] [CrossRef] [PubMed]
- McCullough, K.D.; Martindale, J.L.; Klotz, L.-O.; Aw, T.-Y.; Holbrook, N.J. Gadd153 Sensitizes Cells to Endoplasmic Reticulum Stress by Down-Regulating Bcl2 and Perturbing the Cellular Redox State. Mol. Cell. Biol. 2001, 21, 1249. [Google Scholar] [CrossRef]
- Yoshida, H.; Matsui, T.; Yamamoto, A.; Okada, T.; Mori, K. XBP1 mRNA Is Induced by ATF6 and Spliced by IRE1 in Response to ER Stress to Produce a Highly Active Transcription Factor. Cell 2001, 107, 881–891. [Google Scholar] [CrossRef]
- Ning, J.; Hong, T.; Ward, A.; Pi, J.; Liu, Z.; Liu, H.-Y.; Cao, W. Constitutive Role for IRE1α-XBP1 Signaling Pathway in the Insulin-Mediated Hepatic Lipogenic Program. Endocrinology 2011, 152, 2247. [Google Scholar] [CrossRef]
- Lee, J.M. Nuclear Receptors Resolve Endoplasmic Reticulum Stress to Improve Hepatic Insulin Resistance. Diabetes Metab. J. 2017, 41, 10–19. [Google Scholar] [CrossRef]
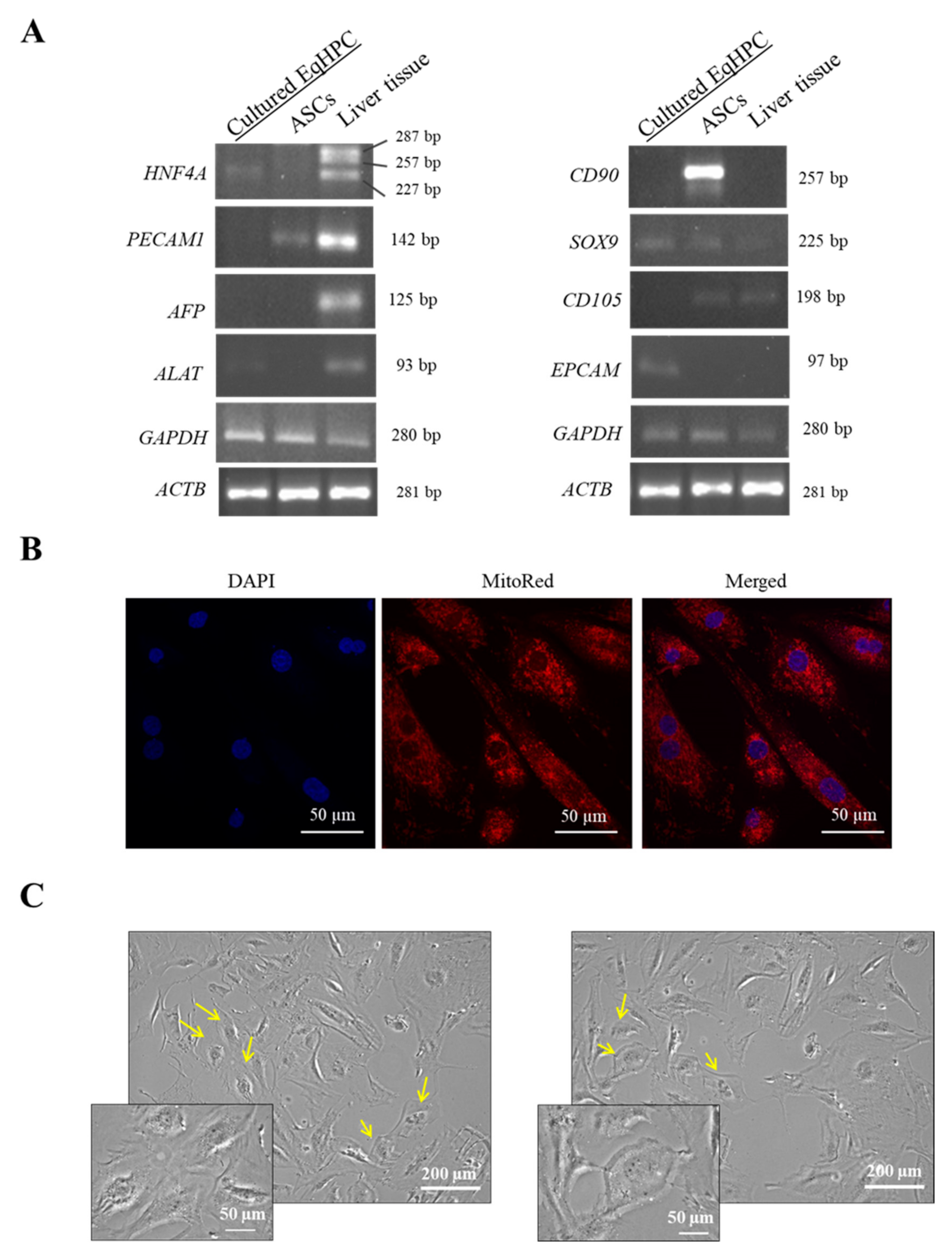

| Gene | Primer Sequence 5′→3′ | Amplicon Length (bp) | Accession No. |
|---|---|---|---|
| HNF4A | F: CAGGAGATGCTGCTGGGAG R: ATTGTGGTGATGGCTCCTGG | 257 | XM_014735168.2 |
| PECAM1 | F: ACACGGAAGTGGAAGTGACC R: CCATCAAGGGAGCCTTCCG | 142 | NM_001101655.2 |
| AFP | F: CAGCCACTTGTTGCCAACTC R: CTGGCCAACACCAGGGTTTA | 125 | NM_001081952.1 |
| ALAT | F: GGGAAGGCACCTACCACTTC R: ACTTGGCATGGAACTGGCTT | 93 | XM_005613421.2 |
| CD90 | F: CTCCCACCCCTGGTGAAAACT R: CGGTGGTATTCTCATGGCGG | 257 | XM_001503225.4 |
| SOX9 | F: GAACGCCTTCATGGTGTGGG R: TTCTTCACCGACTTCCTCCG | 225 | XM_014736619.1 |
| CD105 | F: GACTGCCTTTGTGCAGTTGG R: ATGCTTTCGGGGTCCTTCAG | 198 | XM_003364144.4 |
| EPCAM | F: TTGCCGTCATTGTGGTTGTG R: TCAGCCTTCTCGTACTTCGC | 97 | XM_001917795.4 |
| P53 | F: TACTCCCCTGCCCTCAACAA R: AGGAATCAGGGCCTTGAGGA | 252 | U37120.1 |
| BCl-2 | F: TTCTTTGAGTTCGGTGGGGT R: GGGCCGTACAGTTCCACAA | 164 | XM_014843802.1 |
| BAX | F: CGAGTGGCAGCTGAGATGTT R: AAGGAAGTCCAGTGTCCAGC | 153 | XM_023650077.1 |
| BECN1 | F: GATGCGTTATGCCCAGATGC R: ATCCAGCGAACACTCTTGGG | 147 | XM_014729146.1 |
| SQSTM1 | F: CATCGGAGGATCCCAGTGTG R: CCGGTTTGTTAGGGTCGGAA | 207 | XM_005599173.3 |
| LC3 | F: TTCTGAGACACAGTCGGAGC R: CTTTGTTCGAAGGTGTGGCG | 128 | XM_001493613.6 |
| HSC70 | F: GATTAACAAGAGGGCTGTCCGTC R: GCCTGGGTGCTAGAAGAGAGA | 122 | XM_023628864.1 |
| LAMP2 | F: GCACCCCTGGGAAGTTCTTA R: ATCCAGCGAACACTCTTGGG | 147 | XM_014831347.1 |
| MFN1 | F: AAGTGGCATTTTTCGGCAGG R: TCCATATGAAGGGCATGGGC | 217 | XM_014838357.1 |
| FIS1 | F: GGTGCGAAGCAAGTACAACG R: GTTGCCCACAGCCAGATAGA | 118 | XM_014854003.1 |
| PINK1 | F: GCACAATGAGCCAGGAGCTA R: GGGGTATTCACGCGAAGGTA | 298 | XM_014737247.1 |
| PRKN | F: TCCCAGTGGAGGTCGATTCT R: CCCTCCAGGTGTGTTCGTTT | 218 | XM_014858374.1 |
| PGC1A | F: TCTACCTAGGATGCATGG R: GTGCAAGTAGAAACACTGC | 93 | XM_005608845.2 |
| PERK | F: GTGACTGCAATGGACCAGGA R: TCACGTGCTCACGAGGATATT | 283 | XM_023618757.1 |
| CHOP | F: AGCCAAAATCAGAGCCGGAA R: GGGGTCAAGAGTGGTGAAGG | 272 | XM_001488999.4 |
| ATF6 | F: CAGGGTGCACTAGAACAGGG R: AATGTGTCTCCCCTTCTGCG | 164 | XM_023640315.1 |
| HSPA5 | F: CTGTAGCGTATGGTGCTGCT R: CATGACACCTCCCACGGTTT | 122 | XM_023628864.1 |
| IRE1A | F: GAATCAGACGAGCACCCGAA R: TTTCTTGCAGAGGCCGAAGT | 300 | XM_023652216.1 |
| XBP1 | F: TTACGCGAGAAAACTCATGGCC R: GGGTCCAAGTTGAACAGAATGC | 281 (unspliced) 255 (spliced) | XM_014742035.2 |
| GAPDH | F: GATGCCCCAATGTTTGTGA R: AAGCAGGGATGATGTTCTGG | 250 | NM 001163856.1 |
| ACTB | F: GATGATGATATCGCCGCGCTC R: CGCAGCTCGTTGTAGAAGGT | 281 | NM_001081838.1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alicka, M.; Kornicka-Garbowska, K.; Roecken, M.; Marycz, K. Inhibition of the Low Molecular Weight Protein Tyrosine Phosphatase (LMPTP) as a Potential Therapeutic Strategy for Hepatic Progenitor Cells Lipotoxicity—Short Communication. Int. J. Mol. Sci. 2019, 20, 5873. https://doi.org/10.3390/ijms20235873
Alicka M, Kornicka-Garbowska K, Roecken M, Marycz K. Inhibition of the Low Molecular Weight Protein Tyrosine Phosphatase (LMPTP) as a Potential Therapeutic Strategy for Hepatic Progenitor Cells Lipotoxicity—Short Communication. International Journal of Molecular Sciences. 2019; 20(23):5873. https://doi.org/10.3390/ijms20235873
Chicago/Turabian StyleAlicka, Michalina, Katarzyna Kornicka-Garbowska, Michael Roecken, and Krzysztof Marycz. 2019. "Inhibition of the Low Molecular Weight Protein Tyrosine Phosphatase (LMPTP) as a Potential Therapeutic Strategy for Hepatic Progenitor Cells Lipotoxicity—Short Communication" International Journal of Molecular Sciences 20, no. 23: 5873. https://doi.org/10.3390/ijms20235873
APA StyleAlicka, M., Kornicka-Garbowska, K., Roecken, M., & Marycz, K. (2019). Inhibition of the Low Molecular Weight Protein Tyrosine Phosphatase (LMPTP) as a Potential Therapeutic Strategy for Hepatic Progenitor Cells Lipotoxicity—Short Communication. International Journal of Molecular Sciences, 20(23), 5873. https://doi.org/10.3390/ijms20235873





