A Stress-Associated Protein, PtSAP13, From Populus trichocarpa Provides Tolerance to Salt Stress
Abstract
1. Introduction
2. Results
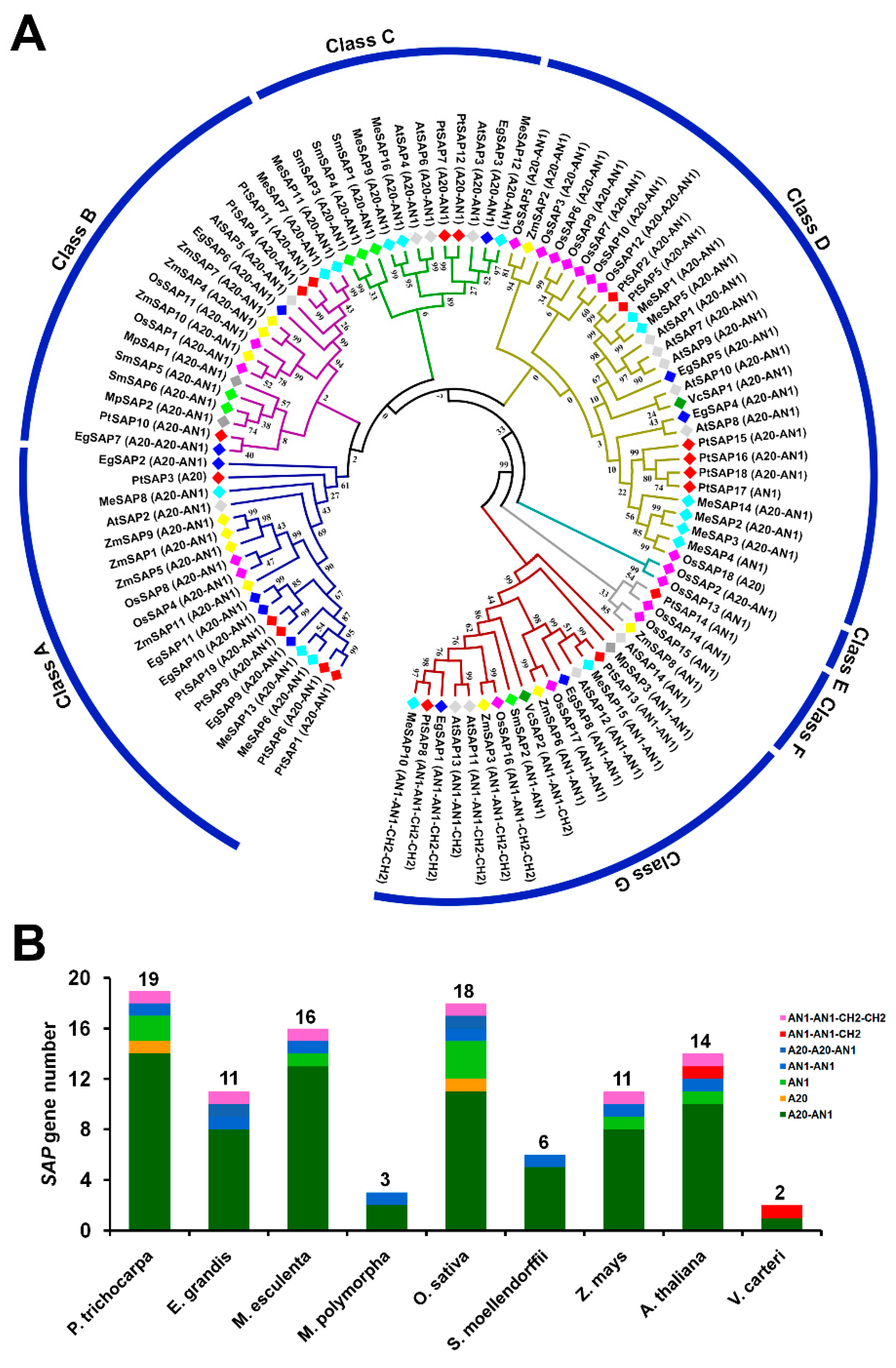
2.1. Phylogenetic Analysis of SAPs in P. trichocarpa and Other Species
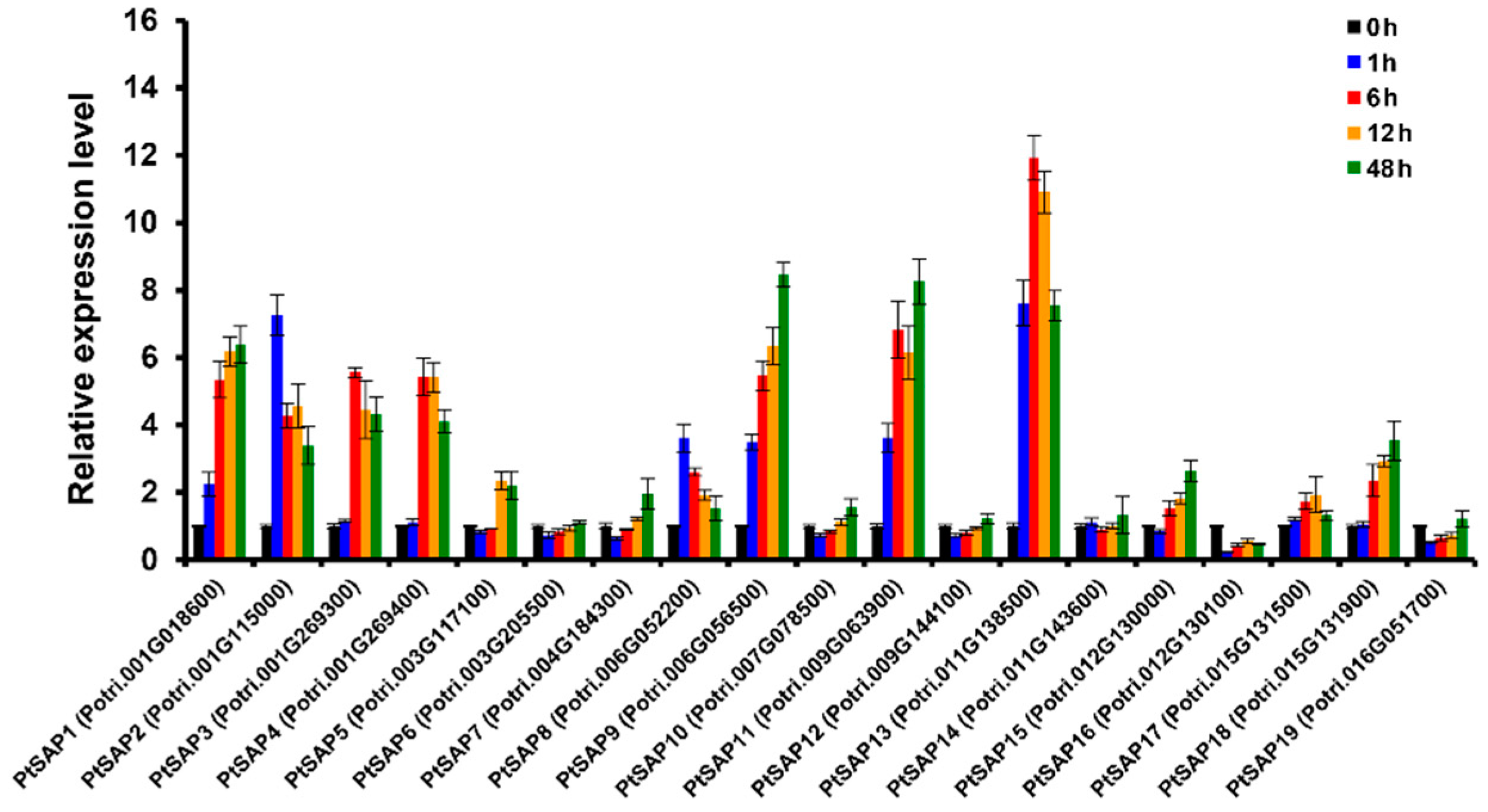
2.2. Expression Profiles of PtSAP Genes
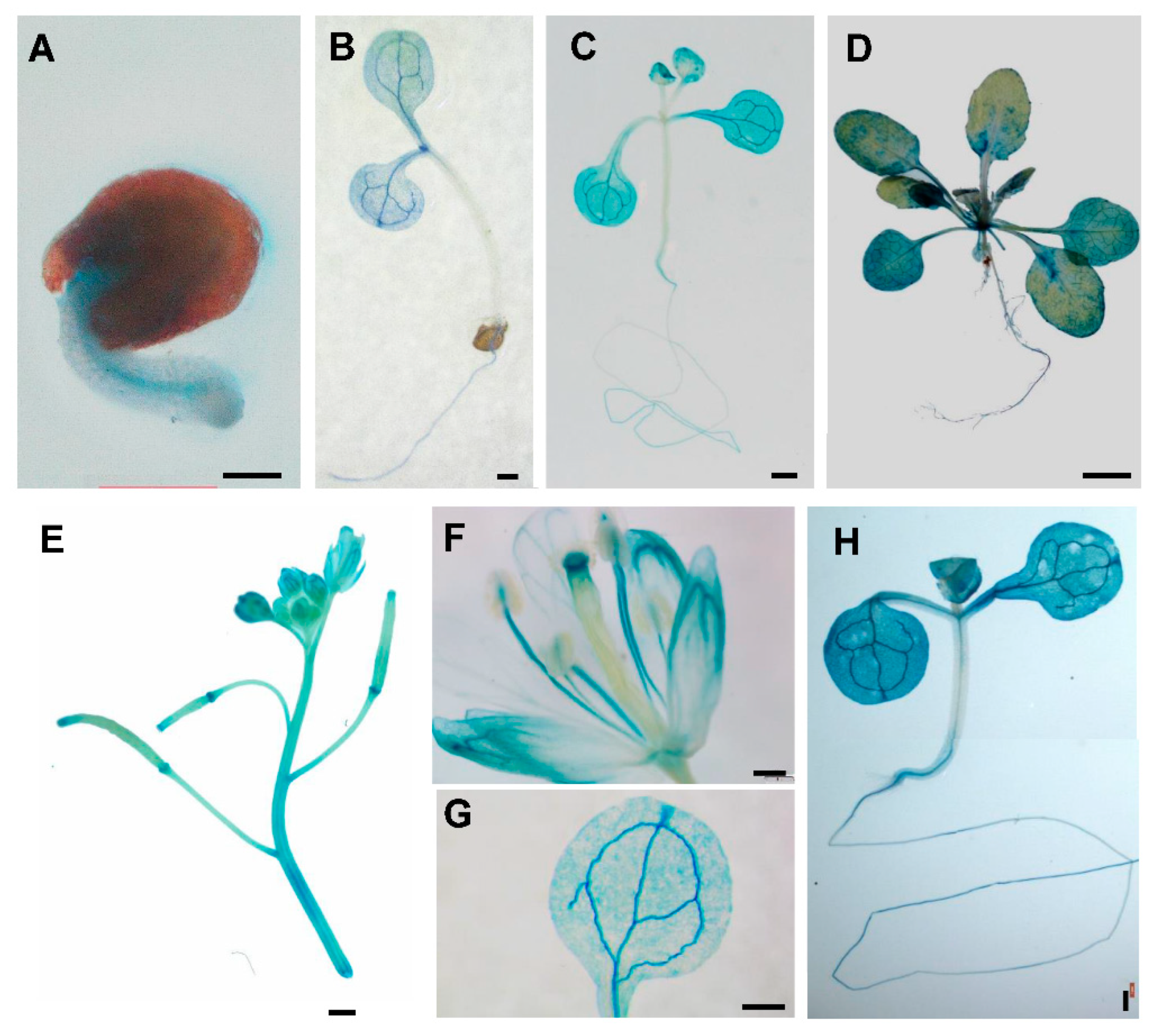
2.3. Expression Patterns of PtSAP13
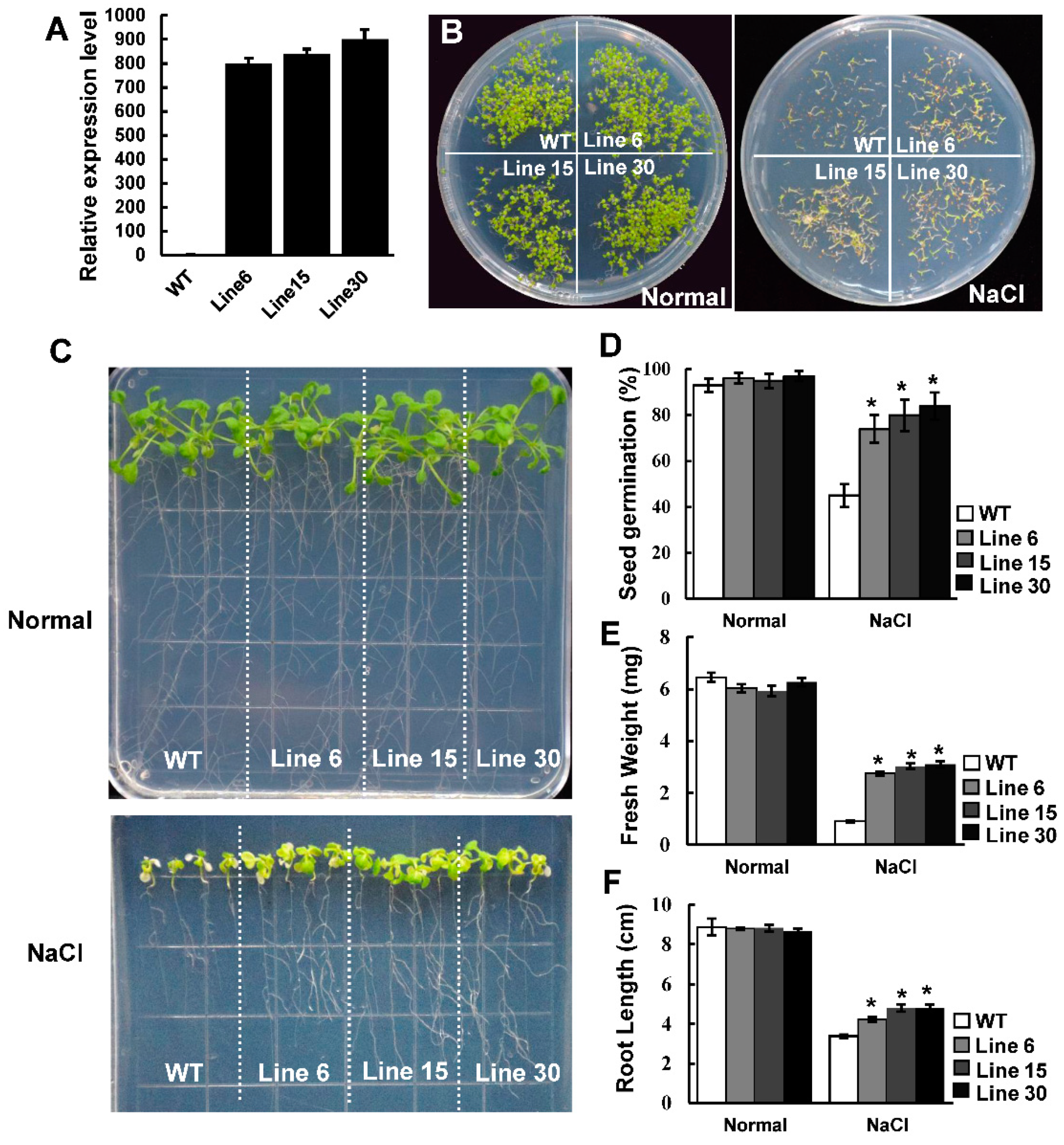
2.4. Effect of Salt Stress on Seed Germination in Transgenic PtSAP13 Arabidopsis Lines
2.5. PtSAP13 Transgenic Arabidopsis Seedling Resistance to Salt Stress
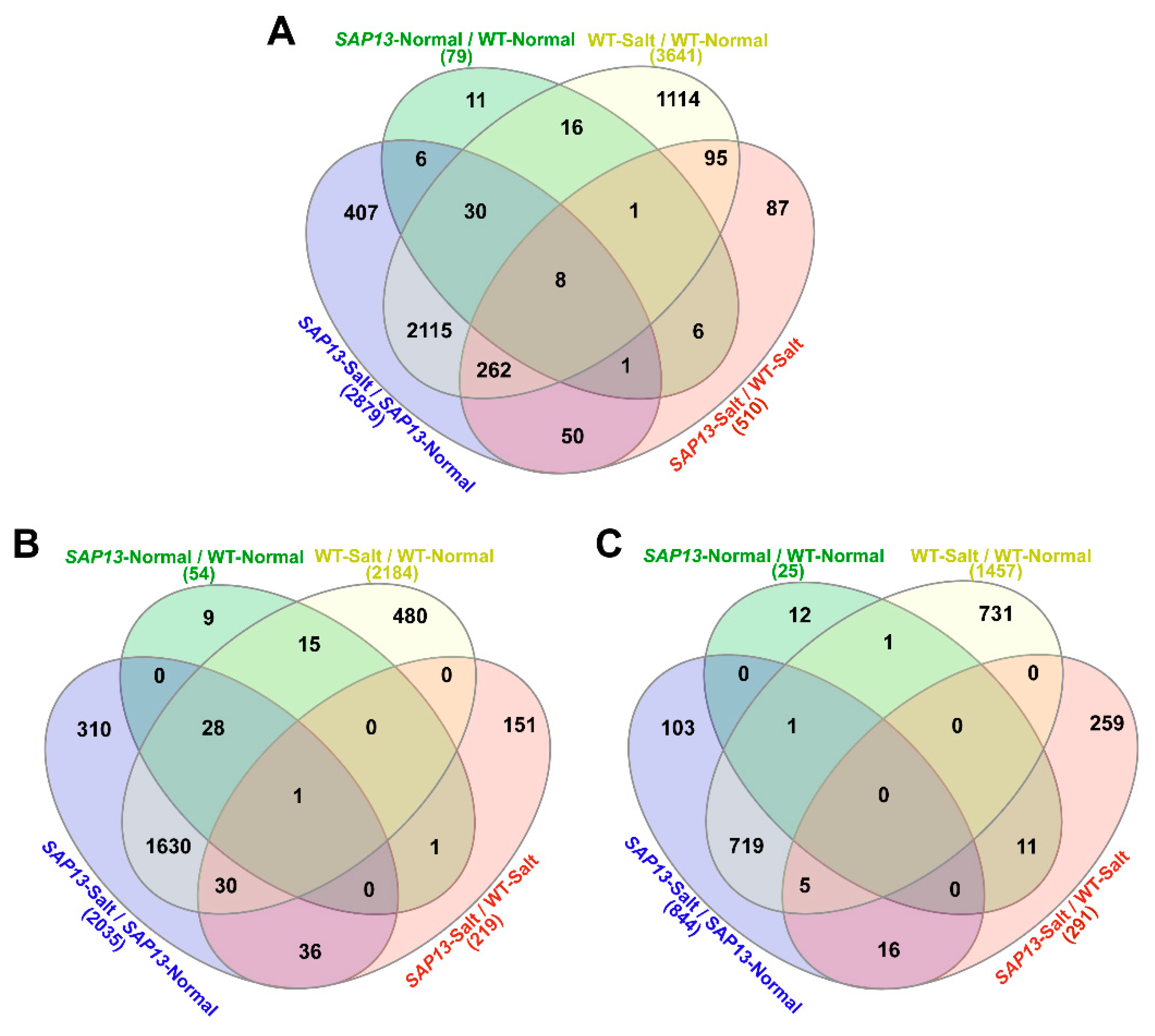
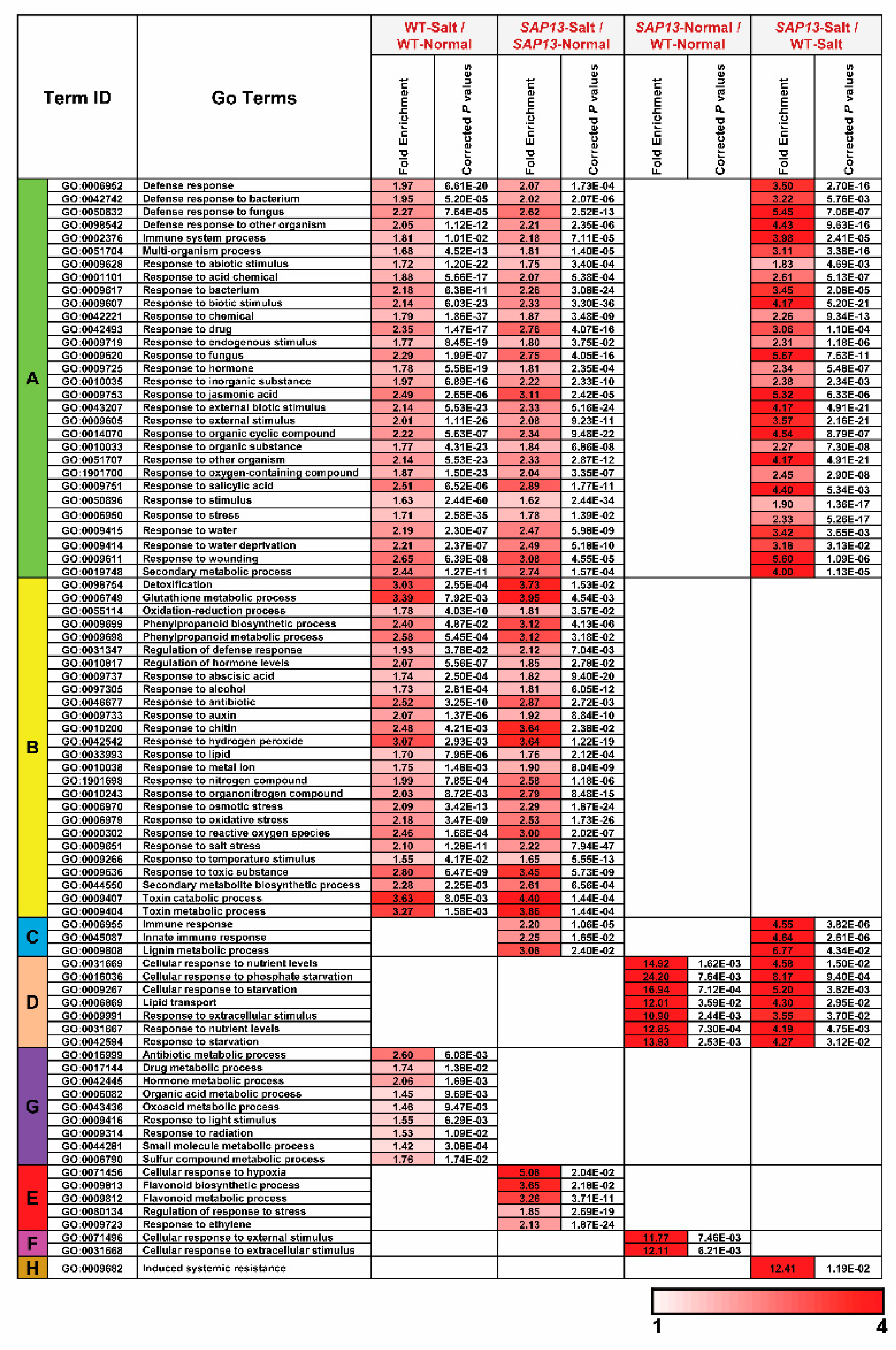
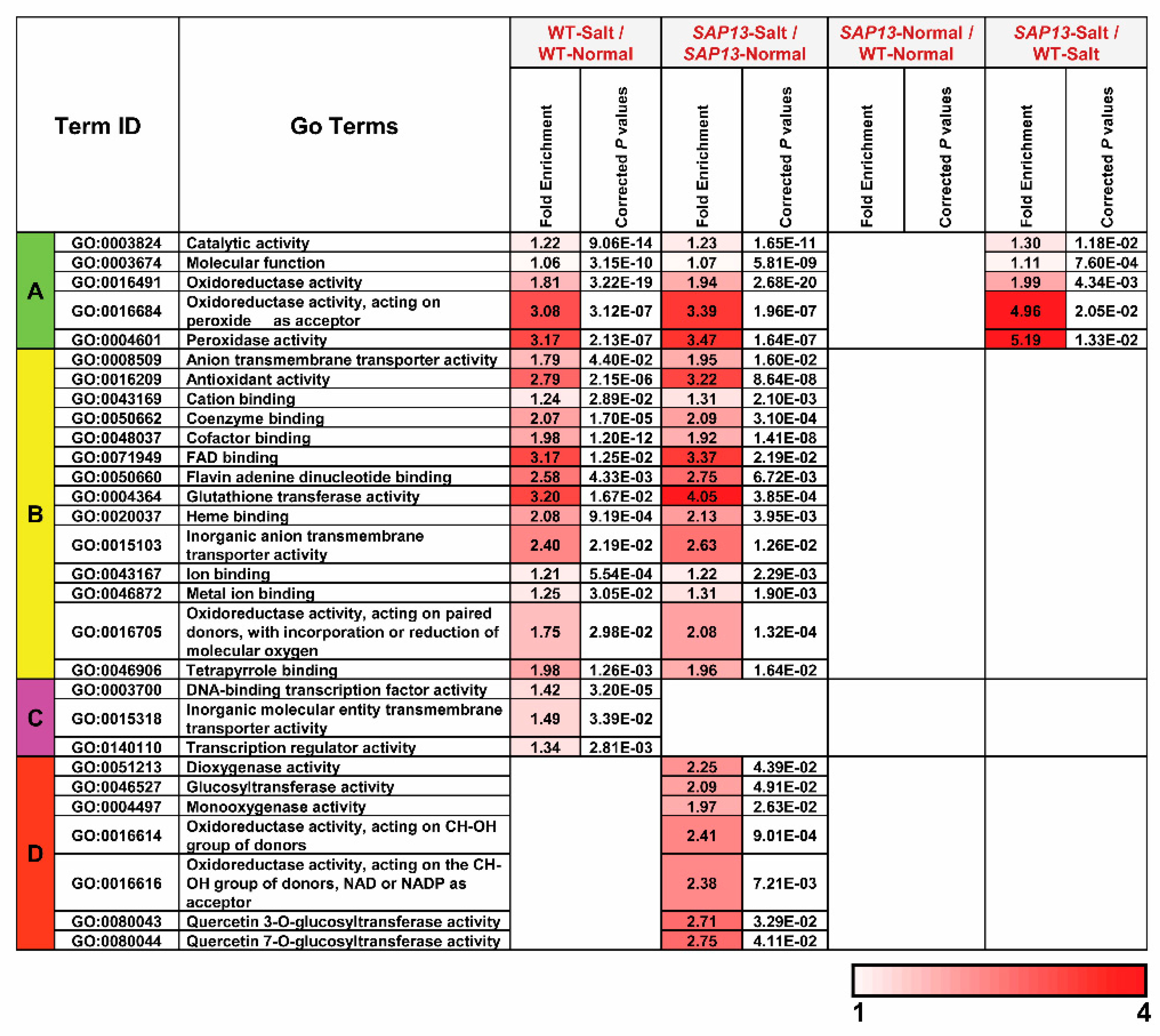
2.6. Overexpression of PtSAP13 Induced the Expression of Stress-Related Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Salt Treatments
4.2. Gene Expression Analysis of PtSAPs
4.3. RNA Isolation and Quantitative Real-Time PCR (qRT-PCR) Analysis
4.4. Generation of Transgenic A. thaliana Plants
4.5. Analysis of Salt Tolerance
4.6. RNA-Seq Analysis of Transgenic Arabidopsis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Yamaguchi, T.; Blumwald, E. Developing salt-tolerant crop plants: Challenges and opportunities. Trends Plant Sci. 2005, 10, 615–620. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Bekturganov, Z.; Tussupova, K.; Berndtsson, R.; Sharapatova, N.; Aryngazin, K.; Zhanasova, M. Water related health problems in central Asia-A review. Water 2016, 8, 219. [Google Scholar] [CrossRef]
- Quintero, F.J.; Ohta, M.; Shi, H.; Zhu, J.; Pardo, J.M. Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc. Natl. Acad. Sci. USA 2002, 99, 9061–9066. [Google Scholar] [CrossRef]
- Apse, M.P.; Aharon, G.S.; Snedden, W.A.; Blumwald, E. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 1999, 285, 1256–1258. [Google Scholar] [CrossRef] [PubMed]
- Yokoi, S.; Quintero, F.J.; Cubero, B.; Ruiz, M.T.; Bressan, R.A.; Hasegawa, P.M.; Pardo, J.M. Differential expression and function of Arabidopsis thaliana NHX Na+/H+ antiporters in the salt stress response. Plant J. 2002, 30, 529–539. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Schachtman, D.P.; Zhang, W. Partial deletion of a loop region in the high affinity K+ transporter HKT1 changes ionic permeability leading to increased salt tolerance. J. Biol. Chem. 2000, 275, 27924. [Google Scholar] [PubMed]
- Zhang, X.; Cheng, Z.; Zhao, K.; Yao, W.; Sun, X.; Jiang, T.; Zhou, B. Functional characterization of poplar NAC13 gene in salt tolerance. Plant Sci. 2019, 281, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Zhang, D.; Lv, K.; Zhang, X.; Cheng, Z.; Li, R.; Zhou, B.; Jiang, T. Functional characterization of poplar WRKY75 in salt and osmotic tolerance. Plant Sci. 2019, 289, 110259. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Cheng, J.; Sun, X.; Zhao, T.; Li, M.; Wang, Q.; Li, S.; Xin, H. Overexpression of VaWRKY14 increases drought tolerance in Arabidopsis by modulating the expression of stress-related genes. Plant Cell Rep. 2018, 37, 1159–1172. [Google Scholar] [CrossRef]
- Wang, S.; Zhou, B.; Yao, W.; Jiang, T. PsnERF75 transcription factor from Populus simonii x P. nigra confers salt tolerance in transgenic Arabidopsis. J. Plant Biol. 2018, 61, 61–71. [Google Scholar] [CrossRef]
- An, J.P.; Zhang, X.W.; Xu, R.R.; You, C.X.; Wang, X.F.; Hao, Y.J. Apple MdERF4 negatively regulates salt tolerance by inhibiting MdERF3 transcription. Plant Sci. 2018, 276, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Garg, R.; Verma, M.; Agrawal, S.; Shankar, R.; Majee, M.; Jain, M. Deep transcriptome sequencing of wild halophyte rice, Porteresia coarctata, provides novel insights into the salinity and submergence tolerance factors. DNA Res. 2013, 21, 69–84. [Google Scholar] [CrossRef] [PubMed]
- Vij, S.; Tyagi, A.K. Genome-wide analysis of the stress associated protein (SAP) gene family containing A20/AN1 zinc-finger(s) in rice and their phylogenetic relationship with Arabidopsis. Mol. Genet. Genom. 2006, 276, 565–575. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Li, J.; Zhang, J.; Ren, Y.; Hu, J.; Lu, M. Genome-wide survey and expression analysis of the stress-associated protein gene family in desert poplar, Populus euphratica. Tree Genet. Genomes 2016, 12, 78. [Google Scholar] [CrossRef]
- Zhou, Y.; Zeng, L.; Chen, R.; Wang, Y.; Song, J. Genome-wide identification and characterization of stress-associated protein (SAP) gene family encoding A20/AN1 zinc-finger proteins in Medicago truncatula. Arch. Biol. Sci. 2017, 70, 87–98. [Google Scholar] [CrossRef]
- He, X.; Xie, S.; Xie, P.; Yao, M.; Liu, W.; Qin, L.; Liu, Z.; Zheng, M.; Liu, H.; Guan, M. Genome-wide identification of stress-associated proteins (SAP) with A20/AN1 zinc finger domains associated with abiotic stresses responses in Brassica napus. Environ. Exp. Bot. 2019, 165, 108–119. [Google Scholar] [CrossRef]
- Opipari, A.; Boguski, M.; Dixit, V. The A20 cDNA induced by tumor necrosis factor alpha encodes a novel type of zinc finger protein. J. Biol. Chem. 1990, 265, 14705–14708. [Google Scholar]
- Jin, Y.; Wang, M.; Fu, J.; Xuan, N.; Zhu, Y.; Lian, Y.; Jia, Z.; Zheng, J.; Wang, G. Phylogenetic and expression analysis of ZnF-AN1 genes in plants. Genomics 2007, 90, 265–275. [Google Scholar] [CrossRef]
- Dansana, P.K.; Kothari, K.S.; Vij, S.; Tyagi, A.K. OsiSAP1 overexpression improves water-deficit stress tolerance in transgenic rice by affecting expression of endogenous stress-related genes. Plant Cell Rep. 2014, 33, 1425–1440. [Google Scholar] [CrossRef]
- Arnab, M.; Shubha, V.; Akhilesh, K.T. Overexpression of a zinc-finger protein gene from rice confers tolerance to cold, dehydration, and salt stress in transgenic tobacco. Proc. Natl. Acad. Sci. USA 2004, 101, 6309–6314. [Google Scholar]
- Tyagi, H.; Jha, S.; Sharma, M.; Giri, J.; Tyagi, A.K. Rice SAPs are responsive to multiple biotic stresses and overexpression of OsSAP1, an A20/AN1 zinc-finger protein, enhances the basal resistance against pathogen infection in tobacco. Plant Sci. 2014, 225, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Kanneganti, V.; Gupta, A.K. Overexpression of OsiSAP8, a member of stress associated protein (SAP) gene family of rice confers tolerance to salt, drought and cold stress in transgenic tobacco and rice. Plant Mol. Biol. 2008, 66, 445. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Wang, M.M.; Jiang, Y.; Bao, Y.M.; Huang, X.; Sun, H.; Xu, D.Q.; Lan, H.X.; Zhang, H.S. Expression analysis of rice A20/AN1-type zinc finger genes and characterization of ZFP177 that contributes to temperature stress tolerance. Gene 2008, 420, 135–144. [Google Scholar] [CrossRef]
- Giri, J.; Vij, S.; Dansana, P.K.; Tyagi, A.K. Rice A20/AN1 zinc-finger containing stress-associated proteins (SAP1/11) and a receptor-like cytoplasmic kinase (OsRLCK253) interact via A20 zinc-finger and confer abiotic stress tolerance in transgenic Arabidopsis plants. New Phytol. 2011, 191, 721–732. [Google Scholar] [CrossRef]
- Hozain, M.D.; Abdelmageed, H.; Lee, J.; Kang, M.; Fokar, M.; Allen, R.D.; Holaday, A.S. Expression of AtSAP5 in cotton up-regulates putative stress-responsive genes and improves the tolerance to rapidly developing water deficit and moderate heat stress. J. Plant Physiol. 2012, 169, 1261–1270. [Google Scholar] [CrossRef]
- Kang, M.; Fokar, M.; Abdelmageed, H.; Allen, R.D. Arabidopsis SAP5 functions as a positive regulator of stress responses and exhibits E3 ubiquitin ligase activity. Plant Mol. Biol. 2011, 75, 451–466. [Google Scholar] [CrossRef]
- Ben, S.R.; Fabre, D.; Mieulet, D.; Meynard, D.; Dingkuhn, M.; Al-Doss, A.; Guiderdoni, E.; Hassairi, A. Expression of the Aeluropus littoralis AlSAP gene in rice confers broad tolerance to abiotic stresses through maintenance of photosynthesis. Plant Cell Environ. 2012, 35, 626–643. [Google Scholar]
- Ghneimherrera, T.; Selvaraj, M.G.; Meynard, D.; Fabre, D.; Peña, A.; Ben, R.W.; Ben, S.R.; Ogawa, S.; Rebolledo, M.C.; Ishitani, M. Expression of the Aeluropus littoralis AlSAP gene enhances rice yield under field drought at the reproductive stage. Front. Plant Sci. 2017, 8, 994. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, L.; Zhang, L.; Xing, T.; Peng, J.; Sun, S.; Chen, G.; Wang, X. A novel stress-associated protein SbSAP14 from Sorghum bicolor confers tolerance to salt stress in transgenic rice. Mol. Breeding 2013, 32, 437–449. [Google Scholar] [CrossRef]
- Sreedharan, S.; Shekhawat, U.K.S.; Ganapathi, T.R. MusaSAP1, a A20/AN1 zinc finger gene from banana functions as a positive regulator in different stress responses. Plant Mol. Biol. 2012, 80, 503–517. [Google Scholar] [CrossRef] [PubMed]
- Dixit, A.; Tomar, P.; Vaine, E.; Abdullah, H.; Hazen, S.; Dhankher, O.P. A stress--associated protein, AtSAP13, from Arabidopsis thaliana provides tolerance to multiple abiotic stresses. Plant Cell Environ. 2018, 41, 1171–1185. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.F.; Mao, X.G.; Wang, Y.X.; Wang, J.Y.; Xi, Y.J.; Jing, R.L. A wheat gene TaSAP17-D encoding an AN1/AN1 zinc finger protein improves salt stress tolerance in transgenic Arabidopsis. J. Integr. Agric. 2018, 17, 507–516. [Google Scholar] [CrossRef]
- Gordon, J.C. Poplars: Trees of the people, trees of the future. For. Chron. 2001, 77, 217–219. [Google Scholar] [CrossRef]
- Sannigrahi, P.; Ragauskas, A.J.; Tuskan, G.A. Poplar as a feedstock for biofuels: A review of compositional characteristics. Biofuels Bioprod. Biorefin. 2010, 4, 209–226. [Google Scholar] [CrossRef]
- Yoon, S.; Bae, E.; Lee, H.; Choi, Y.; Han, M.; Choi, H.; Kang, K.; Park, E. Downregulation of stress-associated protein 1 (PagSAP1) increases salt stress tolerance in poplar (Populus alba × P. glandulosa). Trees-Struct. Funct. 2018, 32, 823–833. [Google Scholar] [CrossRef]
- Ma, T.; Wang, J.; Zhou, G.; Yue, Z.; Hu, Q.; Chen, Y.; Liu, B.; Qiu, Q.; Wang, Z.; Zhang, J. Genomic insights into salt adaptation in a desert poplar. Nat. Commun. 2013, 4, 2797. [Google Scholar] [CrossRef]
- Liu, B.; Hu, J.; Zhang, J. Evolutionary divergence of duplicated Hsf genes in Populus. Cells 2019, 8, 438. [Google Scholar] [CrossRef]
- McKown, A.D.; Guy, R.D.; Klápště, J.; Geraldes, A.; Friedmann, M.; Cronk, Q.C.; El--Kassaby, Y.A.; Mansfield, S.D.; Douglas, C.J. Geographical and environmental gradients shape phenotypic trait variation and genetic structure in Populus trichocarpa. New Phytol. 2014, 201, 1263–1276. [Google Scholar] [CrossRef]
- Munns, R. Comparative physiology of salt and water stress. Plant Cell Environ. 2002, 25, 239–250. [Google Scholar] [CrossRef]
- Denby, K.; Gehring, C. Engineering drought and salinity tolerance in plants: Lessons from genome-wide expression profiling in Arabidopsis. Trends Biotechnol. 2005, 23, 547–552. [Google Scholar] [CrossRef] [PubMed]
- Moreno, A.A.; Mukhtar, M.S.; Blanco, F.; Boatwright, J.L.; Moreno, I.; Jordan, M.R.; Chen, Y.; Brandizzi, F.; Dong, X.; Orellana, A. IRE1/bZIP60-mediated unfolded protein response plays distinct roles in plant immunity and abiotic stress responses. PLoS ONE 2012, 7, e31944. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, L.; Xia, C.; Zhao, G.; Liu, J.; Jia, J.; Kong, X. A novel wheat bZIP transcription factor, TabZIP60, confers multiple abiotic stress tolerances in transgenic Arabidopsis. Physiol. Plantarum 2015, 153, 538–554. [Google Scholar] [CrossRef] [PubMed]
- Milla, M.A.R.; Townsend, J.; Chang, I.F.; Cushman, J.C. The Arabidopsis AtDi19 gene family encodes a novel type of Cys2/His2 Zinc-finger protein implicated in ABA-independent dehydration, high-salinity stress and light signaling pathways. Plant Mol. Biol. 2006, 61, 13–30. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yu, C.; Chen, C.; He, C.; Zhu, Y.; Huang, W. Identification of rice Di19 family reveals OsDi19-4 involved in drought resistance. Plant Cell Rep. 2014, 33, 2047–2062. [Google Scholar] [CrossRef] [PubMed]
- Gaber, A.; Ogata, T.; Maruta, T.; Yoshimura, K.; Tamoi, M.; Shigeoka, S. The involvement of Arabidopsis Glutathione Peroxidase 8 in the suppression of oxidative damage in the nucleus and cytosol. Plant Cell Physiol. 2012, 53, 1596–1606. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Cheng, Y.; Zhang, X.; Guan, Q.; Nishiuchi, S.; Hase, K.; Takano, T. Expression of an NADP-malic enzyme gene in rice (Oryza sativa. L) is induced by environmental stresses; over-expression of the gene in Arabidopsis confers salt and osmotic stress tolerance. Plant Mol. Biol. 2007, 64, 49. [Google Scholar] [CrossRef]
- Guo, Y.; Song, Y.; Zheng, H.; Zhang, Y.; Guo, J.; Sui, N. NADP-malate dehydrogenase of sweet sorghum improves salt tolerance of Arabidopsis thaliana. J. Agric. Food Chem. 2018, 66, 5992–6002. [Google Scholar] [CrossRef]
- Ren, Z.; Zheng, Z.; Chinnusamy, V.; Zhu, J.; Cui, X.; Iida, K.; Zhu, J.K. RAS1, a quantitative trait locus for salt tolerance and ABA sensitivity in Arabidopsis. Proc. Natl. Acad. Sci. USA 2010, 107, 5669–5674. [Google Scholar] [CrossRef]
- Jiang, Y.Q.; Deyholos, M.K. Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 2009, 69, 91–105. [Google Scholar] [CrossRef]
- Ali, M.A.; Azeem, F.; Nawaz, M.A.; Acet, T.; Abbas, A.; Imran, Q.M.; Shah, K.H.; Rehman, H.M.; Chung, G.; Yang, S.H. Transcription factors WRKY11 and WRKY17 are involved in abiotic stress responses in Arabidopsis. J. Plant Physiol. 2018, 226, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Wang, N.N.; Gong, S.Y.; Lu, R.; Li, Y.; Li, X.B. Overexpression of a cotton (Gossypium hirsutum) WRKY gene, GhWRKY34, in Arabidopsis enhances salt-tolerance of the transgenic plants. Plant Physiol. Biochem. 2015, 96, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Chu, X.; Wang, C.; Chen, X.; Lu, W.; Li, H.; Wang, X.; Hao, L.; Guo, X. The cotton WRKY gene GhWRKY41 positively regulates salt and drought stress tolerance in transgenic Nicotiana benthamiana. PLoS ONE 2015, 10, e0143022. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Jia, H.; Chen, X.; Hao, L.; An, H.; Guo, X. The cotton WRKY transcription factor GhWRKY17 functions in drought and salt stress in transgenic Nicotiana benthamiana through ABA signaling and the modulation of reactive oxygen species production. Plant Cell Physiol. 2014, 55, 2060–2076. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Hao, L.; Li, J.; Liu, D.; Guo, X.; Li, H. The Gossypium hirsutum WRKY gene GhWRKY39-1 promotes pathogen infection defense responses and mediates salt stress tolerance in transgenic Nicotiana benthamiana. Plant Cell Rep. 2014, 33, 483–498. [Google Scholar] [CrossRef]
- Miller, G.; Suzuki, N.; Ciftci-Yilmaz, S.; Mittler, R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010, 33, 453–467. [Google Scholar] [CrossRef]
- Ahmad, P.; Sarwat, M.; Sharma, S. Reactive oxygen species, antioxidants and signaling in plants. J. Plant Biol. 2008, 51, 167–173. [Google Scholar] [CrossRef]
- Agati, G.; Azzarello, E.; Pollastri, S.; Tattinic, M. Flavonoids as antioxidants in plants: Location and functional significance. Plant Sci. 2012, 196, 67–76. [Google Scholar] [CrossRef]
- Wang, F.; Zhu, H.; Kong, W.; Peng, R.; Liu, Q.; Yao, Q. The Antirrhinum AmDEL gene enhances flavonoids accumulation and salt and drought tolerance in transgenic Arabidopsis. Planta 2016, 244, 59–73. [Google Scholar] [CrossRef]
- Fang, L.; Su, L.; Sun, X.; Li, X.; Sun, M.; Karungo, S.K.; Fang, S.; Chu, J.; Li, S.; Xin, H. Expression of Vitis amurensis NAC26 in Arabidopsis enhances drought tolerance by modulating jasmonic acid synthesis. J. Exp. Bot. 2016, 67, 2829–2845. [Google Scholar] [CrossRef]
- Nahar, K.; Hasanuzzaman, M.; Rahman, A.; Alam, M.; Mahmud, J.; Suzuki, T.; Fujita, M. Polyamines confer salt tolerance in mung bean (Vigna radiata L.) by reducing sodium uptake, improving nutrient homeostasis, antioxidant defense, and methylglyoxal detoxification systems. Front. Plant Sci. 2016, 7, 1104. [Google Scholar] [CrossRef] [PubMed]
- Ravikovitch, S.; Porath, A. The effect of nutrients on the salt tolerance of crops. Plant Soil 1967, 26, 49–71. [Google Scholar] [CrossRef]
- Li, J.; Zhang, J.; Jia, H.; Li, Y.; Xu, X.; Wang, L.; Lu, M. The Populus trichocarpa PtHSP17.8 involved in heat and salt stress tolerances. Plant Cell Rep. 2016, 35, 1587–1599. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Li, J.; Zhang, J.; Jia, H.; Liu, B.; Sun, P.; Hu, J.; Wang, L.; Lu, M. The WUSCHEL-related homeobox 5a (PtoWOX5a) is involved in adventitious root development in poplar. Tree Physiol. 2018, 38, 139–153. [Google Scholar] [CrossRef]
- Dubois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F. Colorimetric method for determination of sugars and related substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int. J. Genom. 2008, 2008, 619832. [Google Scholar] [CrossRef] [PubMed]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Sun, P.; Xia, Y.; Zheng, G.; Sun, J.; Jia, H. A Stress-Associated Protein, PtSAP13, From Populus trichocarpa Provides Tolerance to Salt Stress. Int. J. Mol. Sci. 2019, 20, 5782. https://doi.org/10.3390/ijms20225782
Li J, Sun P, Xia Y, Zheng G, Sun J, Jia H. A Stress-Associated Protein, PtSAP13, From Populus trichocarpa Provides Tolerance to Salt Stress. International Journal of Molecular Sciences. 2019; 20(22):5782. https://doi.org/10.3390/ijms20225782
Chicago/Turabian StyleLi, Jianbo, Pei Sun, Yongxiu Xia, Guangshun Zheng, Jingshuang Sun, and Huixia Jia. 2019. "A Stress-Associated Protein, PtSAP13, From Populus trichocarpa Provides Tolerance to Salt Stress" International Journal of Molecular Sciences 20, no. 22: 5782. https://doi.org/10.3390/ijms20225782
APA StyleLi, J., Sun, P., Xia, Y., Zheng, G., Sun, J., & Jia, H. (2019). A Stress-Associated Protein, PtSAP13, From Populus trichocarpa Provides Tolerance to Salt Stress. International Journal of Molecular Sciences, 20(22), 5782. https://doi.org/10.3390/ijms20225782



