Y-chromosome and Surname Analyses for Reconstructing Past Population Structures: The Sardinian Population as a Test Case
Abstract
1. Introduction
2. Results
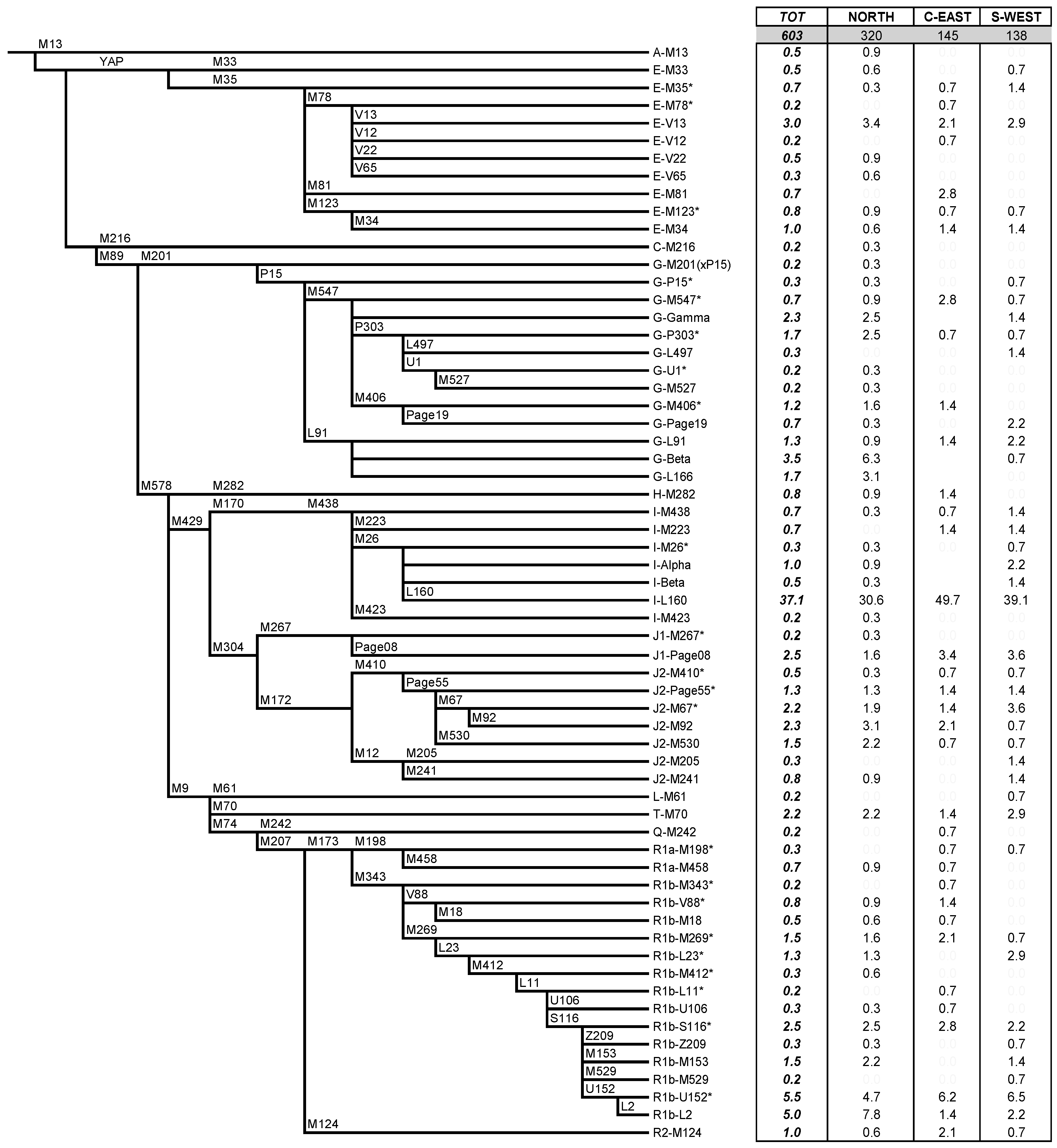
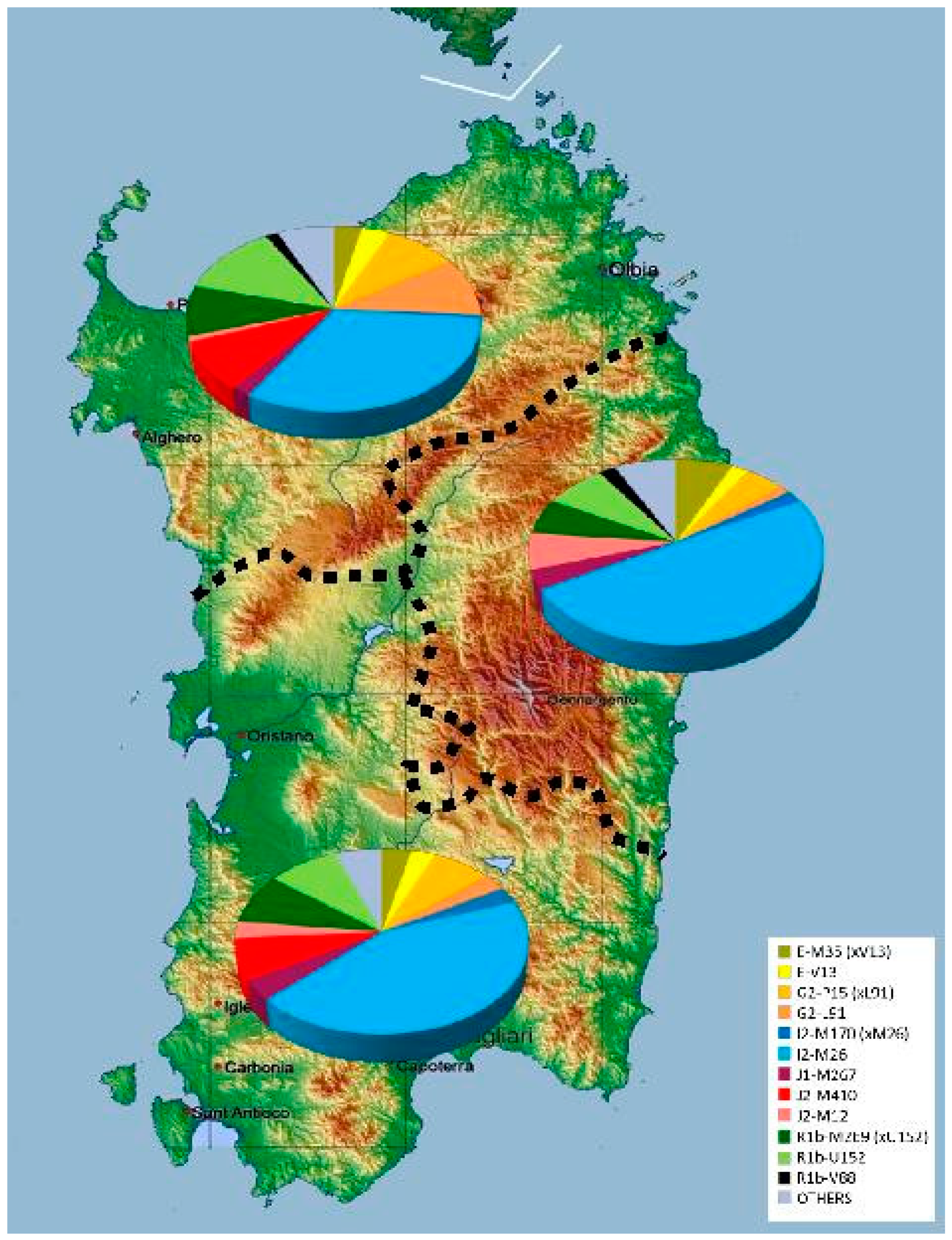
2.1. Classification and Distribution of Y-chromosome Haplogroups in Ancient Isolates of Sardinia
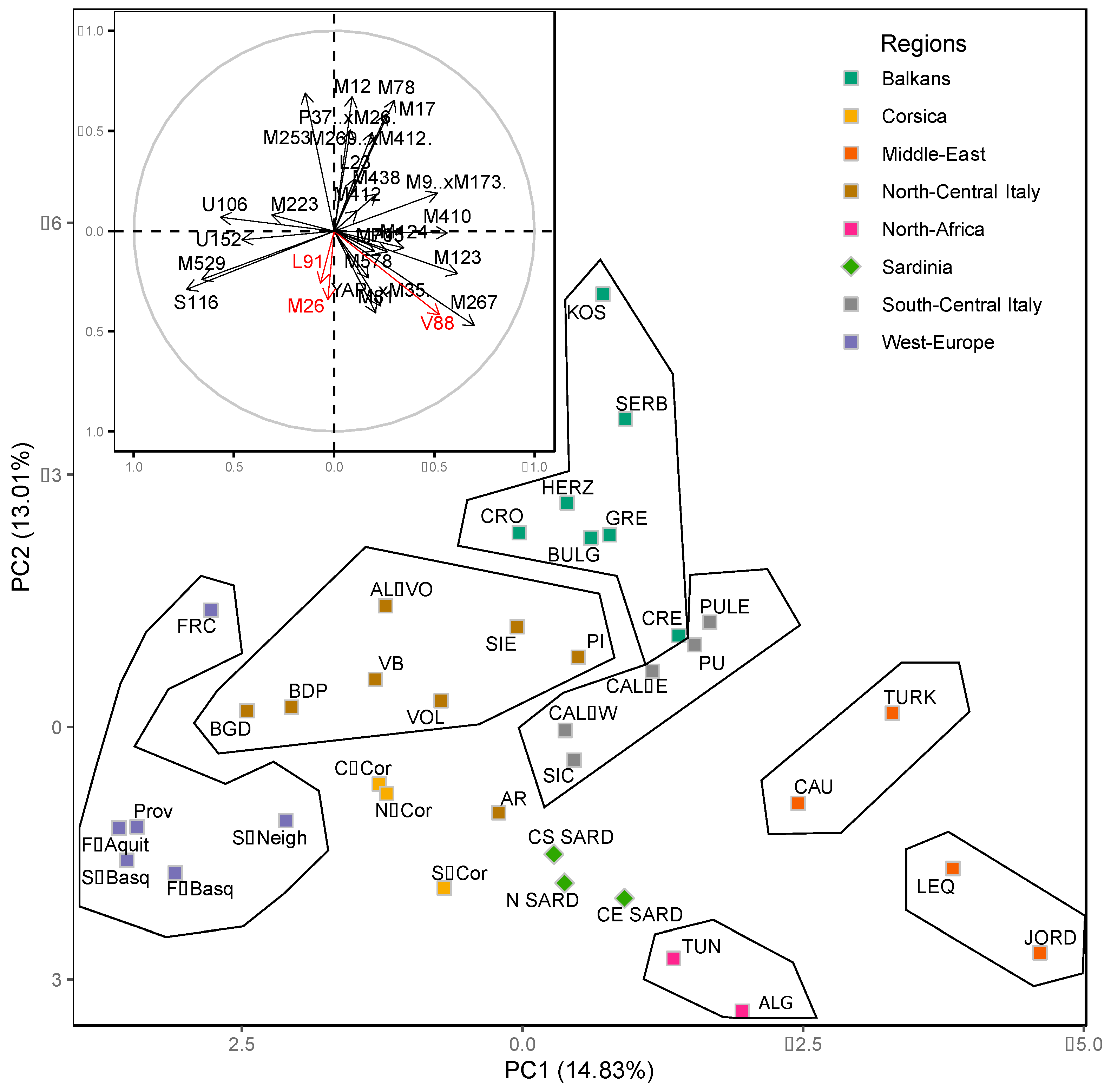
2.2. Sardinian Populations in the Mediterranean Context
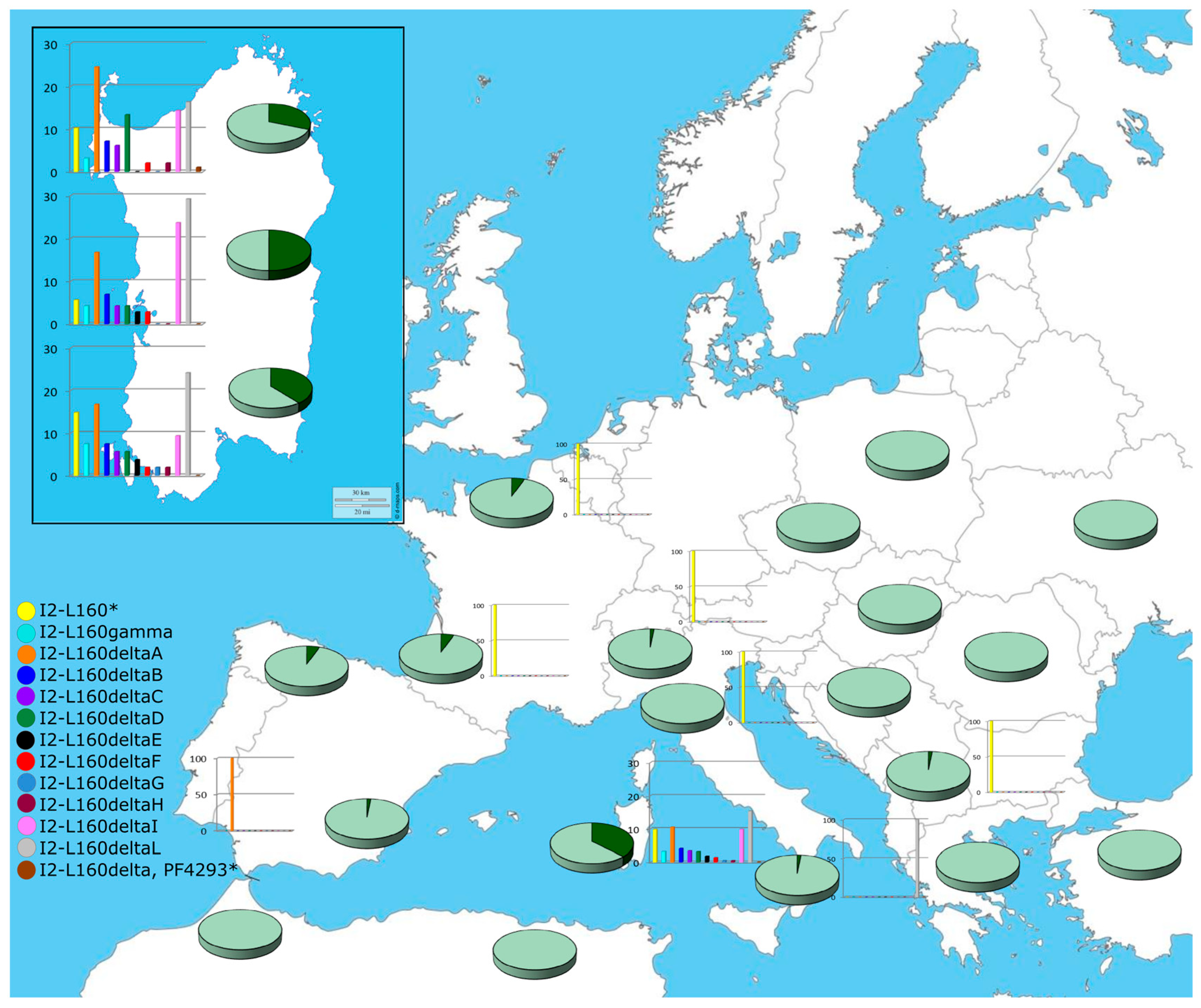
2.3. The Founder Paternal Lineages G2-L91, I2-M26 and R1b-V88
2.4. Haplogroup Distribution in Pre-Historic Sardinian Samples
3. Discussion
4. Materials and Methods
4.1. The Sample
4.2. DNA Analysis
4.3. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BCE | Before common era |
| CE | Common era |
| DHPLC | Denaturating high performance liquid chromatography |
| ky | Kilo years |
| kya | Kilo years ago |
| MDS | Multi-dimensional scaling analysis |
| MSY | Male specific region of the Y chromosome |
| PCA | Principal component analysis |
| RFLP | Restriction fragment length polymorphism |
| STR | Short tandem repeat |
References
- Cavalli-Sforza, L.L.; Piazza, A. Human genomic diversity in Europe: A summary of recent research and prospects for the future. Eur. J. Hum. Genet. 1993, 1, 3–18. [Google Scholar] [CrossRef]
- Di Gaetano, C.; Voglino, F.; Guarrera, S.; Fiorito, G.; Rosa, F.; Di Blasio, A.M.; Manzini, P.; Dianzani, I.; Betti, M.; Cusi, D. An overview of the genetic structure within the Italian population from genome-wide data. PLoS ONE 2012, 7, e43759. [Google Scholar] [CrossRef]
- Grugni, V.; Battaglia, V.; Hooshiar Kashani, B.; Parolo, S.; Al-Zahery, N.; Achilli, A.; Olivieri, A.; Gandini, F.; Houshmand, M.; Sanati, M.H. Ancient migratory events in the Middle East: New clues from the Y-chromosome variation of modern Iranians. PLoS ONE 2012, 7, e41252. [Google Scholar] [CrossRef]
- Pardo, L.M.; Piras, G.; Asproni, R.; Van Der Gaag, K.J.; Gabbas, A.; Ruiz-Linares, A.; De Knijff, P.; Monne, M.; Rizzu, P.; Heutink, P. Dissecting the genetic make-up of North-East Sardinia using a large set of haploid and autosomal markers. Eur. J. Hum. Genet. 2012, 20, 956–964. [Google Scholar] [CrossRef]
- Raveane, A.; Aneli, S.; Montinaro, F.; Athanasiadis, G.; Barlera, S.; Birolo, G.; Boncoraglio, G.; Di Blasio, A.M.; Di Gaetano, C.; Pagani, L. Population structure of modern-day Italians reveals patterns of ancient and archaic ancestries in Southern Europe. Sci. Adv. 2019, 5, eaaw3492. [Google Scholar] [CrossRef]
- Wagner, M.L. Historische Lautlehre des Sardischen, 1st ed.; Halle: Niemeyer, Germany, 1941; pp. 1–344. [Google Scholar]
- Spoor, C.; Sondaar, P. Human fossils from the endemic island fauna of Sardinia. J. Hum. Evol. 1986, 15, 399–408. [Google Scholar] [CrossRef]
- Contini, M.; Cappello, N.; Griffo, R.; Rendine, S.; Piazza, A. Géolinguistique et géogénétique: Une démarche interdisciplnaire. Géolinguistique 1989, 4, 129–197. [Google Scholar]
- Hofmeijer, G.K.; Alderliesten, C.; Van Der Borg, K.; Houston, C.; De Jong, A.; Martini, F.; Sanges, M.; Sondaar, P.; De Visser, J. Dating of the upper Pleistocene lithic industry of Sardinia. Radiocarbon 1989, 31, 986–991. [Google Scholar] [CrossRef]
- Sondaar, P.; Elhurg, R.; Holmeijer, G.K.; Martini, F.; Sanges, M.; Spaan, A.; de Visser, H. The human colonization of Sardinia: A Late-Pleistocene human fossil from Corbeddu cave. CR Acad. Sci. Paris 1995, 320, 145–150. [Google Scholar]
- Dyson, S.L.; Rowland, R.J., Jr. Archaeology and History in Sardinia from the Stone Age to the Middle Ages: Shepherds, Sailors, and Conquerors, 1st ed.; University of Pennsylvania Press: Pennsylvania, NJ, USA, 2007; pp. 1–248. [Google Scholar]
- Broodbank, C. “Minding the gap”: Thinking about change in Early Cycladic island societies from a comparative perspective. Am. J. Archaeol. 2013, 117, 535–543. [Google Scholar] [CrossRef]
- Barreca, F. La Sardegna Fenicia e Punica, 1st ed.; Sassari: Chiarella Editore, Italy, 1979; pp. 1–303. [Google Scholar]
- Cavalli-Sforza, L.L.; Menozzi, P.; Piazza, A. The History and Geography of Human Genes; Princeton University Press: Pennsylvania, NJ, USA, 1994; pp. 273–275. [Google Scholar]
- Chiang, C.W.K.; Marcus, J.H.; Sidore, C.; Biddanda, A.; Al-Asadi, H.; Zoledziewska, M.; Pitzalis, M.; Busonero, F.; Maschio, A.; Pistis, G. Genomic history of the Sardinian population. Nat. Genet. 2018, 50, 1426–1434. [Google Scholar] [CrossRef] [PubMed]
- Calò, C.; Melis, A.; Vona, G.; Piras, I. Review synthetic article: Sardinian population (Italy): A genetic review. AJOL 2008, 1, 39–64. [Google Scholar] [CrossRef]
- Morelli, L.; Grosso, M.; Vona, G.; Varesi, L.; Torroni, A.; Francalacci, P. Frequency distribution of mitochondrial DNA haplogroups in Corsica and Sardinia. Hum. Biol. 2000, 72, 585–595. [Google Scholar] [PubMed]
- Semino, O.; Passarino, G.; Oefner, P.J.; Lin, A.A.; Arbuzova, S.; Beckman, L.E.; De Benedictis, G.; Francalacci, P.; Kouvatsi, A.; Limborska, S. The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: A Y chromosome perspective. Science 2000, 290, 1155–1159. [Google Scholar] [CrossRef] [PubMed]
- Zei, G.; Lisa, A.; Fiorani, O.; Magri, C.; Quintana-Murci, L.; Semino, O.; Santachiara-Benerecetti, A.S. From surnames to the history of Y chromosomes: The Sardinian population as a paradigm. Eur. J. Hum. Genet. 2003, 11, 802–807. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Contu, D.; Morelli, L.; Santoni, F.; Foster, J.W.; Francalacci, P.; Cucca, F. Y-chromosome based evidence for pre-neolithic origin of the genetically homogeneous but diverse Sardinian population: Inference for association scans. PLoS ONE 2008, 3, e1430. [Google Scholar] [CrossRef]
- Pala, M.; Achilli, A.; Olivieri, A.; Hooshiar Kashani, B.; Perego, U.A.; Sanna, D.; Metspalu, E.; Tambets, K.; Tamm, E.; Accetturo, M. Mitochondrial haplogroup U5b3: A distant echo of the Epipaleolithic in Italy and the legacy of the early Sardinians. Am. J. Hum. Genet. 2009, 84, 814–821. [Google Scholar] [CrossRef]
- Francalacci, P.; Morelli, L.; Angius, A.; Berutti, R.; Reinier, F.; Atzeni, R.; Pilu, R.; Busonero, F.; Maschio, A.; Zara, I. Low-pass DNA sequencing of 1200 Sardinians reconstructs European Y-chromosome phylogeny. Science 2013, 341, 565–569. [Google Scholar] [CrossRef]
- Olivieri, A.; Sidore, C.; Achilli, A.; Angius, A.; Posth, C.; Furtwängler, A.; Brandini, S.; Capodiferro, M.R.; Gandini, F.; Zoledziewska, M. Mitogenome diversity in Sardinians: A genetic window onto an island’s past. Mol. Biol. Evol. 2017, 34, 1230–1239. [Google Scholar] [CrossRef]
- Tenesa, A.; Wright, A.; Knott, S.; Carothers, A.; Hayward, C.; Angius, A.; Persico, I.; Maestrale, G.; Hastie, N.; Pirastu, M. Extent of linkage disequilibrium in a Sardinian sub-isolate: Sampling and methodological considerations. Hum. Mol. Genet. 2004, 13, 25–33. [Google Scholar] [CrossRef]
- Tamm, E.; Cristofaro, J.D.; Mazières, S.; Pennarun, E.; Kushniarevich, A.; Raveane, A.; Semino, O.; Chiaroni, J.; Pereira, L.; Metspalu, M. Genome-wide analysis of Corsican population reveals a close affinity with Northern and Central Italy. Sci. Rep. 2019, 9, 13581. [Google Scholar] [CrossRef] [PubMed]
- Lazaridis, I.; Patterson, N.; Mittnik, A.; Renaud, G.; Mallick, S.; Kirsanow, K.; Sudmant, P.H.; Schraiber, J.G.; Castellano, S.; Lipson, M. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 2014, 513, 409–413. [Google Scholar] [CrossRef] [PubMed]
- Haak, W.; Lazaridis, I.; Patterson, N.; Rohland, N.; Mallick, S.; Llamas, B.; Brandt, G.; Nordenfelt, S.; Harney, E.; Stewardson, K. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 2015, 522, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Keller, A.; Graefen, A.; Ball, M.; Matzas, M.; Boisguerin, V.; Maixner, F.; Leidinger, P.; Backes, C.; Khairat, R.; Forster, M. New insights into the Tyrolean Iceman’s origin and phenotype as inferred by whole-genome sequencing. Nat. Commun. 2012, 3, 698. [Google Scholar] [CrossRef] [PubMed]
- Sikora, M.; Carpenter, M.L.; Moreno-Estrada, A.; Henn, B.M.; Underhill, P.A.; Sánchez-Quinto, F.; Zara, I.; Pitzalis, M.; Sidore, C.; Busonero, F. Population genomic analysis of ancient and modern genomes yields new insights into the genetic ancestry of the Tyrolean Iceman and the genetic structure of Europe. PLoS Genet. 2014, 10, e1004353. [Google Scholar] [CrossRef] [PubMed]
- Lazaridis, I.; Nadel, D.; Rollefson, G.; Merrett, D.C.; Rohland, N.; Mallick, S.; Fernandes, D.; Novak, M.; Gamarra, B.; Sirak, K. Genomic insights into the origin of farming in the ancient Near East. Nature 2016, 536, 419–424. [Google Scholar] [CrossRef]
- Workman, P.; Lucarelli, P.; Agostino, R.; Scarabino, R.; Scacchi, R.; Carapella, E.; Palmarino, R.; Bottini, E. Genetic differentiation among Sardinian villages. Am. J. Phys. Anthropol. 1975, 43, 165–176. [Google Scholar] [CrossRef]
- Wagner, M.L. La lingua sarda. Storia spirito e forma. In Max Leopold Wagner (Book Review); Brepols Publishers: Chicago, IL, USA, 1950; pp. 1–438. [Google Scholar]
- Barbujani, G.; Sokal, R.R. Genetic population structure of Italy. I. Geographic patterns of gene frequencies. Hum. Biol. 1991, 63, 253–272. [Google Scholar]
- Cappello, N.; Rendine, S.; Griffo, R.; Mameli, G.; Succa, V.; Vona, G.; Piazza, A. Genetic analysis of Sardinia: I. data on 12 polymorphisms in 21 linguistic domains. Ann. Hum. Genet. 1996, 60, 125–141. [Google Scholar] [CrossRef]
- Zavattari, P.; Deidda, E.; Whalen, M.; Lampis, R.; Mulargia, A.; Loddo, M.; Eaves, I.; Mastio, G.; Todd, J.A.; Cucca, F. Major factors influencing linkage disequilibrium by analysis of different chromosome regions in distinct populations: Demography, chromosome recombination frequency and selection. Hum. Mol. Genet. 2000, 9, 2947–2957. [Google Scholar] [CrossRef][Green Version]
- Angius, A.; Melis, P.M.; Morelli, L.; Petretto, E.; Casu, G.; Maestrale, G.; Fraumene, C.; Bebbere, D.; Forabosco, P.; Pirastu, M. Archival, demographic and genetic studies define a Sardinian sub-isolate as a suitable model for mapping complex traits. Hum. Genet. 2001, 109, 198–209. [Google Scholar] [CrossRef] [PubMed]
- Scozzari, R.; Cruciani, F.; Pangrazio, A.; Santolamazza, P.; Vona, G.; Moral, P.; Latini, V.; Varesi, L.; Memmi, M.M.; Romano, V. Human Y-chromosome variation in the western Mediterranean area: Implications for the peopling of the region. Hum. Immunol. 2001, 62, 871–884. [Google Scholar] [CrossRef]
- Fraumene, C.; Petretto, E.; Angius, A.; Pirastu, M. Striking differentiation of sub-populations within a genetically homogeneous isolate (Ogliastra) in Sardinia as revealed by mtDNA analysis. Hum. Genet. 2003, 114, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Destro-Bisol, G.; Anagnostou, P.; Batini, C.; Battaggia, C.; Bertoncini, S.; Boattini, A.; Caciagli, L.; Caló, M.; Capelli, C.; Capocasa, M. Italian isolates today: Geographic and linguistic factors shaping human biodiversity. J. Anthropol. Sci. JASS 2008, 86, 179–188. [Google Scholar] [PubMed]
- Pistis, G.; Piras, I.; Pirastu, N.; Persico, I.; Sassu, A.; Picciau, A.; Prodi, D.; Fraumene, C.; Mocci, E.; Manias, M.T. High differentiation among eight villages in a secluded area of Sardinia revealed by genome-wide high density SNPs analysis. PLoS ONE 2009, 4, e4654. [Google Scholar] [CrossRef]
- Montinaro, F.; Boschi, I.; Trombetta, F.; Merigioli, S.; Anagnostou, P.; Battaggia, C.; Capocasa, M.; Crivellaro, F.; Destro-Bisol, G.; Coia, V. Using forensic microsatellites to decipher the genetic structure of linguistic and geographic isolates: A survey in the eastern Italian Alps. Forensic Sci. Int. Gen. 2012, 6, 827–833. [Google Scholar] [CrossRef]
- Capocasa, M.; Anagnostou, P.; Bachis, V.; Battaggia, C.; Bertoncini, S.; Biondi, G.; Boattini, A.; Boschi, I.; Brisighelli, F.; Calò, C.M. Linguistic, geographic and genetic isolation: A collaborative study of Italian populations. J. Anthropol. Sci. JASS 2014, 92, 201–231. [Google Scholar]
- Francalacci, P.; Sanna, D.; Useli, A.; Berutti, R.; Barbato, M.; Whalen, M.B.; Angius, A.; Sidore, C.; Alonso, S.; Tofanelli, S. Detection of phylogenetically informative polymorphisms in the entire euchromatic portion of human Y chromosome from a Sardinian sample. BMC Res. Notes 2015, 8, 174. [Google Scholar] [CrossRef][Green Version]
- Underhill, P.A.; Shen, P.; Lin, A.A.; Jin, L.; Passarino, G.; Yang, W.H.; Kauffman, E.; Bonné-Tamir, B.; Bertranpetit, J.; Francalacci, P. Y chromosome sequence variation and the history of human populations. Nat. Genet. 2000, 26, 358–361. [Google Scholar] [CrossRef]
- Passarino, G.; Underhill, P.A.; Cavalli-Sforza, L.L.; Semino, O.; Pes, G.M.; Carru, C.; Ferrucci, L.; Bonafè, M.; Franceschi, C.; Deiana, L. Y chromosome binary markers to study the high prevalence of males in Sardinian centenarians and the genetic structure of the Sardinian population. Hum. Hered. 2001, 52, 136–139. [Google Scholar] [CrossRef]
- Francalacci, P.; Morelli, L.; Underhill, P.A.; Lillie, A.S.; Passarino, G.; Useli, A.; Madeddu, R.; Paoli, G.; Tofanelli, S.; Calò, C.M. Peopling of three Mediterranean islands (Corsica, Sardinia, and Sicily) inferred by Y-chromosome biallelic variability. Am. J. Phys. Anthropol. 2003, 121, 270–279. [Google Scholar] [CrossRef] [PubMed]
- Cruciani, F.; Trombetta, B.; Sellitto, D.; Massaia, A.; Destro-Bisol, G.; Watson, E.; Colomb, E.B.; Dugoujon, J.-M.; Moral, P.; Scozzari, R. Human Y chromosome haplogroup R-V88: A paternal genetic record of early mid Holocene trans-Saharan connections and the spread of Chadic languages. Eur. J. Hum. Genet. 2010, 18, 800–807. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rootsi, S.; Myres, N.M.; Lin, A.A.; Järve, M.; King, R.J.; Kutuev, I.; Cabrera, V.M.; Khusnutdinova, E.K.; Varendi, K.; Sahakyan, H. Distinguishing the co-ancestries of haplogroup G Y-chromosomes in the populations of Europe and the Caucasus. Eur. J. Hum. Genet. 2012, 20, 1275–1282. [Google Scholar] [CrossRef] [PubMed]
- Berger, B.; Niederstätter, H.; Erhart, D.; Gassner, C.; Schennach, H.; Parson, W. High resolution mapping of Y haplogroup G in Tyrol (Austria). Forensic Sci. Int. Genet. 2013, 7, 529–536. [Google Scholar] [CrossRef]
- Price, T.D. Europe’s First Farmers; Cambridge University Press: Cambridge, UK, 2000; pp. 1–412. [Google Scholar]
- Rootsi, S.; Kivisild, T.; Benuzzi, G.; Help, H.; Bermisheva, M.; Kutuev, I.; Barać, L.; Peričić, M.; Balanovsky, O.; Pshenichnov, A. Phylogeography of Y-chromosome haplogroup I reveals distinct domains of prehistoric gene flow in Europe. Am. J. Hum. Genet. 2004, 75, 128–137. [Google Scholar] [CrossRef]
- YFull Tree, I-L158. Available online: https://www.yfull.com/tree/I-L158/ (accessed on 16 October 2019).
- Lacan, M.; Keyser, C.; Ricaut, F.-X.; Brucato, N.; Duranthon, F.; Guilaine, J.; Crubézy, E.; Ludes, B. Ancient DNA reveals male diffusion through the Neolithic Mediterranean route. Proc. Natl. Acad. Sci. USA 2011, 108, 9788–9791. [Google Scholar] [CrossRef]
- YFull Tree. Available online: https://www.yfull.com/tree/ (accessed on 16 October 2019).
- González, M.; Gomes, V.; López-Parra, A.M.; Amorim, A.; Carracedo, A.; Sánchez-Diz, P.; Arroyo-Pardo, E.; Gusmão, L. The genetic landscape of Equatorial Guinea and the origin and migration routes of the Y chromosome haplogroup R-V88. Eur. J. Hum. Genet. 2013, 21, 324–331. [Google Scholar] [CrossRef]
- D’Atanasio, E.; Trombetta, B.; Bonito, M.; Finocchio, A.; Di Vito, G.; Seghizzi, M.; Romano, R.; Russo, G.; Paganotti, G.M.; Watson, E. The peopling of the last Green Sahara revealed by high-coverage resequencing of trans-Saharan patrilineages. Genome Biol. 2018, 19, 20. [Google Scholar] [CrossRef]
- Zalloua, P.A.; Platt, D.E.; El Sibai, M.; Khalife, J.; Makhoul, N.; Haber, M.; Xue, Y.; Izaabel, H.; Bosch, E.; Adams, S.M. Identifying genetic traces of historical expansions: Phoenician footprints in the Mediterranean. Am. J. Hum. Genet. 2008, 83, 633–642. [Google Scholar] [CrossRef]
- Kivisild, T. The study of human Y chromosome variation through ancient DNA. Hum. Genet. 2017, 136, 529–546. [Google Scholar] [CrossRef]
- Haber, M.; Mezzavilla, M.; Xue, Y.; Comas, D.; Gasparini, P.; Zalloua, P.; Tyler-Smith, C. Genetic evidence for an origin of the Armenians from Bronze Age mixing of multiple populations. Eur. J. Hum. Genet. 2016, 24, 931–936. [Google Scholar] [CrossRef] [PubMed]
- Lipson, M.; Cheronet, O.; Mallick, S.; Rohland, N.; Oxenham, M.; Pietrusewsky, M.; Pryce, T.O.; Willis, A.; Matsumura, H.; Buckley, H. Ancient genomes document multiple waves of migration in Southeast Asian prehistory. Science 2018, 361, 92–95. [Google Scholar] [CrossRef] [PubMed]
- Mathieson, I.; Alpaslan-Roodenberg, S.; Posth, C.; Szécsényi-Nagy, A.; Rohland, N.; Mallick, S.; Olalde, I.; Broomandkhoshbacht, N.; Candilio, F.; Cheronet, O. The genomic history of southeastern Europe. Nature 2018, 555, 197. [Google Scholar] [CrossRef] [PubMed]
- Marcus, J.H.; Posth, C.; Ringbauer, H.; Lai, L.; Skeates, R.; Sidore, C.; Beckett, J.; Furtwängler, A.; Olivieri, A.; Chiang, C.W.K. Population history from the Neolithic to present on the Mediterranean island of Sardinia: An ancient DNA perspective. BioRxiv 2019, 583104. [Google Scholar] [CrossRef]
- Fernandes, D.M.; Mittnik, A.; Olalde, I.; Lazaridis, I.; Cheronet, O.; Rohland, N.; Mallick, S.; Bernardos, R.; Broomandkhoshbacht, N.; Carlsson, J. The arrival of Steppe and iranian related ancestry in the islands of the Western Mediterranean. BioRxiv 2019, 584714. [Google Scholar] [CrossRef]
- Mathieson, I.; Reich, D. Differences in the rare variant spectrum among human populations. PLoS Genet. 2017, 13, e1006581. [Google Scholar] [CrossRef]
- Kılınç, G.M.; Omrak, A.; Özer, F.; Günther, T.; Büyükkarakaya, A.M.; Bıçakçı, E.; Baird, D.; Dönertaş, H.M.; Ghalichi, A.; Yaka, R. The demographic development of the first farmers in Anatolia. Curr. Biol. 2016, 26, 2659–2666. [Google Scholar] [CrossRef]
- Mathieson, I.; Lazaridis, I.; Rohland, N.; Mallick, S.; Patterson, N.; Roodenberg, S.A.; Harney, E.; Stewardson, K.; Fernandes, D.; Novak, M. Genome-wide patterns of selection in 230 ancient Eurasians. Nature 2015, 528, 499–503. [Google Scholar] [CrossRef]
- Olalde, I.; Brace, S.; Allentoft, M.E.; Armit, I.; Kristiansen, K.; Booth, T.; Rohland, N.; Mallick, S.; Szécsényi-Nagy, A.; Mittnik, A. The Beaker phenomenon and the genomic transformation of northwest Europe. Nature 2018, 555, 190–196. [Google Scholar] [CrossRef]
- Fu, Q.; Posth, C.; Hajdinjak, M.; Petr, M.; Mallick, S.; Fernandes, D.; Furtwängler, A.; Haak, W.; Meyer, M.; Mittnik, A. The genetic history of ice age Europe. Nature 2016, 534, 200–205. [Google Scholar] [CrossRef]
- Haber, M.; Doumet-Serhal, C.; Scheib, C.; Xue, Y.; Danecek, P.; Mezzavilla, M.; Youhanna, S.; Martiniano, R.; Prado-Martinez, J.; Szpak, M. Continuity and admixture in the last five millennia of Levantine history from Ancient Canaanite and present-day Lebanese genome sequences. Am. J. Hum. Genet. 2017, 101, 274–282. [Google Scholar] [CrossRef] [PubMed]
- Semino, O.; Magri, C.; Benuzzi, G.; Lin, A.A.; Al-Zahery, N.; Battaglia, V.; Maccioni, L.; Triantaphyllidis, C.; Shen, P.; Oefner, P.J. Origin, diffusion, and differentiation of Y-chromosome haplogroups E and J: Inferences on the neolithization of Europe and later migratory events in the Mediterranean area. Am. J. Hum. Genet. 2004, 74, 1023–1034. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, V.; Fornarino, S.; Al-Zahery, N.; Olivieri, A.; Pala, M.; Myres, N.M.; King, R.J.; Rootsi, S.; Marjanovic, D.; Primorac, D. Y-chromosomal evidence of the cultural diffusion of agriculture in Southeast Europe. Eur. J. Hum. Genet. 2009, 17, 820–830. [Google Scholar] [CrossRef] [PubMed]
- Van de Loosdrecht, M.; Bouzouggar, A.; Humphrey, L.; Posth, C.; Barton, N.; Aximu-Petri, A.; Nickel, B.; Nagel, S.; Talbi, E.H.; El Hajraoui, M.A. Pleistocene North African genomes link Near Eastern and sub-Saharan African human populations. Science 2018, 360, 548–552. [Google Scholar] [CrossRef] [PubMed]
- Cruciani, F.; La Fratta, R.; Trombetta, B.; Santolamazza, P.; Sellitto, D.; Colomb, E.B.; Dugoujon, J.-M.; Crivellaro, F.; Benincasa, T.; Pascone, R. Tracing past human male movements in northern/eastern Africa and western Eurasia: New clues from Y-chromosomal haplogroups E-M78 and J-M12. Mol. Biol. Evol. 2007, 24, 1300–1311. [Google Scholar] [CrossRef] [PubMed]
- Fiorito, G.; Di Gaetano, C.; Guarrera, S.; Rosa, F.; Feldman, M.W.; Piazza, A.; Matullo, G. The Italian genome reflects the history of Europe and the Mediterranean basin. Eur. J. Hum. Genet. 2016, 24, 1056–1062. [Google Scholar] [CrossRef] [PubMed]
- Parolo, S.; Lisa, A.; Gentilini, D.; Di Blasio, A.M.; Barlera, S.; Nicolis, E.B.; Boncoraglio, G.B.; Parati, E.A.; Bione, S. Characterization of the biological processes shaping the genetic structure of the Italian population. BMC Genet. 2015, 16, 132. [Google Scholar] [CrossRef]
- Sazzini, M.; Ruscone, G.A.G.; Giuliani, C.; Sarno, S.; Quagliariello, A.; De Fanti, S.; Boattini, A.; Gentilini, D.; Fiorito, G.; Catanoso, M. Complex interplay between neutral and adaptive evolution shaped differential genomic background and disease susceptibility along the Italian peninsula. Sci. Rep. 2016, 6, 32513. [Google Scholar] [CrossRef]
- Contini, M. Classification phonologique des langages sardes. Bull. Inst. Phonetique Grenoble 1979, 8, 57–96. [Google Scholar]
- Zei, G.; Piazza, A.; Moroni, A.; Cavalli-Sforza, L.L. Surnames in Sardinia| III. The spatial distribution of surnames for testing neutrality of genes. Ann. Hum. Genet. 1986, 50, 169–180. [Google Scholar] [CrossRef]
- Skorecki, K.; Selig, S.; Blazer, S.; Bradman, R.; Bradman, N.; Waburton, P.; Ismajlowicz, M.; Hammer, M.F. Y chromosomes of Jewish priests. Nature 1997, 385, 32. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.G.; Skoreckiad, K.; Ben-Amid, H.; Parfitt, T.; Bradman, N.; Goldstein, D.B. Origins of old testament priests. Nature 1998, 394, 138–140. [Google Scholar] [CrossRef] [PubMed]
- Sykes, B.; Irven, C. Surnames and the Y chromosome. Am. J. Hum. Genet. 2000, 66, 1417–1419. [Google Scholar] [CrossRef] [PubMed]
- King, T.E.; Jobling, M.A. Founders, drift, and infidelity: The relationship between Y chromosome diversity and patrilineal surnames. Mol. Biol. Evol. 2009, 26, 1093–1102. [Google Scholar] [CrossRef] [PubMed]
- Van Oven, M.; Van Geystelen, A.; Kayser, M.; Decorte, R.; Larmuseau, M.H. Seeing the wood for the trees: A minimal reference phylogeny for the human Y chromosome. Hum. Mut. 2014, 35, 187–191. [Google Scholar] [CrossRef] [PubMed]
- ISOGG, International Society of Genetic Genealogy. ISOGG Version 14.177. 2019. Available online: https://isogg.org/tree/ (accessed on 16 October 2019).
- Karachanak, S.; Grugni, V.; Fornarino, S.; Nesheva, D.; Al-Zahery, N.; Battaglia, V.; Carossa, V.; Yordanov, Y.; Torroni, A.; Galabov, A.S. Y-chromosome diversity in modern Bulgarians: New clues about their ancestry. PLoS ONE 2013, 8, e56779. [Google Scholar] [CrossRef] [PubMed]
- Family Tree DNA, Y Haplotree. Available online: https://www.familytreedna.com/public/y-dna-haplotree/A (accessed on 16 October 2019).
- Underhill, P.A.; Jin, L.; Lin, A.A.; Mehdi, S.Q.; Jenkins, T.; Vollrath, D.; Davis, R.W.; Cavalli-Sforza, L.L.; Oefner, P.J. Detection of numerous Y chromosome biallelic polymorphisms by denaturing high-performance liquid chromatography. Genome Res. 1997, 7, 996–1005. [Google Scholar] [CrossRef]
- STRBase. Available online: https://strbase.nist.gov/index.htm (accessed on 16 October 2019).
- Nei, M. Phylogenetic analysis in molecular evolutionary genetics. Annu. Rev. Genet. 1996, 30, 371–403. [Google Scholar] [CrossRef]
- Slatkin, M. A measure of population subdivision based on microsatellite allele frequencies. Genetics 1995, 139, 457–462. [Google Scholar]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 1992, 131, 479–491. [Google Scholar]
- Bandelt, H.-J.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef]
- Fluxus Engineering. Available online: http://www.fluxus-engineering.com/sharenet.htm (accessed on 16 October 2019).
- Bandelt, H.-J.; Forster, P.; Sykes, B.C.; Richards, M.B. Mitochondrial portraits of human populations using median networks. Genetics 1995, 141, 743–753. [Google Scholar]
- Zhivotovsky, L.A.; Underhill, P.A.; Cinnioğlu, C.; Kayser, M.; Morar, B.; Kivisild, T.; Scozzari, R.; Cruciani, F.; Destro-Bisol, G.; Spedini, G. The effective mutation rate at Y chromosome short tandem repeats, with application to human population-divergence time. Am. J. Hum. Genet. 2004, 74, 50–61. [Google Scholar] [CrossRef][Green Version]
- Sengupta, S.; Zhivotovsky, L.A.; King, R.; Mehdi, S.Q.; Edmonds, C.A.; Chow, C.E.; Lin, A.A.; Mitra, M.; Sil, S.K.; Ramesh, A. Polarity and temporality of high-resolution y-chromosome distributions in India identify both indigenous and exogenous expansions and reveal minor genetic influence of Central Asian pastoralists. Am. J. Hum. Genet. 2006, 78, 202–221. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grugni, V.; Raveane, A.; Colombo, G.; Nici, C.; Crobu, F.; Ongaro, L.; Battaglia, V.; Sanna, D.; Al-Zahery, N.; Fiorani, O.; et al. Y-chromosome and Surname Analyses for Reconstructing Past Population Structures: The Sardinian Population as a Test Case. Int. J. Mol. Sci. 2019, 20, 5763. https://doi.org/10.3390/ijms20225763
Grugni V, Raveane A, Colombo G, Nici C, Crobu F, Ongaro L, Battaglia V, Sanna D, Al-Zahery N, Fiorani O, et al. Y-chromosome and Surname Analyses for Reconstructing Past Population Structures: The Sardinian Population as a Test Case. International Journal of Molecular Sciences. 2019; 20(22):5763. https://doi.org/10.3390/ijms20225763
Chicago/Turabian StyleGrugni, Viola, Alessandro Raveane, Giulia Colombo, Carmen Nici, Francesca Crobu, Linda Ongaro, Vincenza Battaglia, Daria Sanna, Nadia Al-Zahery, Ornella Fiorani, and et al. 2019. "Y-chromosome and Surname Analyses for Reconstructing Past Population Structures: The Sardinian Population as a Test Case" International Journal of Molecular Sciences 20, no. 22: 5763. https://doi.org/10.3390/ijms20225763
APA StyleGrugni, V., Raveane, A., Colombo, G., Nici, C., Crobu, F., Ongaro, L., Battaglia, V., Sanna, D., Al-Zahery, N., Fiorani, O., Lisa, A., Ferretti, L., Achilli, A., Olivieri, A., Francalacci, P., Piazza, A., Torroni, A., & Semino, O. (2019). Y-chromosome and Surname Analyses for Reconstructing Past Population Structures: The Sardinian Population as a Test Case. International Journal of Molecular Sciences, 20(22), 5763. https://doi.org/10.3390/ijms20225763





