SiMYB3 in Foxtail Millet (Setaria italica) Confers Tolerance to Low-Nitrogen Stress by Regulating Root Growth in Transgenic Plants
Abstract
1. Introduction
2. Results
2.1. RNA-Seq Analyses of Low-Nitrogen-Induced Foxtail Millet
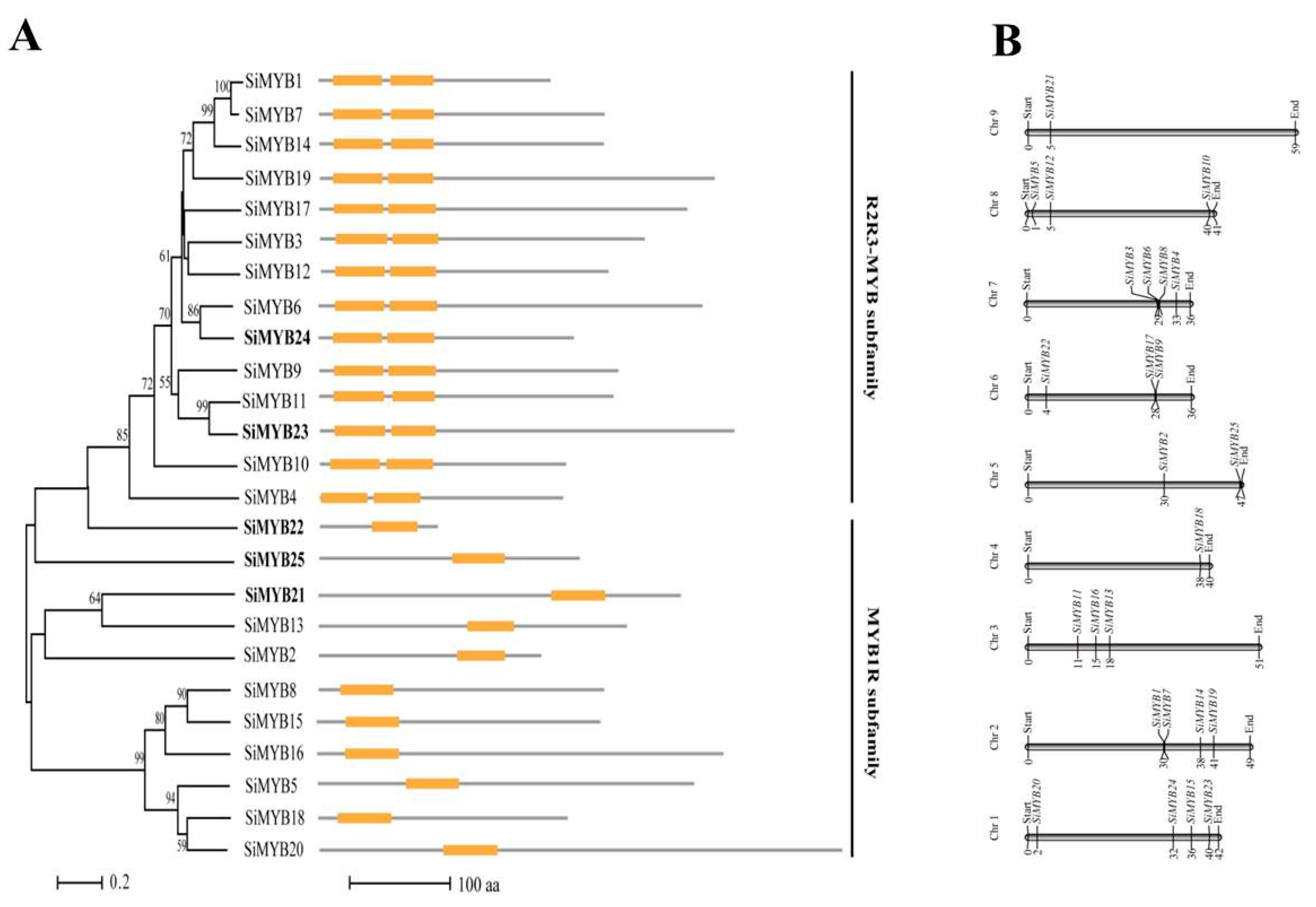
2.2. Characteristics Analysis of 25 MYB-Like Transcription Factors in Response to Low-Nitrogen Stress in Foxtail Millet
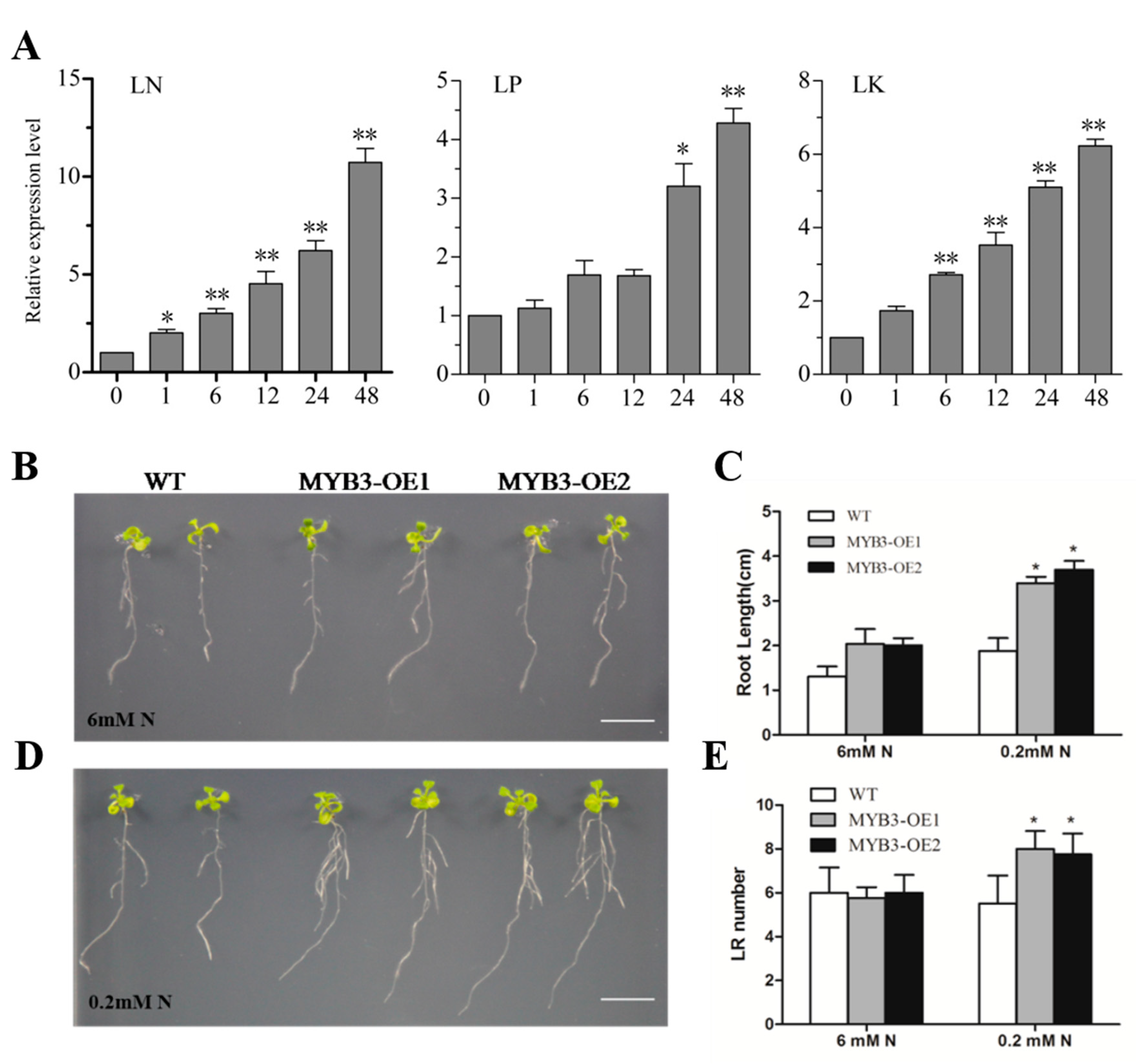
2.3. Subcellular Localization, Expression Pattern, and Gene Functions of SiMYB3
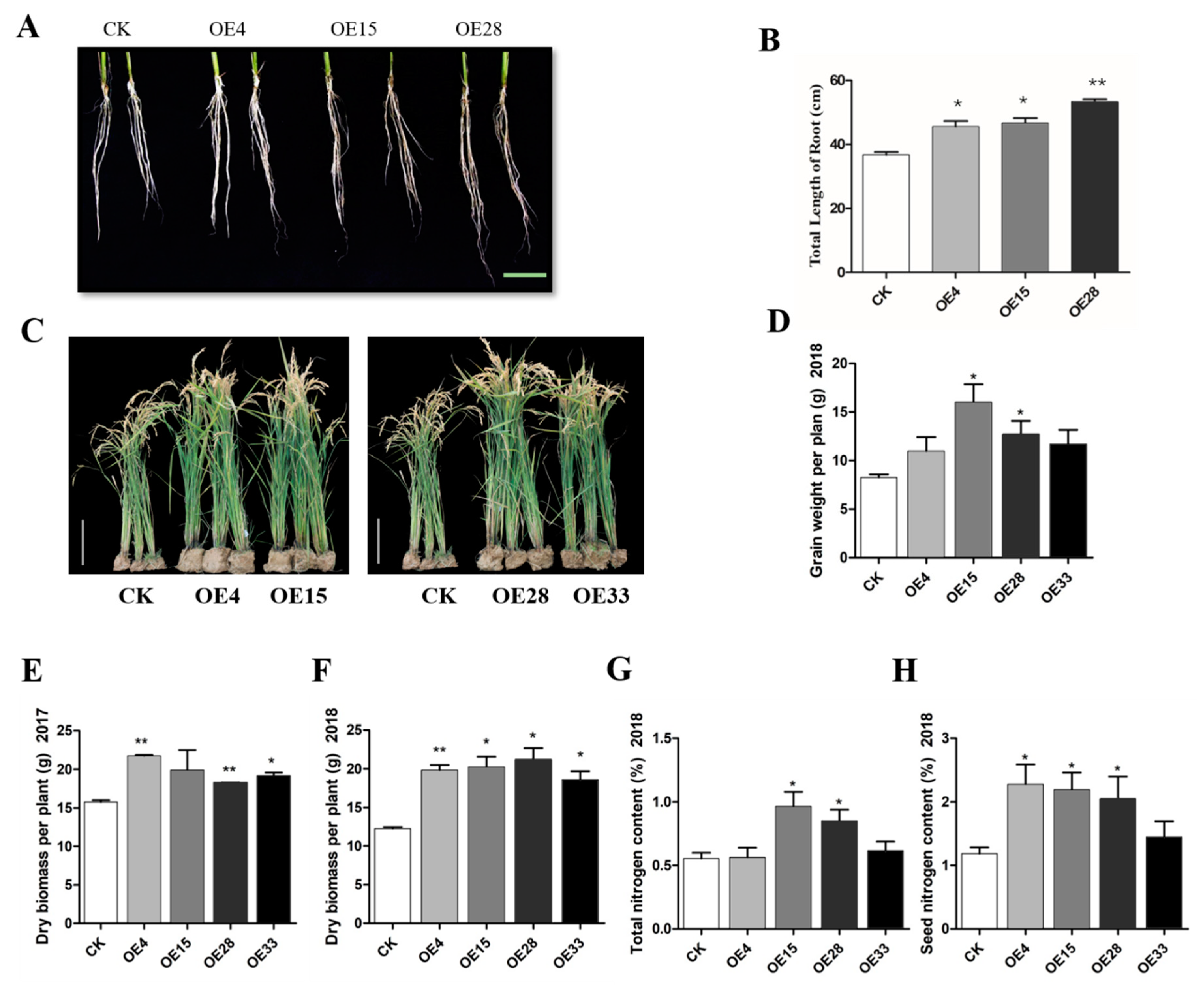
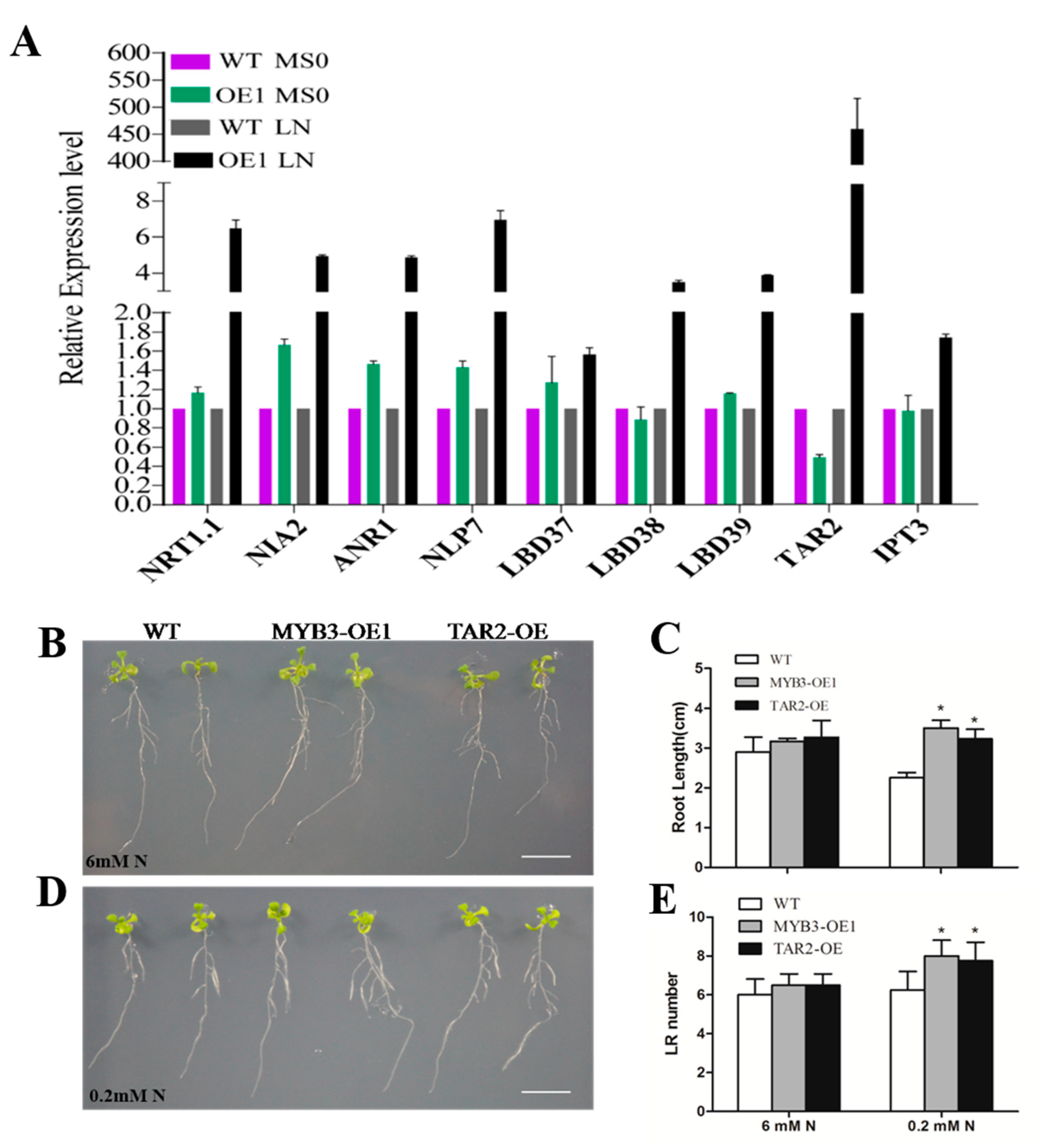
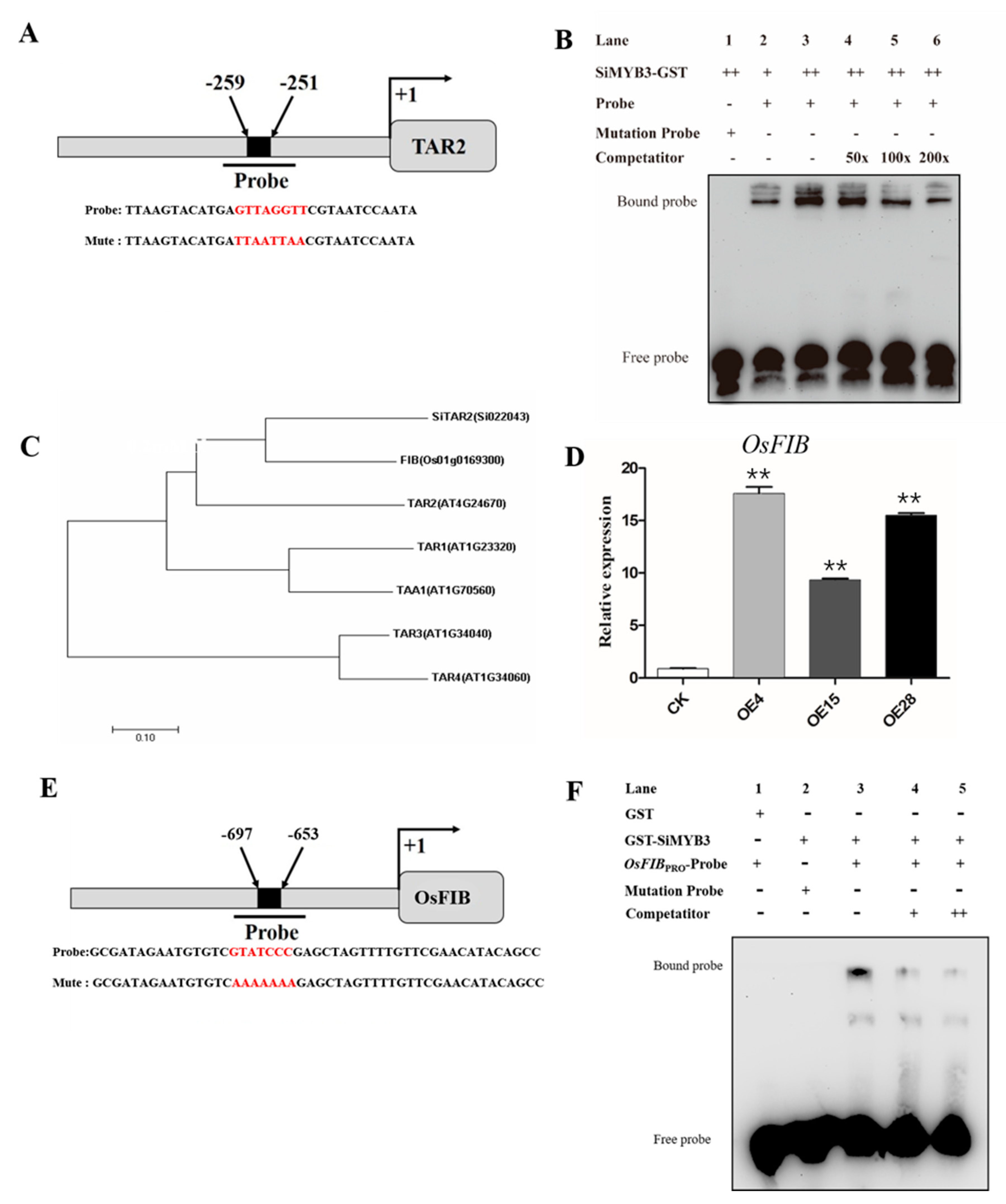
2.4. SiMYB3 Enhanced Tolerance to Low-Nitrogen Stress by Regulating Auxin Synthesis-Related Genes, TAR2, and Low-Nitrogen Stress-Related Genes
3. Discussion
4. Materials and Methods
4.1. Swiss-Prot, GO, and KEGG Pathway Annotation
4.2. Gene Structure and cis-Acting Elements
4.3. Plant Materials and Growth Conditions
4.4. Nitrogen Treatments on Media
4.5. Field Low-Nitrogen Stress Testing of Transgenic Rice
4.6. Subcellular Localization
4.7. RNA Isolation and Quantitative Real-Time RT-PCR
4.8. Protein Purification and Electrophoretic Mobility Shift Assays (EMSA)
5. Conclusions
Supplementary Materials
Availability of data and materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| GO | Gene Ontology |
| bp | Base Pair |
| cDNA | Complementary DNA |
| CK | Control Check |
| CDS | Coding Sequence |
| RT-PCR | Reverse Transcription-Polymerase Chain Reaction |
| WT | Wild Type |
| OE | Overexpression Line |
| LR | Lateral Root |
| LN | Low Nitrogen |
| LP | Low Phosphorus |
| LK | Low Potassium |
| TF | Transcription Factor |
References
- Miller, A.J.; Cramer, M.D. Root nitrogen acquisition and assimilation. Plant Soil 2005, 274, 1–36. [Google Scholar] [CrossRef]
- Gilroy, S.; Jones, D.L. Through form to function: Root hair development and nutrient uptake. Trends Plant Sci. 2000, 5, 56–60. [Google Scholar] [CrossRef]
- Strahm, B.D.; Harrison, R.B. Nitrate Sorption in A Variable-Charge Forest Soil of the Pacific Northwest. Soil Sci. 2006, 171, 313–321. [Google Scholar] [CrossRef]
- Lima, J.E.; Kojima, S.; Takahashi, H.; von Wirén, N. Ammonium triggers lateral root branching in Arabidopsis in an AMMONIUM TRANSPORTER1;3-dependent manner. Plant Cell 2010, 22, 3621–3633. [Google Scholar] [CrossRef]
- Remans, T.; Nacry, P.; Pervent, M.; Filleur, S.; Diatloff, E.; Mounier, E.; Tillard, P.; Forde, B.G.; Gojon, A. The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc. Natl. Acad. Sci. USA 1920, 103, 19206–19211. [Google Scholar] [CrossRef]
- Lynch, J. Root Architecture and Plant Productivity. Plant Physiol. 1995, 109, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Tian, Q.; Chen, F.; Zhang, F.; Mi, G. Possible Involvement of Cytokinin in Nitrate-mediated Root Growth in Maize. Plant Soil 2005, 277, 185. [Google Scholar] [CrossRef]
- Rubio, V.; Bustos, R.; Irigoyen, M.L.; Cardona-López, X.; Rojas-Triana, M.; Paz-Ares, J. Plant hormones and nutrient signaling. Plant Mol. Biol. 2008, 69, 361. [Google Scholar] [CrossRef]
- Krouk, G.; Ruffel, S.; Gutiérrez, R.A.; Gojon, A.; Crawford, N.M.; Coruzzi, G.M.; Lacombe, B. A framework integrating plant growth with hormones and nutrients. Trends Plant Sci. 2011, 16, 178–182. [Google Scholar] [CrossRef]
- Li, S.; Tian, Y.; Wu, K.; Ye, Y.; Yu, J.; Zhang, J.; Liu, Q.; Hu, M.; Li, H.; Tong, Y.; et al. Modulating plant growth-metabolism coordination for sustainable agriculture. Nature 2018, 560, 595–600. [Google Scholar] [CrossRef]
- Caba, J.M.; Centeno, M.L.; Fernandez, B.; Gresshoff, P.M.; Ligero, F. Inoculation and nitrate alter phytohormone levels in soybean roots: Differences between a supernodulating mutant and the wild type. Planta 2000, 211, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Walch-Liu, P.; Ivanov, I.I.; Filleur, S.; Gan, Y.; Remans, T.; Forde, B.G. Nitrogen regulation of root branching. Ann. Bot. 2006, 97, 875–881. [Google Scholar] [CrossRef] [PubMed]
- Tian, Q.; Chen, F.; Liu, J.; Zhang, F.; Mi, G. Inhibition of maize root growth by high nitrate supply is correlated with reduced IAA levels in roots. J. Plant Physiol. 2008, 165, 942–951. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Jennings, A.; Barlow, P.W.; Forde, B.G. Dual pathways for regulation of root branching by nitrate. Proc. Natl. Acad. Sci. USA 1999, 96, 6529–6534. [Google Scholar] [CrossRef]
- Korasick, D.A.; Enders, T.A.; Strader, L.C. Auxin biosynthesis and storage forms. J. Exp. Bot. 2013, 64, 2541–2555. [Google Scholar] [CrossRef]
- Mano, Y.; Nemoto, K. The pathway of auxin biosynthesis in plants. J. Exp. Bot. 2012, 63, 2853–2872. [Google Scholar] [CrossRef]
- Stepanova, A.N.; Robertson-Hoyt, J.; Yun, J.; Benavente, L.M.; Xie, D.Y.; Dolezal, K.; Schlereth, A.; Jurgens, G.; Alonso, J.M. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 2008, 133, 177–191. [Google Scholar] [CrossRef]
- Tao, Y.; Ferrer, J.L.; Ljung, K.; Pojer, F.; Hong, F.; Long, J.A.; Li, L.; Moreno, J.E.; Bowman, M.E.; Ivans, L.J.; et al. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 2008, 133, 164–176. [Google Scholar] [CrossRef]
- Mashiguchi, K.; Tanaka, K.; Sakai, T.; Sugawara, S.; Kawaide, H.; Natsume, M.; Hanada, A.; Yaeno, T.; Shirasu, K.; Yao, H.; et al. The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 18512–18517. [Google Scholar] [CrossRef]
- Ma, W.; Li, J.; Qu, B.; He, X.; Zhao, X.; Li, B.; Fu, X.; Tong, Y. Auxin biosynthetic gene TAR2 is involved in low nitrogen-mediated reprogramming of root architecture in Arabidopsis. Plant J. 2014, 78, 70–79. [Google Scholar] [CrossRef]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Molecular responses to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 2000, 3, 217–223. [Google Scholar] [CrossRef]
- Kasuga, M.; Liu, Q.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 1999, 17, 287. [Google Scholar] [CrossRef] [PubMed]
- Jaglo-Ottosen, K.R.; Gilmour, S.J.; Zarka, D.G.; Schabenberger, O.; Thomashow, M.F. Arabidopsis CBF1 Overexpression Induces COR Genes and Enhances Freezing Tolerance. Science 1998, 280, 104–106. [Google Scholar] [CrossRef]
- Jin, H.; Martin, C. Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol. 1999, 41, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Osbourn, A.; Ma, P. MYB Transcription Factors as Regulators of Phenylpropanoid Metabolism in Plants. Mol. Plant 2015, 8, 689–708. [Google Scholar] [CrossRef]
- Albert, N.W.; Davies, K.M.; Lewis, D.H.; Zhang, H.; Montefiori, M.; Brendolise, C.; Boase, M.R.; Ngo, H.; Jameson, P.E.; Schwinn, K.E. A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 2014, 26, 962–980. [Google Scholar] [CrossRef]
- Xie, Q.; Hu, Z.; Zhang, Y.; Tian, S.; Wang, Z.; Zhao, Z.; Yang, Y.; Chen, G. Accumulation and Molecular Regulation of Anthocyanin in Purple Tumorous Stem Mustard (Brassica juncea var. tumida Tsen et Lee). J. Agric. Food Chem. 2014, 62, 7813–7821. [Google Scholar] [CrossRef]
- Zhang, Q.; Hao, R.; Xu, Z.; Yang, W.; Wang, J.; Cheng, T.; Pan, H.; Zhang, Q. Isolation and functional characterization of a R2R3-MYB regulator of Prunus mume anthocyanin biosynthetic pathway. Plant Cell Tissue Organ Cult. 2017, 131, 417–429. [Google Scholar] [CrossRef]
- Patzlaff, A.; Newman, L.J.; Dubos, C.; Whetten, R.W.; Smith, C.; McInnis, S.; Bevan, M.W.; Sederoff, R.R.; Campbell, M.M. Characterisation of PtMYB1, an R2R3-MYB from pine xylem. Plant Mol. Biol. 2003, 53, 597–608. [Google Scholar] [CrossRef]
- Gómez-Maldonado, J.; Avila, C.; Torre, F.; Cañas, R.; Cánovas, F.M.; Campbell, M.M. Functional interactions between a glutamine synthetase promoter and MYB proteins. Plant J. 2004, 39, 513–526. [Google Scholar] [CrossRef]
- Raffaele, S.; Rivas, S.; Roby, D. An essential role for salicylic acid in AtMYB30-mediated control of the hypersensitive cell death program in Arabidopsis. FEBS Lett. 2006, 580, 3498–3504. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) Function as Transcriptional Activators in Abscisic Acid Signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef] [PubMed]
- Murray, F.; Kalla, R.; Jacobsen, J.; Gubler, F. A role for HvGAMYB in another development. Plant J. 2003, 33, 481–491. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.-W.; Qi, M.; Yang, Y. A Novel Jasmonic Acid-Inducible Rice myb Gene Associates with Fungal Infection and Host Cell Death. Mol. Plant Microbe Interact. 2001, 14, 527–535. [Google Scholar] [CrossRef] [PubMed]
- Lea, U.S.; Slimestad, R.; Smedvig, P.; Lillo, C. Nitrogen deficiency enhances expression of specific MYB and bHLH transcription factors and accumulation of end products in the flavonoid pathway. Planta 2007, 225, 1245–1253. [Google Scholar] [CrossRef] [PubMed]
- Bernhardt, C.; Lee, M.M.; Gonzalez, A.; Zhang, F.; Lloyd, A.; Schiefelbein, J. The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 2003, 130, 6431–6439. [Google Scholar] [CrossRef] [PubMed]
- Miyake, K.; Ito, T.; Senda, M.; Ishikawa, R.; Harada, T.; Niizeki, M.; Akada, S.J.P.M.B. Isolation of a subfamily of genes for R2R3-MYB transcription factors showing up-regulated expression under nitrogen nutrient-limited conditions. Plant Mol. Biol. 2003, 53, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Bender, J.; Fink, G.R. A Myb homologue, ATR1, activates tryptophan gene expression in Arabidopsis. Proc. Natl. Acad. Sci. USA 1998, 95, 5655–5660. [Google Scholar] [CrossRef]
- Yang, X.; Wan, Z.; Perry, L.; Lu, H.; Wang, Q.; Zhao, C.; Li, J.; Xie, F.; Yu, J.; Cui, T.; et al. Early millet use in northern China. Proc. Natl. Acad. Sci. USA 2012, 109, 3726–3730. [Google Scholar] [CrossRef]
- Nadeem, F.; Ahmad, Z.; Wang, R.; Han, J.; Shen, Q.; Chang, F.; Diao, X.; Zhang, F.; Li, X. Foxtail Millet [Setaria italica (L.) Beauv.] Grown under Low Nitrogen Shows a Smaller Root System, Enhanced Biomass Accumulation, and Nitrate Transporter Expression. Front. Plant Sci. 2018, 9, 205. [Google Scholar] [CrossRef]
- Bennetzen, J.L.; Schmutz, J.; Wang, H.; Percifield, R.; Hawkins, J.; Pontaroli, A.C.; Estep, M.; Feng, L.; Vaughn, J.N.; Grimwood, J.; et al. Reference genome sequence of the model plant Setaria. Nat. Biotechnol. 2012, 30, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Liu, X.; Quan, Z.; Cheng, S.; Xu, X.; Pan, S.; Xie, M.; Zeng, P.; Yue, Z.; Wang, W.; et al. Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nat. Biotechnol. 2012, 30, 549–554. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Huang, X.; Zhi, H.; Zhao, Y.; Zhao, Q.; Li, W.; Chai, Y.; Yang, L.; Liu, K.; Lu, H.; et al. A haplotype map of genomic variations and genome-wide association studies of agronomic traits in foxtail millet (Setaria italica). Nat. Genet. 2013, 45, 957–961. [Google Scholar] [CrossRef] [PubMed]
- Lata, C.; Gupta, S.; Prasad, M. Foxtail millet: A model crop for genetic and genomic studies in bioenergy grasses. Crit. Rev. Biotechnol. 2013, 33, 328–343. [Google Scholar] [CrossRef] [PubMed]
- Haling, R.E.; Brown, L.K.; Bengough, A.G.; Valentine, T.A.; White, P.J.; Young, I.M.; George, T.S. Root hair length and rhizosheath mass depend on soil porosity, strength and water content in barley genotypes. Planta 2014, 239, 643–651. [Google Scholar] [CrossRef]
- Qi, X.; Xie, S.; Liu, Y.; Yi, F.; Yu, J. Genome-wide annotation of genes and noncoding RNAs of foxtail millet in response to simulated drought stress by deep sequencing. Plant Mol. Biol. 2013, 83, 459–473. [Google Scholar] [CrossRef]
- Muthamilarasan, M.; Khandelwal, R.; Yadav, C.B.; Bonthala, V.S.; Khan, Y.; Prasad, M. Identification and molecular characterization of MYB Transcription Factor Superfamily in C4 model plant foxtail millet (Setaria italica L.). PLoS ONE 2014, 3, e109920. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Abu-Zaitoon, Y.M.; Bennett, K.; Normanly, J.; Nonhebel, H.M. A large increase in IAA during development of rice grains correlates with the expression of tryptophan aminotransferase OsTAR1 and a grain-specific YUCCA. Physiol. Plant. 2012, 146, 487–499. [Google Scholar] [CrossRef]
- Scheible, W.R.; Morcuende, R.; Czechowski, T.; Fritz, C.; Osuna, D.; Palacios-Rojas, N.; Schindelasch, D.; Thimm, O.; Udvardi, M.K.; Stitt, M. Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol. 2004, 136, 2483–2499. [Google Scholar] [CrossRef]
- Liu, J.; Moore, S.; Chen, C.; Lindsey, K. Crosstalk Complexities between Auxin, Cytokinin, and Ethylene in Arabidopsis Root Development: From Experiments to Systems Modeling, and Back Again. Mol. Plant 2017, 10, 1480–1496. [Google Scholar] [CrossRef] [PubMed]
- Tsay, Y.F.; Schroeder, J.I.; Feldmann, K.A.; Crawford, N.M. The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 1993, 72, 705–713. [Google Scholar] [CrossRef]
- Bouguyon, E.; Perrine-Walker, F.; Pervent, M.; Rochette, J.; Cuesta, C.; Benkova, E.; Martiniere, A.; Bach, L.; Krouk, G.; Gojon, A.; et al. Nitrate Controls Root Development through Posttranscriptional Regulation of the NRT1.1/NPF6.3 Transporter/Sensor. Plant Physiol. 2016, 172, 1237–1248. [Google Scholar]
- Bouguyon, E.; Brun, F.; Meynard, D.; Kubes, M.; Pervent, M.; Leran, S.; Lacombe, B.; Krouk, G.; Guiderdoni, E.; Zazimalova, E.; et al. Multiple mechanisms of nitrate sensing by Arabidopsis nitrate transceptor NRT1.1. Nat. Plants 2015, 1, 15015. [Google Scholar] [CrossRef]
- Knudsen, S. Promoter2.0: For the recognition of PolII promoter sequences. Bioinformatics 1999, 15, 356–361. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Cui, X.Y.; Du, Y.T.; Fu, J.D.; Yu, T.F.; Wang, C.T.; Chen, M.; Chen, J.; Ma, Y.Z.; Xu, Z.S. Wheat CBL-interacting protein kinase 23 positively regulates drought stress and ABA responses. BMC Plant Biol. 2018, 18, 93. [Google Scholar] [CrossRef]
- Bechtold, N.; Pelletier, G. In Planta AgrobacteriumMediated Transformation of Adult Arabidopsis thaliana Plants by Vacuum Infiltration. Methods Mol. Biol. 1998, 82, 259–266. [Google Scholar]
- Asai, T.; Tena, G.; Plotnikova, J.; Willmann, M.R.; Chiu, W.L.; Gomez-Gomez, L.; Boller, T.; Ausubel, F.M.; Sheen, J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 2002, 415, 977–983. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ge, L.; Dou, Y.; Li, M.; Qu, P.; He, Z.; Liu, Y.; Xu, Z.; Chen, J.; Chen, M.; Ma, Y. SiMYB3 in Foxtail Millet (Setaria italica) Confers Tolerance to Low-Nitrogen Stress by Regulating Root Growth in Transgenic Plants. Int. J. Mol. Sci. 2019, 20, 5741. https://doi.org/10.3390/ijms20225741
Ge L, Dou Y, Li M, Qu P, He Z, Liu Y, Xu Z, Chen J, Chen M, Ma Y. SiMYB3 in Foxtail Millet (Setaria italica) Confers Tolerance to Low-Nitrogen Stress by Regulating Root Growth in Transgenic Plants. International Journal of Molecular Sciences. 2019; 20(22):5741. https://doi.org/10.3390/ijms20225741
Chicago/Turabian StyleGe, Linhao, Yining Dou, Maomao Li, Pengju Qu, Zhang He, Y Liu, Zhaoshi Xu, Jun Chen, Ming Chen, and Youzhi Ma. 2019. "SiMYB3 in Foxtail Millet (Setaria italica) Confers Tolerance to Low-Nitrogen Stress by Regulating Root Growth in Transgenic Plants" International Journal of Molecular Sciences 20, no. 22: 5741. https://doi.org/10.3390/ijms20225741
APA StyleGe, L., Dou, Y., Li, M., Qu, P., He, Z., Liu, Y., Xu, Z., Chen, J., Chen, M., & Ma, Y. (2019). SiMYB3 in Foxtail Millet (Setaria italica) Confers Tolerance to Low-Nitrogen Stress by Regulating Root Growth in Transgenic Plants. International Journal of Molecular Sciences, 20(22), 5741. https://doi.org/10.3390/ijms20225741






