Carboxypeptidase E-∆N Promotes Proliferation and Invasion of Pancreatic Cancer Cells via Upregulation of CXCR2 Gene Expression
Abstract
:1. Introduction
2. Results
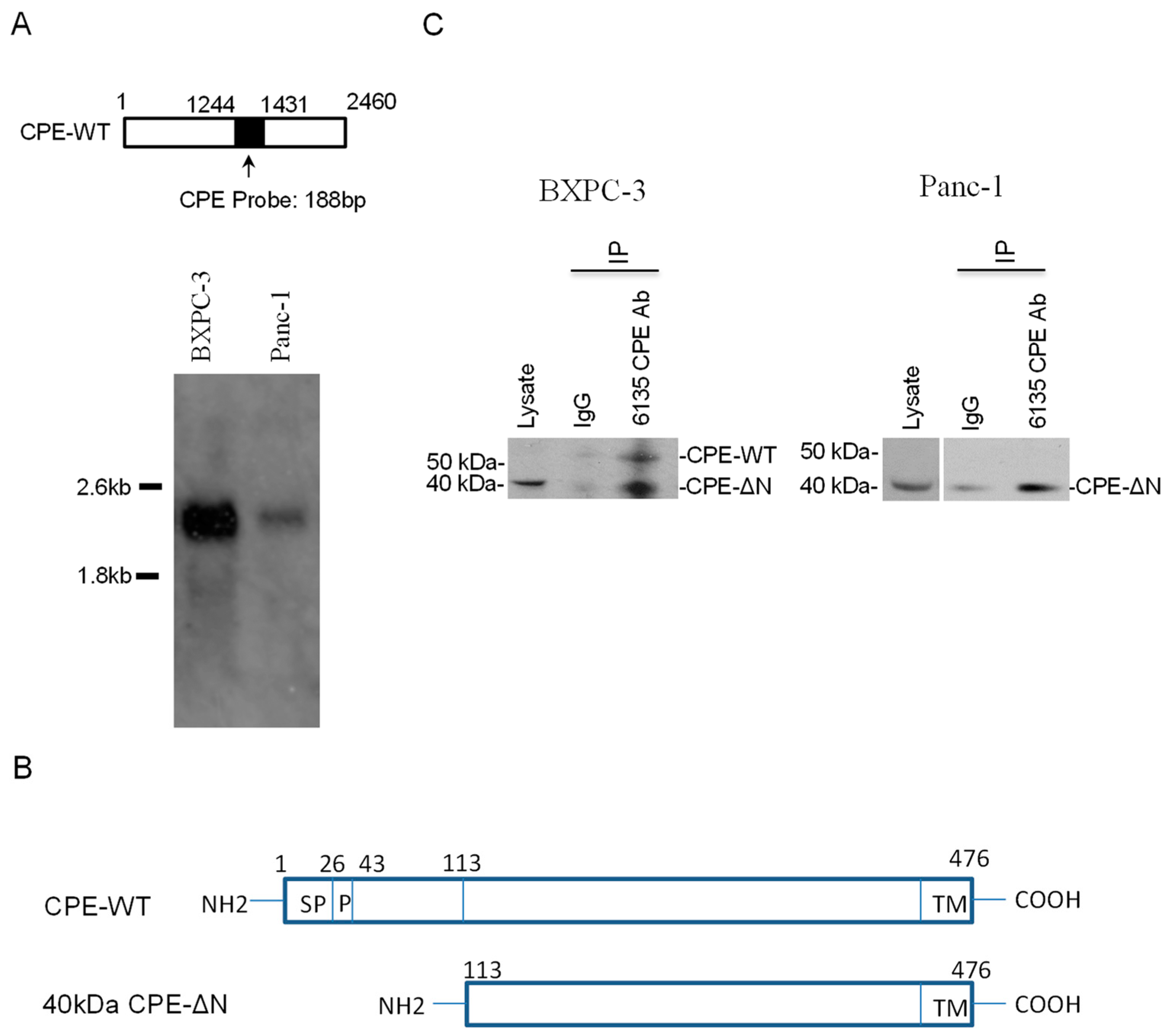
2.1. CPE Transcripts and Proteins Expressed in Human Pancreatic Cancer Cell Lines
2.2. Subcellular Distribution of CPE and CPE-ΔN in Panc-1 Cells
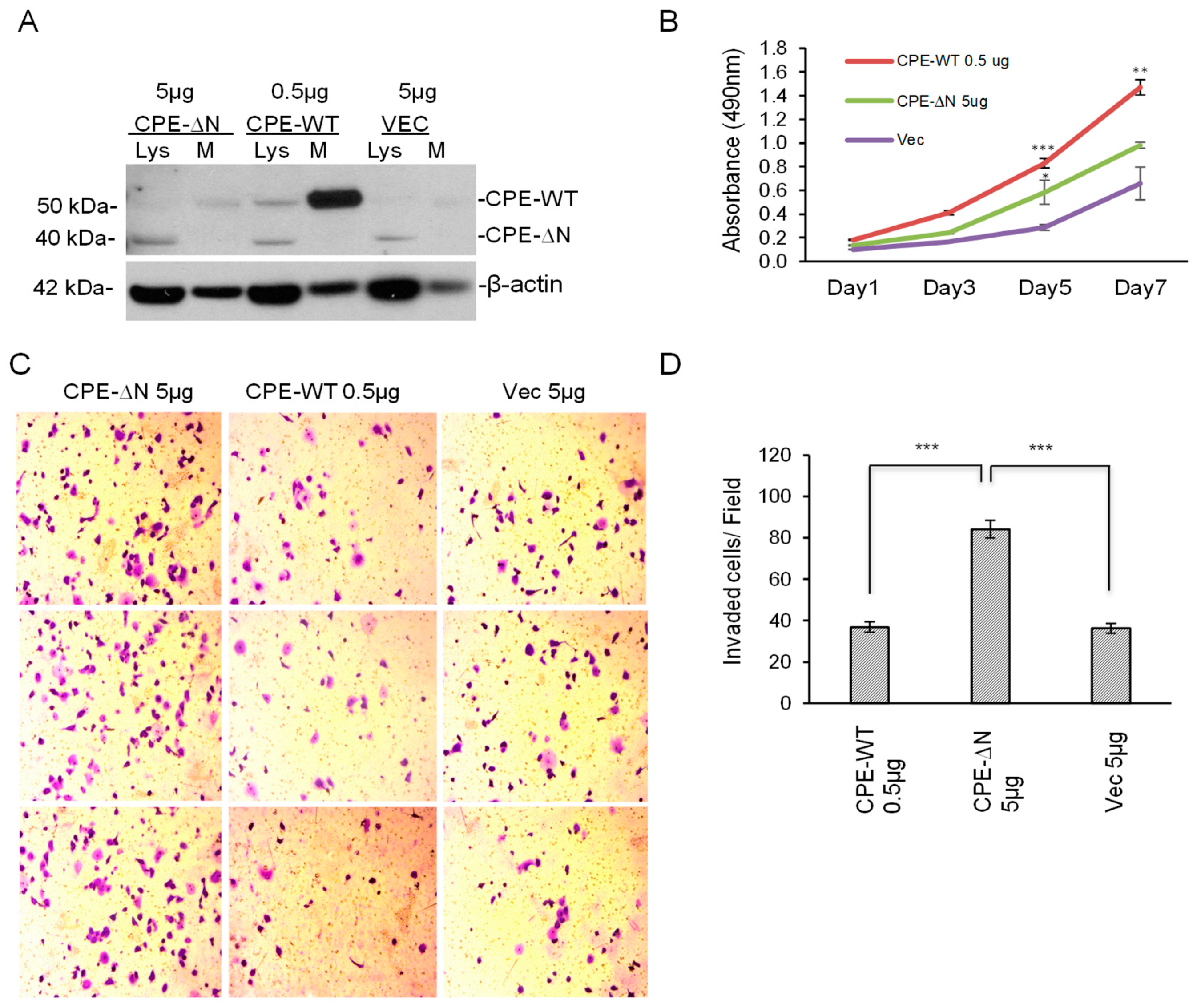
2.3. Overexpression of CPE-ΔN in Panc-1 Cells Increases Proliferation and Invasion
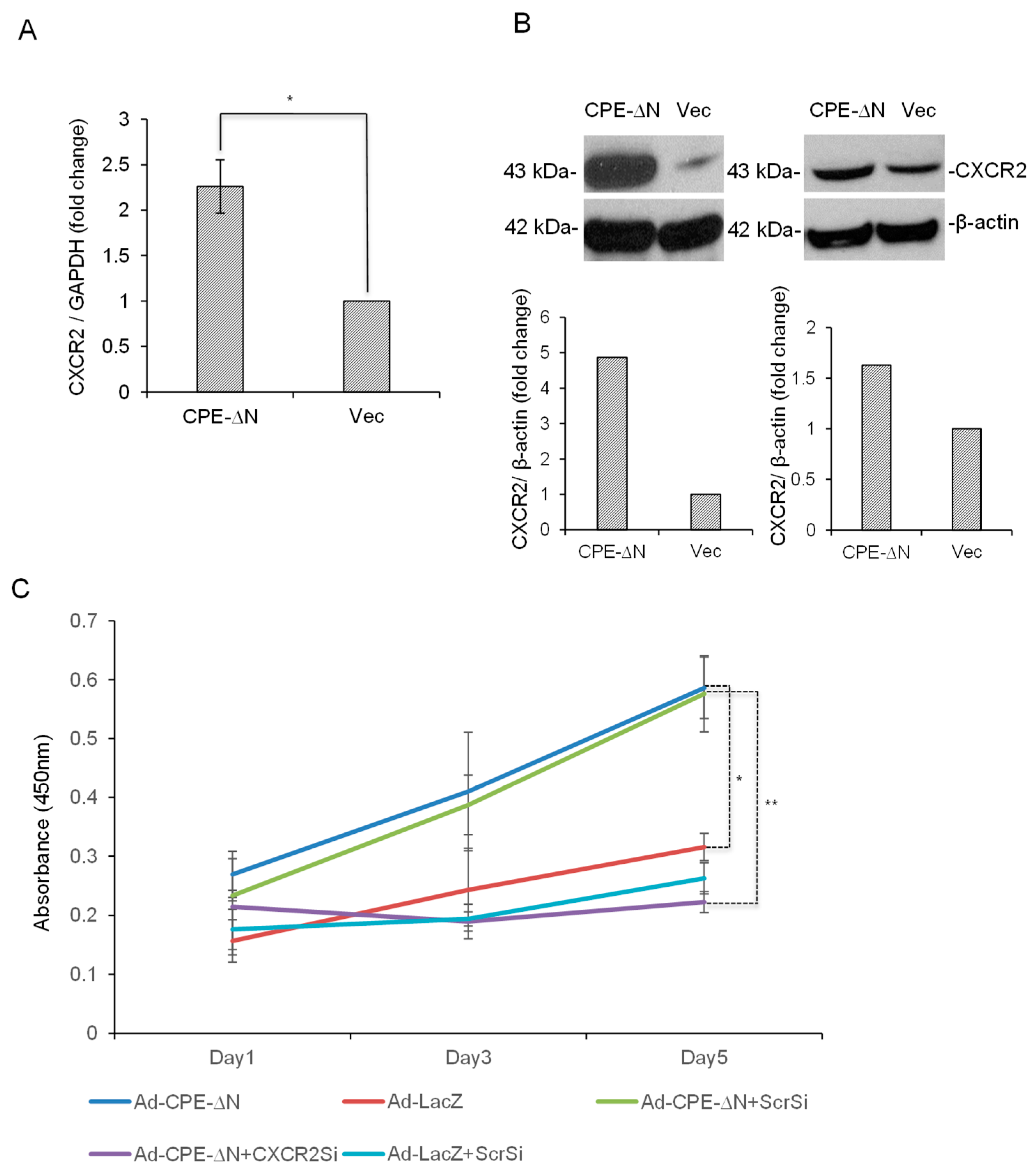
2.4. CPE-ΔN Upregulates CXCR2 Expression in Panc-1 Cells
2.5. CPE-ΔN Induces Proliferation of Panc-1 Cells in a CXCR2-Dependent Manner
3. Discussion
4. Materials and Methods
4.1. Cell Lines
4.2. Northern Blot
4.3. Transfection of Cells with CPE-ΔN and CPE-WT Constructs
4.4. Immunoprecipitation of CPE/CPE-ΔN
4.5. Western Blot
4.6. Immunocytochemistry
4.7. Quantitative Real-Time RT-PCR
4.8. MTT Cell Proliferation Assay
4.9. Invasion Assay
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CPE | Carboxypeptidase E |
| PC | Pancreatic cancer |
| HCC | Hepatocellular carcinoma |
| WT | Wild-type |
| PDAC | Pancreatic ductal adenocarcinoma |
| OS | Osteosarcoma |
References
- Khan, M.A.; Azim, S.; Zubair, H.; Bhardwaj, A.; Patel, G.K.; Khushman, M.; Singh, S.; Singh, A.P. Molecular Drivers of Pancreatic Cancer Pathogenesis: Looking Inward to Move Forward. Int. J. Mol. Sci. 2017, 18, 779–804. [Google Scholar] [CrossRef] [PubMed]
- Hines, O.J.; Reber, H.A. Pancreatic surgery. Curr. Opin. Gastroenterol. 2008, 24, 603–611. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Cawley, N.X.; Loh, Y.P. Carboxypeptidase E (NF-alpha1): A new trophic factor in neuroprotection. Neurosci. Bull. 2014, 30, 692–696. [Google Scholar] [CrossRef] [PubMed]
- Selvaraj, P.; Xiao, L.; Lee, C.; Murthy, S.R.; Cawley, N.X.; Lane, M.; Merchenthaler, I.; Ahn, S.; Loh, Y.P. Neurotrophic Factor-alpha1: A Key Wnt-beta-Catenin Dependent Anti-Proliferation Factor and ERK-Sox9 Activated Inducer of Embryonic Neural Stem Cell Differentiation to Astrocytes in Neurodevelopment. Stem Cells 2017, 35, 557–571. [Google Scholar] [CrossRef] [PubMed]
- Armento, A.; Ilina, E.I.; Kaoma, T.; Muller, A.; Vallar, L.; Niclou, S.P.; Kruger, M.A.; Mittelbronn, M.; Naumann, U. Carboxypeptidase E transmits its anti-migratory function in glioma cells via transcriptional regulation of cell architecture and motility regulating factors. Int. J. Oncol. 2017, 51, 702–714. [Google Scholar] [CrossRef] [PubMed]
- Cawley, N.X.; Wetsel, W.C.; Murthy, S.R.; Park, J.J.; Pacak, K.; Loh, Y.P. New roles of carboxypeptidase E in endocrine and neural function and cancer. Endocr. Rev. 2012, 33, 216–253. [Google Scholar] [CrossRef]
- Murthy, S.R.K.; Dupart, E.; Al-Sweel, N.; Chen, A.; Cawley, N.X.; Loh, Y.P. Carboxypeptidase E promotes cancer cell survival, but inhibits migration and invasion. Cancer Lett. 2013, 341, 204–213. [Google Scholar] [CrossRef]
- Liang, X.H.; Li, L.L.; Wu, G.G.; Xie, Y.C.; Zhang, G.X.; Chen, W.; Yang, H.F.; Liu, Q.L.; Li, W.H.; He, W.G.; et al. Upregulation of CPE promotes cell proliferation and tumorigenicity in colorectal cancer. BMC Cancer 2013, 13, 412. [Google Scholar] [CrossRef]
- Huang, S.F.; Wu, H.D.; Chen, Y.T.; Murthy, S.R.; Chiu, Y.T.; Chang, Y.; Chang, I.C.; Yang, X.; Loh, Y.P. Carboxypeptidase E is a prediction marker for tumor recurrence in early-stage hepatocellular carcinoma. Tumour Biol. 2016, 37, 9745–9753. [Google Scholar] [CrossRef]
- Shen, H.W.; Tan, J.F.; Shang, J.H.; Hou, M.Z.; Liu, J.; He, L.; Yao, S.Z.; He, S.Y. CPE overexpression is correlated with pelvic lymph node metastasis and poor prognosis in patients with early-stage cervical cancer. Arch. Gynecol. Obstet. 2016, 294, 333–342. [Google Scholar] [CrossRef]
- Wang, X.C.; Xu, S.Y.; Wu, X.Y.; Song, H.D.; Mao, Y.F.; Fan, H.Y.; Yu, F.; Mou, B.; Gu, Y.Y.; Xu, L.Q.; et al. Gene expression profiling in human insulinoma tissue: Genes involved in the insulin secretion pathway and cloning of novel full-length cDNAs. Endocr. Relat. Cancer. 2004, 11, 295–303. [Google Scholar] [CrossRef] [PubMed]
- He, P.; Varticovski, L.; Bowman, E.D.; Fukuoka, J.; Welsh, J.A.; Miura, K.; Jen, J.; Gabrielson, E.; Brambilla, E.; Travis, W.D.; et al. Identification of carboxypeptidase E and gamma-glutamyl hydrolase as biomarkers for pulmonary neuroendocrine tumors by cDNA microarray. Hum. Pathol. 2004, 35, 1196–1209. [Google Scholar] [CrossRef] [PubMed]
- Grimwood, B.G.; Plummer, T.H., Jr.; Tarentino, A.L.; Carboxypeptidase, H. A regulatory peptide-processing enzyme produced by human hepatoma Hep G2 cells. J. Biol. Chem. 1989, 264, 15662–15667. [Google Scholar] [PubMed]
- Manser, E.; Fernandez, D.; Lim, L. Processing and secretion of human carboxypeptidase E by C6 glioma cells. Biochem. J. 1991, 280 Pt 3, 695–701. [Google Scholar] [CrossRef]
- Horing, E.; Harter, P.N.; Seznec, J.; Schittenhelm, J.; Buhring, H.J.; Bhattacharyya, S.; von Hattingen, E.; Zachskorn, C.; Mittelbronn, M.; Naumann, U. The “go or grow” potential of gliomas is linked to the neuropeptide processing enzyme carboxypeptidase E and mediated by metabolic stress. Acta Neuropathol. 2012, 124, 83–97. [Google Scholar] [CrossRef]
- Purohit, A.; Varney, M.; Rachagani, S.; Ouellette, M.M.; Batra, S.K.; Singh, R.K. CXCR2 signaling regulates KRAS(G(1)(2)D)—Induced autocrine growth of pancreatic cancer. Oncotarget 2016, 7, 7280–7296. [Google Scholar] [CrossRef]
- Steele, C.W.; Karim, S.A.; Leach, J.D.G.; Bailey, P.; Upstill-Goddard, R.; Rishi, L.; Foth, M.; Bryson, S.; McDaid, K.; Wilson, Z.; et al. CXCR2 Inhibition Profoundly Suppresses Metastases and Augments Immunotherapy in Pancreatic Ductal Adenocarcinoma. Cancer Cell 2016, 29, 832–845. [Google Scholar] [CrossRef]
- Yang, L.; Huang, J.; Ren, X.; Gorska, A.E.; Chytil, A.; Aakre, M.; Carbone, D.P.; Matrisian, L.M.; Richmond, A.; Lin, P.C.; et al. Abrogation of TGF beta signaling in mammary carcinomas recruits Gr-1+CD11b+ myeloid cells that promote metastasis. Cancer Cell 2008, 13, 23–35. [Google Scholar] [CrossRef]
- Yang, X.; Lou, L.; Chen, Y.; Huang, S.; Loh, Y.P. A novel 40kDa N-terminal truncated Carboxypeptidase E splice variant: Cloning, cDNA sequence analysis and role in regulation of metastatic genes in human cancers. Genes Cancer 2019. [Google Scholar] [CrossRef]
- Sun, J.; Meng, D.; Li, L.; Tian, X.; Jia, Y.; Wang, H.; Yu, H.; Sun, T.; Qu, A.; Shen, H.; et al. N-terminal truncated carboxypeptidase E expression is associated with poor prognosis of lung adenocarcinoma. Oncol. Lett. 2016, 12, 4659–4664. [Google Scholar] [CrossRef]
- Fan, S.; Li, X.; Li, L.; Wang, L.; Du, Z.; Yang, Y.; Zhao, J.; Li, Y. Silencing of carboxypeptidase E inhibits cell proliferation, tumorigenicity, and metastasis of osteosarcoma cells. Onco Targets Ther. 2016, 9, 2795–2803. [Google Scholar] [PubMed]
- Liu, A.; Shao, C.; Jin, G.; Liu, R.; Hao, J.; Shao, Z.; Liu, Q.; Hu, X. Downregulation of CPE regulates cell proliferation and chemosensitivity in pancreatic cancer. Tumour Biol. 2014, 35, 12459–12465. [Google Scholar] [CrossRef] [PubMed]
- Oldfield, L.E.; Connor, A.A.; Gallinger, S. Molecular Events in the Natural History of Pancreatic Cancer. Trends Cancer 2017, 3, 336–346. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Zhao, B.; Liu, Z.; Ren, X.; Zhao, W.; Li, Z.; You, L.; Zhao, Y. Molecular Subtyping of Pancreatic Cancer: Translating Genomics and Transcriptomics into the Clinic. J. Cancer 2017, 8, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Jamieson, T.; Clarke, M.; Steele, C.W.; Samuel, M.S.; Neumann, J.; Jung, A.; Huels, D.; Olson, M.F.; Das, S.; Nibbs, R.J.; et al. Inhibition of CXCR2 profoundly suppresses inflammation-driven and spontaneous tumorigenesis. J. Clin. Investig. 2012, 122, 3127–3144. [Google Scholar] [CrossRef]
- Arumugam, T.; Ramachandran, V.; Fournier, K.F.; Wang, H.; Marquis, L.; Abbruzzese, J.L.; Gallick, G.E.; Logsdon, C.D.; McConkey, D.J.; Choi, W. Epithelial to mesenchymal transition contributes to drug resistance in pancreatic cancer. Cancer Res. 2009, 69, 5820–5828. [Google Scholar] [CrossRef]
- Gaianigo, N.; Melisi, D.; Carbone, C. EMT and Treatment Resistance in Pancreatic Cancer. Cancers (Basel) 2017, 9, 122. [Google Scholar] [CrossRef]
- Ying, H.; Dey, P.; Yao, W.; Kimmelman, A.C.; Draetta, G.F.; Maitra, A.; DePinho, R.A. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2016, 30, 355–385. [Google Scholar] [CrossRef]
- Melisi, D.; Ishiyama, S.; Sclabas, G.M.; Fleming, J.B.; Xia, Q.; Tortora, G.; Abbruzzese, J.L.; Chiao, P.J. LY2109761, a novel transforming growth factor beta receptor type I and type II dual inhibitor, as a therapeutic approach to suppressing pancreatic cancer metastasis. Mol. Cancer Ther 2008, 7, 829–840. [Google Scholar] [CrossRef]
- Filios, S.R.; Xu, G.; Chen, J.; Hong, K.; Jing, G.; Shalev, A. MicroRNA-200 is induced by thioredoxin-interacting protein and regulates Zeb1 protein signaling and beta cell apoptosis. J. Biol. Chem 2014, 289, 36275–36283. [Google Scholar] [CrossRef]
- Fan, S.; Gao, X.; Chen, P.; Li, X. Carboxypeptidase E-DeltaN promotes migration, invasiveness, and epithelial-mesenchymal transition of human osteosarcoma cells via the Wnt-beta-catenin pathway. Biochem. Cell Biol. 2019, 97, 446–453. [Google Scholar] [CrossRef] [PubMed]
- Waters, A.M.; Der, C.J. KRAS: The Critical Driver and Therapeutic Target for Pancreatic Cancer. Cold Spring Harb. Perspect. Med. 2018, 8, a031435. [Google Scholar] [CrossRef] [PubMed]
- Kanda, M.; Matthaei, H.; Wu, J.; Hong, S.M.; Yu, J.; Borges, M.; Hruban, R.H.; Maitra, A.; Kinzler, K.; Vogelstein, B.; et al. Presence of somatic mutations in most early-stage pancreatic intraepithelial neoplasia. Gastroenterology 2012, 142, 730–733, 739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, J.; Cox, A.D.; Li, J.; McCready, W.; Ulanova, M. Activation of innate immune responses by Haemophilus influenzae lipooligosaccharide. Clin. Vaccine Immunol. 2014, 21, 769–776. [Google Scholar] [CrossRef] [Green Version]
- Xiao, L.; Yang, X.; Sharma, V.K.; Loh, Y.P. Cloning, gene regulation, and neuronal proliferation functions of novel N-terminal-truncated carboxypeptidase E/neurotrophic factor-alphal variants in embryonic mouse brain. FASEB J. 2019, 33, 808–820. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hareendran, S.; Yang, X.; Lou, H.; Xiao, L.; Loh, Y.P. Carboxypeptidase E-∆N Promotes Proliferation and Invasion of Pancreatic Cancer Cells via Upregulation of CXCR2 Gene Expression. Int. J. Mol. Sci. 2019, 20, 5725. https://doi.org/10.3390/ijms20225725
Hareendran S, Yang X, Lou H, Xiao L, Loh YP. Carboxypeptidase E-∆N Promotes Proliferation and Invasion of Pancreatic Cancer Cells via Upregulation of CXCR2 Gene Expression. International Journal of Molecular Sciences. 2019; 20(22):5725. https://doi.org/10.3390/ijms20225725
Chicago/Turabian StyleHareendran, Sangeetha, Xuyu Yang, Hong Lou, Lan Xiao, and Y. Peng Loh. 2019. "Carboxypeptidase E-∆N Promotes Proliferation and Invasion of Pancreatic Cancer Cells via Upregulation of CXCR2 Gene Expression" International Journal of Molecular Sciences 20, no. 22: 5725. https://doi.org/10.3390/ijms20225725
APA StyleHareendran, S., Yang, X., Lou, H., Xiao, L., & Loh, Y. P. (2019). Carboxypeptidase E-∆N Promotes Proliferation and Invasion of Pancreatic Cancer Cells via Upregulation of CXCR2 Gene Expression. International Journal of Molecular Sciences, 20(22), 5725. https://doi.org/10.3390/ijms20225725




