Conditional Deletion of Dicer in Adult Mice Impairs Skeletal Muscle Regeneration
Abstract
1. Introduction
2. Results
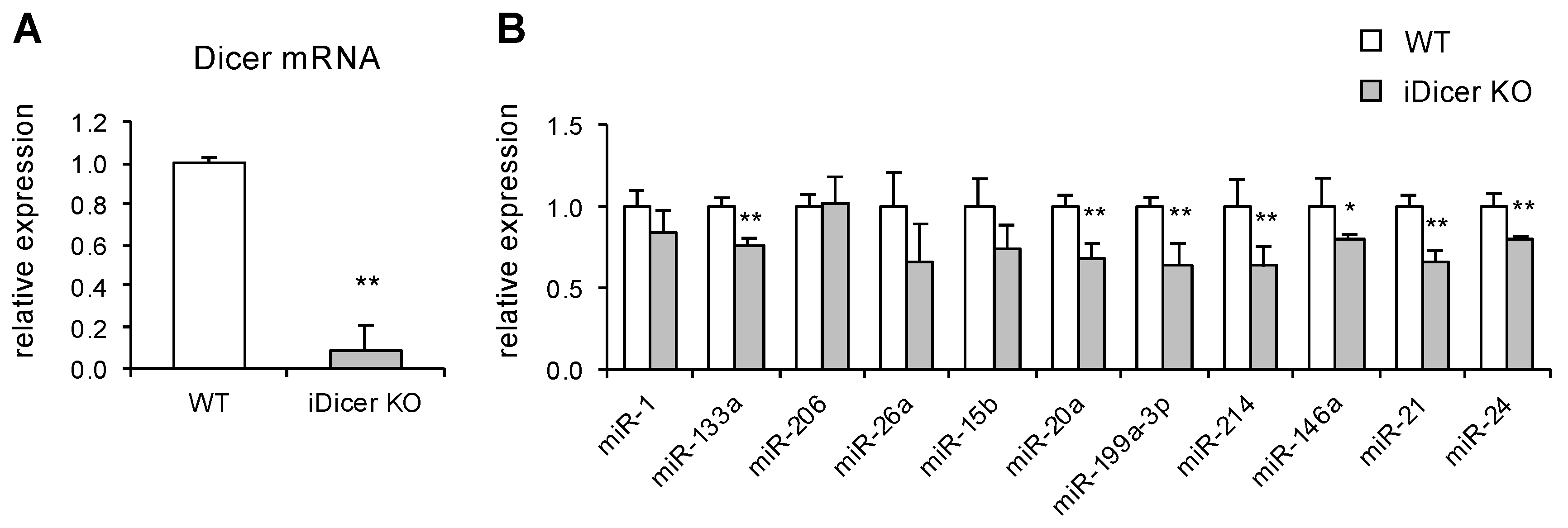
2.1. Expression of Dicer and miRNAs in iDicer KO Mice
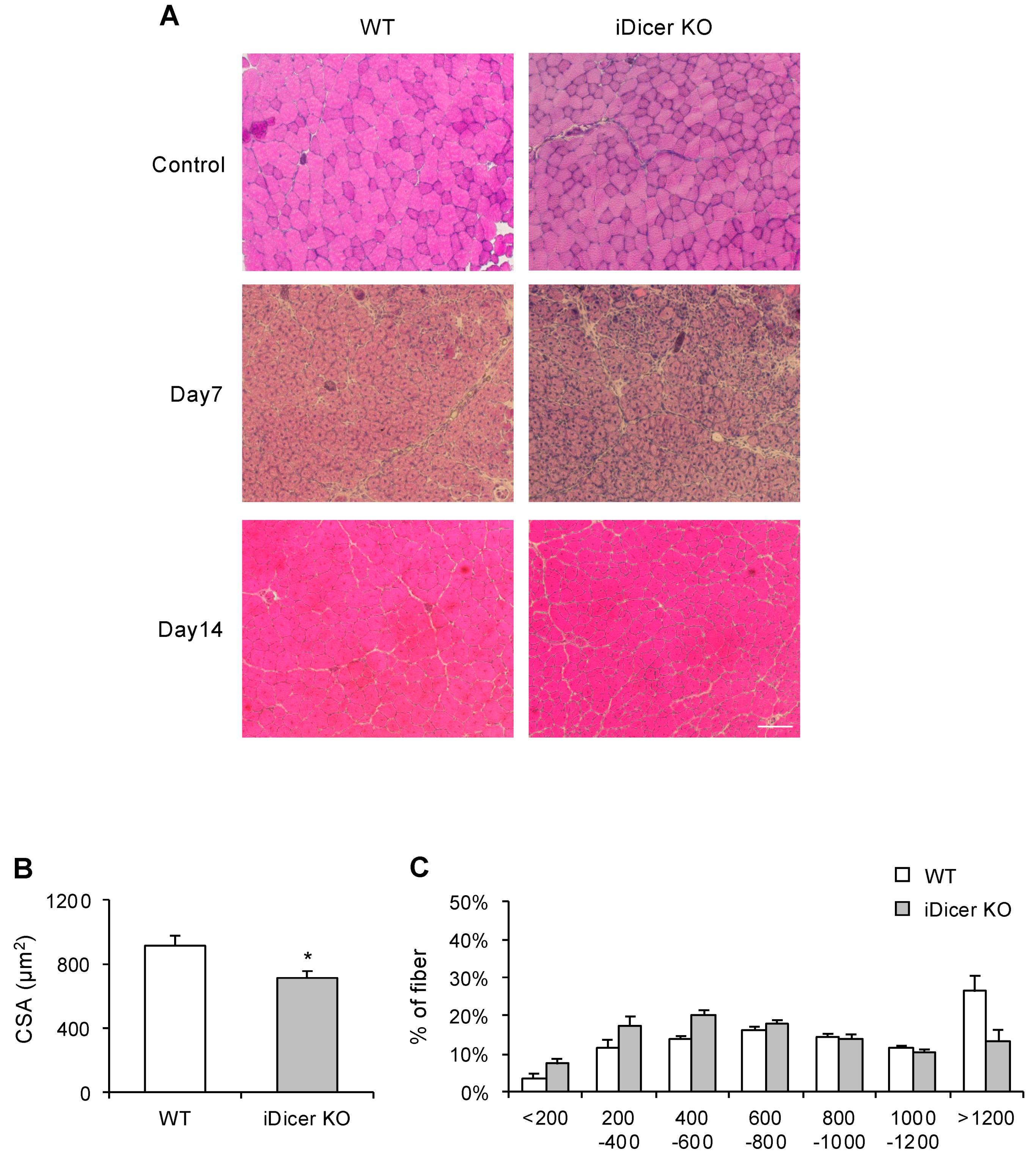
2.2. Skeletal Muscle Regeneration Is Impaired in iDicer KO Mice
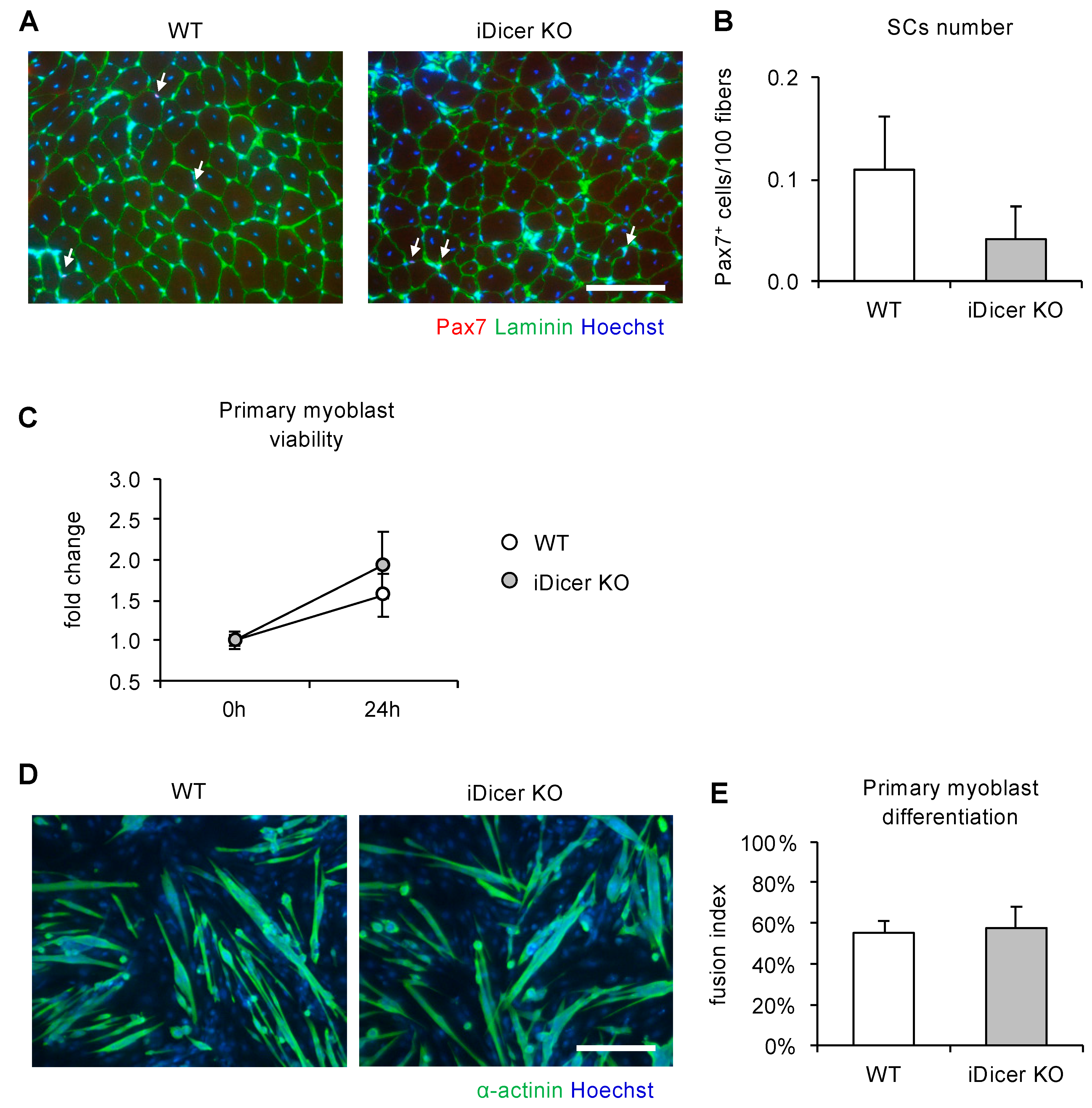
2.3. Inducible Knockout of Dicer Does Not Affect Cell Proliferation or Differentiation of Primary Myoblasts
3. Discussion
4. Materials and Methods
4.1. Animal Experiments
4.2. qPCR
4.3. Histological Analysis
4.4. Isolation and Culture of Primary Myoblasts
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| SC | Satellite cell |
| miRNA | microRNA |
| TA | Tibialis anterior |
| WT | Wild-type |
| CSA | Cross-sectional area |
| CTX | Cardiotoxin |
References
- Chargé, S.B.; Rudnicki, M.A. Cellular and molecular regulation of muscle regeneration. Physiol. Rev. 2004, 84, 209–238. [Google Scholar] [CrossRef] [PubMed]
- Pietsch, P. The effects of colchicine on regeneration of mouse skeletal muscle. Anat. Rec. 1961, 139, 167–172. [Google Scholar] [CrossRef]
- Oustanina, S.; Hause, G.; Braun, T. Pax7 directs postnatal renewal and propagation of myogenic satellite cells but not their specification. Embo. J. 2004, 23, 3430–3439. [Google Scholar] [CrossRef] [PubMed]
- Seale, P.; Sabourin, L.A.; Girgis-Gabardo, A.; Mansouri, A.; Gruss, P.; Rudnicki, M.A. Pax7 is Required for the Specification of Myogenic Satellite Cells. Cell 2000, 102, 777–786. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Ha, M.; Kim, N.V. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Chen, J.-F.; Mandel, E.M.; Thomson, M.J.; Wu, Q.; Callis, T.E.; Hammond, S.M.; Conlon, F.L.; Wang, D.-Z. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2005, 38, 228–233. [Google Scholar] [CrossRef]
- Kim, H.; Lee, Y.; Sivaprasad, U.; Malhotra, A.; Dutta, A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J. Cell Biol. 2006, 174, 677–687. [Google Scholar] [CrossRef]
- Chen, J.-F.; Tao, Y.; Li, J.; Deng, Z.; Yan, Z.; Xiao, X.; Wang, D.-Z. microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J. Cell Biol. 2010, 190, 867–879. [Google Scholar] [CrossRef]
- Wong, C.; Tellam, R.L. MicroRNA-26a Targets the Histone Methyltransferase Enhancer of Zeste homolog 2 during Myogenesis. J. Biol. Chem. 2008, 283, 9836–9843. [Google Scholar] [CrossRef]
- Silva, W.; Graça, F.; Cruz, A.; Silvestre, J.; Labeit, S.; Miyabara, E.; Yan, C.; Wang, D.; Moriscot, A. miR-29c improves skeletal muscle mass and function throughout myocyte proliferation and differentiation and by repressing atrophy-related genes. Acta Physiol. 2019, 226, e13278. [Google Scholar] [CrossRef] [PubMed]
- O’Rourke, J.; Georges, S.; Seay, H.; Tapscott, S.; McManus, M.; Goldhamer, D.; Swanson, M.; Harfe, B. Essential role for Dicer during skeletal muscle development. Dev. Biol. 2007, 311, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Sempere, L.F.; Freemantle, S.; Pitha-Rowe, I.; Moss, E.; Dmitrovsky, E.; Ambros, V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004, 5, R13. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Williams, A.H.; Kim, Y.; McAnally, J.; Bezprozvannaya, S.; Sutherland, L.B.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc. Natl. Acad. Sci. USA 2007, 104, 20844–20849. [Google Scholar] [CrossRef]
- Rao, P.K.; Kumar, R.M.; Farkhondeh, M.; Baskerville, S.; Lodish, H.F. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc. Natl. Acad. Sci. USA 2006, 103, 8721–8726. [Google Scholar] [CrossRef]
- Sweetman, D.; Goljanek, K.; Rathjen, T.; Oustanina, S.; Braun, T.; Dalmay, T.; Munsterberg, A. Specific requirements of MRFs for the expression of muscle specific microRNAs, miR-1, miR-206 and miR-133. Dev. Biol. 2008, 321, 491–499. [Google Scholar] [CrossRef]
- Koutsoulidou, A.; Mastroyiannopoulos, N.P.; Furling, D.; Uney, J.B.; Phylactou, L.A. Expression of miR-1, miR-133a, miR-133b and miR-206 increases during development of human skeletal muscle. BMC Dev. Biol. 2011, 11, 34. [Google Scholar] [CrossRef]
- Hirai, H.; Verma, M.; Watanabe, S.; Tastad, C.; Asakura, Y.; Asakura, A. MyoD regulates apoptosis of myoblasts through microRNA-mediated down-regulation of Pax3. J. Cell Biol. 2010, 191, 347–365. [Google Scholar] [CrossRef]
- Oikawa, S.; Lee, M.; Motohashi, N.; Maeda, S.; Akimoto, T. An inducible knockout of Dicer in adult mice does not affect endurance exercise-induced muscle adaptation. Am. J. Physiol. Cell Physiol. 2019, 316, C285–C292. [Google Scholar] [CrossRef]
- Snyder, C.M.; Rice, A.L.; Estrella, N.L.; Held, A.; Kandarian, S.C.; Naya, F.J. MEF2A regulates the Gtl2-Dio3 microRNA mega-cluster to modulate WNT signaling in skeletal muscle regeneration. Development 2013, 140, 31–42. [Google Scholar] [CrossRef]
- Dey, B.K.; Gagan, J.; Yan, Z.; Dutta, A. miR-26a is required for skeletal muscle differentiation and regeneration in mice. Genes Dev. 2012, 26, 2180–2191. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.-P.; Shin, Y.; Panda, A.C.; Abdelmohsen, K.; Kim, J.; Lee, S.-M.; Bahn, Y.; Choi, J.; Kwon, E.-S.; Baek, S.-J.; et al. miR-431 promotes differentiation and regeneration of old skeletal muscle by targeting Smad4. Genes Dev. 2015, 29, 1605–1617. [Google Scholar] [CrossRef] [PubMed]
- Bronisz-Budzyńska, I.; Chwalenia, K.; Mucha, O.; Podkalicka, P.; Karolina, B.-S.; Józkowicz, A.; Łoboda, A.; Kozakowska, M.; Dulak, J. miR-146a deficiency does not aggravate muscular dystrophy in mdx mice. Skelet. Muscle 2019, 9, 22. [Google Scholar] [CrossRef] [PubMed]
- Qiu, H.; Liu, N.; Luo, L.; Zhong, J.; Tang, Z.; Kang, K.; Qu, J.; Peng, W.; Liu, L.; Li, L.; et al. MicroRNA-17-92 regulates myoblast proliferation and differentiation by targeting the ENH1/Id1 signaling axis. Cell Death Differ. 2016, 23, 1658–1669. [Google Scholar] [CrossRef] [PubMed]
- Cheung, T.H.; Quach, N.L.; Charville, G.W.; Liu, L.; Park, L.; Edalati, A.; Yoo, B.; Hoang, P.; Rando, T.A. Maintenance of muscle stem-cell quiescence by microRNA-489. Nature 2012, 482, 524–528. [Google Scholar] [CrossRef] [PubMed]
- Baghdadi, M.B.; Tajbakhsh, S. Regulation and phylogeny of skeletal muscle regeneration. Dev. Biol. 2018, 433, 200–209. [Google Scholar] [CrossRef]
- Joe, A.W.; Yi, L.; Natarajan, A.; Grand, F.; So, L.; Wang, J.; Rudnicki, M.A.; Rossi, F.M. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat. Cell Biol. 2010, 12, 153–163. [Google Scholar] [CrossRef]
- Uezumi, A.; Fukada, S.; Yamamoto, N.; Takeda, S.; Tsuchida, K. Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nat. Cell Biol. 2010, 12, 143–152. [Google Scholar] [CrossRef]
- Wosczyna, M.N.; Konishi, C.T.; Carbajal, E.E.; Wang, T.T.; Walsh, R.A.; Gan, Q.; Wagner, M.W.; Rando, T.A. Mesenchymal Stromal Cells Are Required for Regeneration and Homeostatic Maintenance of Skeletal Muscle. Cell Rep. 2019, 27, 2029–2035. [Google Scholar] [CrossRef]
- Verma, M.; Asakura, Y.; Murakonda, B.; Pengo, T.; Latroche, C.; Chazaud, B.; McLoon, L.K.; Asakura, A. Muscle Satellite Cell Cross-Talk with a Vascular Niche Maintains Quiescence via VEGF and Notch Signaling. Cell Stem Cell 2018, 23, 530–543. [Google Scholar] [CrossRef]
- Liu, N.; Williams, A.H.; Maxeiner, J.M.; Bezprozvannaya, S.; elton, J.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. microRNA-206 promotes skeletal muscle regeneration and delays progression of Duchenne muscular dystrophy in mice. J. Clin. Investig. 2012, 122, 2054–2065. [Google Scholar] [CrossRef] [PubMed]
- Vechetti, I.J.; Wen, Y.; Chaillou, T.; Murach, K.A.; Alimov, A.P.; Figueiredo, V.C.; Dal-Pai-Silva, M.; McCarthy, J.J. Life-long reduction in myomiR expression does not adversely affect skeletal muscle morphology. Sci. Rep. UK 2019, 9, 5483. [Google Scholar] [CrossRef] [PubMed]
- Gatfield, D.; Martelot, L.G.; Vejnar, C.; Gerlach, D.; Schaad, O.; Fleury-Olela, F.; Ruskeepaa, A.; Oresic, M.; Esau, C.; Zdobnov, E.; et al. Integration of microRNA miR-122 in hepatic circadian gene expression. Genes Dev. 2009, 23, 1313–1326. [Google Scholar] [CrossRef] [PubMed]
- Van Rooij, E.; Sutherland, L.; Qi, X.; Richardson, J.; Hill, J.; Olson, E. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science 2007, 316, 575–579. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, A.; O’Carroll, D.; Tan, C.; Hillman, D.; Sugimori, M.; Llinas, R.; Greengard, P. Cerebellar neurodegeneration in the absence of microRNAs. J. Exp. Med. 2007, 204, 1553–1558. [Google Scholar] [CrossRef]
- Reichholf, B.; Herzog, V.A.; Fasching, N.; Manzenreither, R.A.; Sowemimo, I.; Ameres, S.L. Time-Resolved Small RNA Sequencing Unravels the Molecular Principles of MicroRNA Homeostasis. Mol. Cell 2019, 75, 756–768. [Google Scholar] [CrossRef]
- Marzi, M.J.; Ghini, F.; Cerruti, B.; de Pretis, S.; Bonetti, P.; Giacomelli, C.; Gorski, M.M.; Kress, T.; Pelizzola, M.; Muller, H.; et al. Degradation dynamics of microRNAs revealed by a novel pulse-chase approach. Genome Res. 2016, 26, 554–565. [Google Scholar] [CrossRef]
- Oikawa, S.; Wada, S.; Lee, M.; Maeda, S.; Akimoto, T. Role of endothelial microRNA-23 clusters in angiogenesis in vivo. Am. J. Physiol. Heart C 2018, 315, H838–H846. [Google Scholar] [CrossRef]
- Wada, S.; Kato, Y.; Okutsu, M.; Miyaki, S.; Suzuki, K.; Yan, Z.; Schiaffino, S.; Asahara, H.; Ushida, T.; Akimoto, T. Translational suppression of atrophic regulators by microRNA-23a integrates resistance to skeletal muscle atrophy. J. Biol. Chem. 2011, 286, 38456–38465. [Google Scholar] [CrossRef]
- Rando, T.; Blau, H. Primary mouse myoblast purification, characterization, and transplantation for cell-mediated gene therapy. J. Cell Biol. 1994, 125, 1275–1287. [Google Scholar] [CrossRef]
- Hindi, L.; McMillan, J.; Afroze, D.; Hindi, S.; Kumar, A. Isolation, Culturing, and Differentiation of Primary Myoblasts from Skeletal Muscle of Adult Mice. Bio. Protoc. 2017, 7, 7. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oikawa, S.; Lee, M.; Akimoto, T. Conditional Deletion of Dicer in Adult Mice Impairs Skeletal Muscle Regeneration. Int. J. Mol. Sci. 2019, 20, 5686. https://doi.org/10.3390/ijms20225686
Oikawa S, Lee M, Akimoto T. Conditional Deletion of Dicer in Adult Mice Impairs Skeletal Muscle Regeneration. International Journal of Molecular Sciences. 2019; 20(22):5686. https://doi.org/10.3390/ijms20225686
Chicago/Turabian StyleOikawa, Satoshi, Minjung Lee, and Takayuki Akimoto. 2019. "Conditional Deletion of Dicer in Adult Mice Impairs Skeletal Muscle Regeneration" International Journal of Molecular Sciences 20, no. 22: 5686. https://doi.org/10.3390/ijms20225686
APA StyleOikawa, S., Lee, M., & Akimoto, T. (2019). Conditional Deletion of Dicer in Adult Mice Impairs Skeletal Muscle Regeneration. International Journal of Molecular Sciences, 20(22), 5686. https://doi.org/10.3390/ijms20225686




