Small RNA Sequencing Reveals Regulatory Roles of MicroRNAs in the Development of Meloidogyne incognita
Abstract
1. Introduction
2. Results
2.1. Small RNA Deep Sequencing of Eggs and J2 Juveniles
2.2. Characterization of Small RNAs in RKN Eggs and J2 Larvae
2.3. Identification and Specific Expression of M. Incognita miRNAs at Eggs and J2 Stages
2.4. Identification and Functional Analysis of miRNA Targets
3. Discussion
4. Materials and Methods
4.1. M. incognita Culture and Sample Collection
4.2. RNA Extraction and Deep Sequencing
4.3. miRNA Identification and Expression
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Koenning, S.R.; Wrather, J.A.; Kirkpatrick, T.L.; Walker, N.R.; Starr, J.L.; Mueller, J.D. Plant-Parasitic Nematodes Attacking Cotton in the United States: Old and Emerging Production Challenges. Plant Dis. 2004, 88, 100–113. [Google Scholar] [CrossRef] [PubMed]
- Gardner, M.; Verma, A.; Mitchum, M.G. Chapter Eleven-Emerging Roles of Cyst Nematode Effectors in Exploiting Plant Cellular Processes. In Advances in Botanical Research; Escobar, C., Fenoll, C., Eds.; Academic Press: Amsterdam, The Netherlands, 2015; Volume 73, pp. 259–291. [Google Scholar]
- Rosso, M.; Hussey, R.S.; Davis, E.L.; Smant, G.; Baum, T.J.; Abad, P.; Mitchum, M.G. Nematode Effector Proteins: Targets and Functions in Plant Parasitism. In Effectors in Plant–Microbe Interactions; Martin, F., Kamoun, S., Eds.; Wiley: Hoboken, NJ, USA, 2011; pp. 327–354. Available online: https://doi.org/10.1002/9781119949138.ch13 (accessed on 18 September 2019).
- Mitchum, M.G.; Hussey, R.S.; Baum, T.J.; Wang, X.; Elling, A.A.; Wubben, M.; Davis, E.L. Nematode effector proteins: An emerging paradigm of parasitism. New Phytol. 2013, 199, 879–894. [Google Scholar] [CrossRef] [PubMed]
- Abad, P.; Gouzy, J.; Aury, J.-M.; Castagnone-Sereno, P.; Danchin, E.G.J.; Deleury, E.; Perfus-Barbeoch, L.; Anthouard, V.; Artiguenave, F.; Blok, V.C.; et al. Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat. Biotechnol. 2008, 26, 909. [Google Scholar] [CrossRef] [PubMed]
- Davis, E.L.; Hussey, R.S.; Baum, T.J.; Bakker, J.; Schots, A.; Rosso, M.-N.; Abad, P. Nematode Parasitism Genes. Annu. Rev. Phytopathol. 2000, 38, 365–396. [Google Scholar] [CrossRef]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef]
- Xie, X.H.; Lu, J.; Kulbokas, E.J.; Golub, T.R.; Mootha, V.; Lindblad-Toh, K.; Lander, E.S.; Kellis, M. Systematic discovery of regulatory motifs in human promoters and 3 ‘ UTRs by comparison of several mammals. Nature 2005, 434, 338–345. [Google Scholar] [CrossRef]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [CrossRef]
- Tanno, B.; Cesi, V.; Vitali, R.; Sesti, F.; Giuffrida, M.L.; Mancini, C.; Calabretta, B.; Raschella, G. Silencing of endogenous IGFBP-5 by micro RNA interference affects proliferation, apoptosis and differentiation of neuroblastoma cells. Cell Death Differ. 2005, 12, 213–223. [Google Scholar] [CrossRef]
- Zhang, B.H.; Wang, Q.L.; Pan, X.P. MicroRNAs and their regulatory roles in animals and plants. J. Cell Physiol. 2007, 210, 279–289. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern-formation in C-Elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef]
- Pasquinelli, A.E.; Reinhart, B.J.; Slack, F.; Martindale, M.Q.; Kuroda, M.I.; Maller, B.; Hayward, D.C.; Ball, E.E.; Degnan, B.; Muller, P.; et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000, 408, 86–89. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Y.; Xie, F.; Li, C.; Zhang, B.; Nichols, R.L.; Pan, X. Identification and characterization of microRNAs in the plant parasitic root-knot nematode Meloidogyne incognita using deep sequencing. Funct. Integr. Genom. 2016, 16, 127–142. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Mao, Z.; Yan, J.; Cheng, X.; Liu, F.; Xiao, L.; Dai, L.; Luo, F.; Xie, B. Identification of MicroRNAs in Meloidogyne incognita Using Deep Sequencing. PLoS ONE 2015, 10, e0133491. [Google Scholar] [CrossRef]
- Huang, G.Z.; Allen, R.; Davis, E.L.; Baum, T.J.; Hussey, R.S. Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Proc. Natl. Acad. Sci. USA 2006, 103, 14302–14306. [Google Scholar] [CrossRef]
- Matsunaga, Y.; Kawano, K.; Iwasaki, T.; Kawano, T. RNA Interference-Mediated Growth Control of the Southern Root-Knot Nematode Meloidogyne incognita. Biosci. Biotechnol. Biochem. 2012, 76, 378–380. [Google Scholar] [CrossRef]
- Rosso, M.N.; Dubrana, M.P.; Cimbolini, N.; Jaubert, S.; Abad, P. Application of RNA interference to root-knot nematode genes encoding esophageal gland proteins. Mol. Plant Microbe. Interact. 2005, 18, 615–620. [Google Scholar] [CrossRef]
- Bakhetia, M.; Charlton, W.; Atkinson, H.J.; McPherson, M.J. RNA interference of dual oxidase in the plant nematode Meloidogyne incognita. Mol. Plant Microbe Interact. 2005, 18, 1099–1106. [Google Scholar] [CrossRef]
- Villaverde, J.J.; Sandín-España, P.; Sevilla-Morán, B.; López-Goti, C.; Alonso-Prados, J.L. Biopesticides from natural products: Current development, legislative framework, and future trends. BioRes 2016, 11, 5618–5640. [Google Scholar] [CrossRef]
- Marchand, P.A. Basic substances: An opportunity for approval of low-concern substances under EU pesticide regulation. Pest Manag. Sci. 2015, 71, 1197–1200. [Google Scholar] [CrossRef]
- Subramanian, P.; Choi, I.C.; Mani, V.; Park, J.; Subramaniyam, S.; Choi, K.H.; Sim, J.S.; Lee, C.M.; Koo, J.C.; Hahn, B.S. Stage-Wise Identification and Analysis of miRNA from Root-Knot Nematode Meloidogyne incognita. Int. J. Mol. Sci. 2016, 17. [Google Scholar] [CrossRef] [PubMed]
- Simon, D.J.; Madison, J.M.; Conery, A.L.; Thompson-Peer, K.L.; Soskis, M.; Ruvkun, G.B.; Kaplan, J.M.; Kim, J.K. The microRNA miR-1 regulates a MEF-2-dependent retrograde signal at neuromuscular junctions. Cell 2008, 133, 903–915. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Sun, Y.; Zhang, D.; Dong, M.; Jiang, G.; Zhang, X.; Zhou, J. miR-1 inhibits the progression of colon cancer by regulating the expression of vascular endothelial growth factor. Oncol. Rep. 2018, 40, 589–598. [Google Scholar] [CrossRef] [PubMed]
- Kojima, S.; Chiyomaru, T.; Kawakami, K.; Yoshino, H.; Enokida, H.; Nohata, N.; Fuse, M.; Ichikawa, T.; Naya, Y.; Nakagawa, M.; et al. Tumour suppressors miR-1 and miR-133a target the oncogenic function of purine nucleoside phosphorylase (PNP) in prostate cancer. Br. J. Cancer 2012, 106, 405–413. [Google Scholar] [CrossRef]
- Nasser, M.W.; Datta, J.; Nuovo, G.; Kutay, H.; Motiwala, T.; Majumder, S.; Wang, B.; Suster, S.; Jacob, S.T.; Ghoshal, K. Downregulation of micro-RNA-1 (miR-1) in lung cancer. Suppression of tumorigenic property of lung cancer cells and their sensitization to doxorubicin-induced apoptosis by miR-1. J. Biol. Chem. 2018, 293, 12945. [Google Scholar] [CrossRef]
- Wu, H.; Huang, C.; Taki, F.A.; Zhang, Y.; Dobbins, D.L.; Li, L.; Yan, H.; Pan, X. Benzo-α-pyrene induced oxidative stress in Caenorhabditis elegans and the potential involvements of microRNA. Chemosphere 2015, 139, 496–503. [Google Scholar] [CrossRef]
- Li, N.; Wei, C.; Olena, A.F.; Patton, J.G. Regulation of endoderm formation and left-right asymmetry by miR-92 during early zebrafish development. Dev. (Camb. Engl.) 2011, 138, 1817–1826. [Google Scholar] [CrossRef]
- Yuva-Aydemir, Y.; Xu, X.-L.; Aydemir, O.; Gascon, E.; Sayin, S.; Zhou, W.; Hong, Y.; Gao, F.-B. Downregulation of the Host Gene jigr1 by miR-92 Is Essential for Neuroblast Self-Renewal in Drosophila. Plos Genet. 2015, 11, 1005264. [Google Scholar] [CrossRef]
- Tsuchida, A.; Ohno, S.; Wu, W.; Borjigin, N.; Fujita, K.; Aoki, T.; Ueda, S.; Takanashi, M.; Kuroda, M. miR-92 is a key oncogenic component of the miR-17–92 cluster in colon cancer. Cancer Sci. 2011, 102, 2264–2271. [Google Scholar] [CrossRef]
- Chen, L.; Li, C.; Zhang, R.; Gao, X.; Qu, X.; Zhao, M.; Qiao, C.; Xu, J.; Li, J. miR-17-92 cluster microRNAs confers tumorigenicity in multiple myeloma. Cancer Lett. 2011, 309, 62–70. [Google Scholar] [CrossRef]
- Poole, C.B.; Gu, W.; Kumar, S.; Jin, J.; Davis, P.J.; Bauche, D.; McReynolds, L.A. Diversity and Expression of MicroRNAs in the Filarial Parasite, Brugia malayi. PLoS ONE 2014, 9, 96498. [Google Scholar] [CrossRef] [PubMed]
- Clark, A.M.; Goldstein, L.D.; Tevlin, M.; Tavaré, S.; Shaham, S.; Miska, E.A. The microRNA miR-124 controls gene expression in the sensory nervous system of Caenorhabditis elegans. Nucleic Acids Res. 2010, 38, 3780–3793. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhi, L.; Shakoor, S.; Liao, K.; Wang, D. microRNAs Involved in the Control of Innate Immunity in Candida Infected Caenorhabditis elegans. Sci. Rep. 2016, 6, 36036. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Nichols, K.M.; Poynton, H.C.; Sepúlveda, M.S. MicroRNAs are involved in cadmium tolerance in Daphnia pulex. Aquat. Toxicol. 2016, 175, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Zhou, Y.; Liu, Y.; Deng, Y.; Puthiyakunnon, S.; Chen, X. miR-252 of the Asian tiger mosquito Aedes albopictus regulates dengue virus replication by suppressing the expression of the dengue virus envelope protein. J. Med. Virol. 2014, 86, 1428–1436. [Google Scholar] [CrossRef]
- Xie, F.; Wang, Q.; Sun, R.; Zhang, B. Deep sequencing reveals important roles of microRNAs in response to drought and salinity stress in cotton. J. Exp. Bot. 2015, 66, 789–804. [Google Scholar] [CrossRef]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- Gardner, P.P.; Daub, J.; Tate, J.G.; Nawrocki, E.P.; Kolbe, D.L.; Lindgreen, S.; Wilkinson, A.C.; Finn, R.D.; Griffiths-Jones, S.; Eddy, S.R.; et al. Rfam: Updates to the RNA families database. Nucleic Acids Res. 2009, 37, D136–D140. [Google Scholar] [CrossRef]
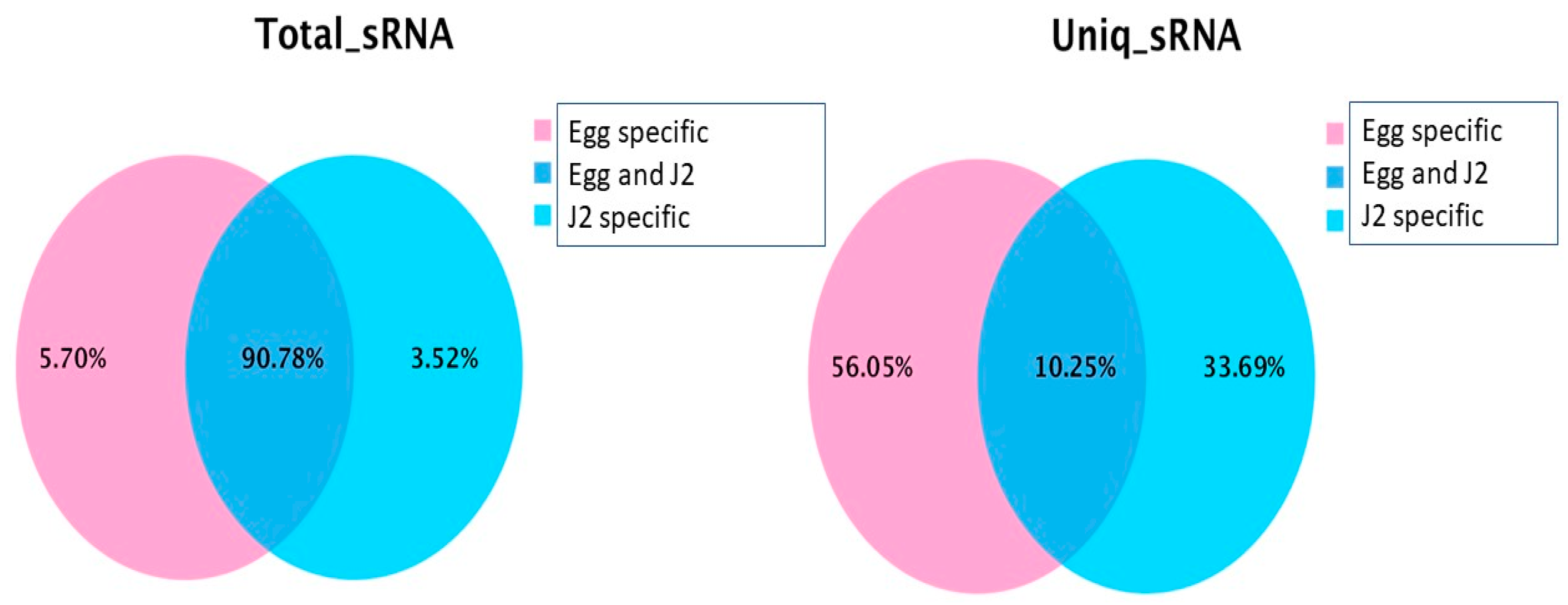
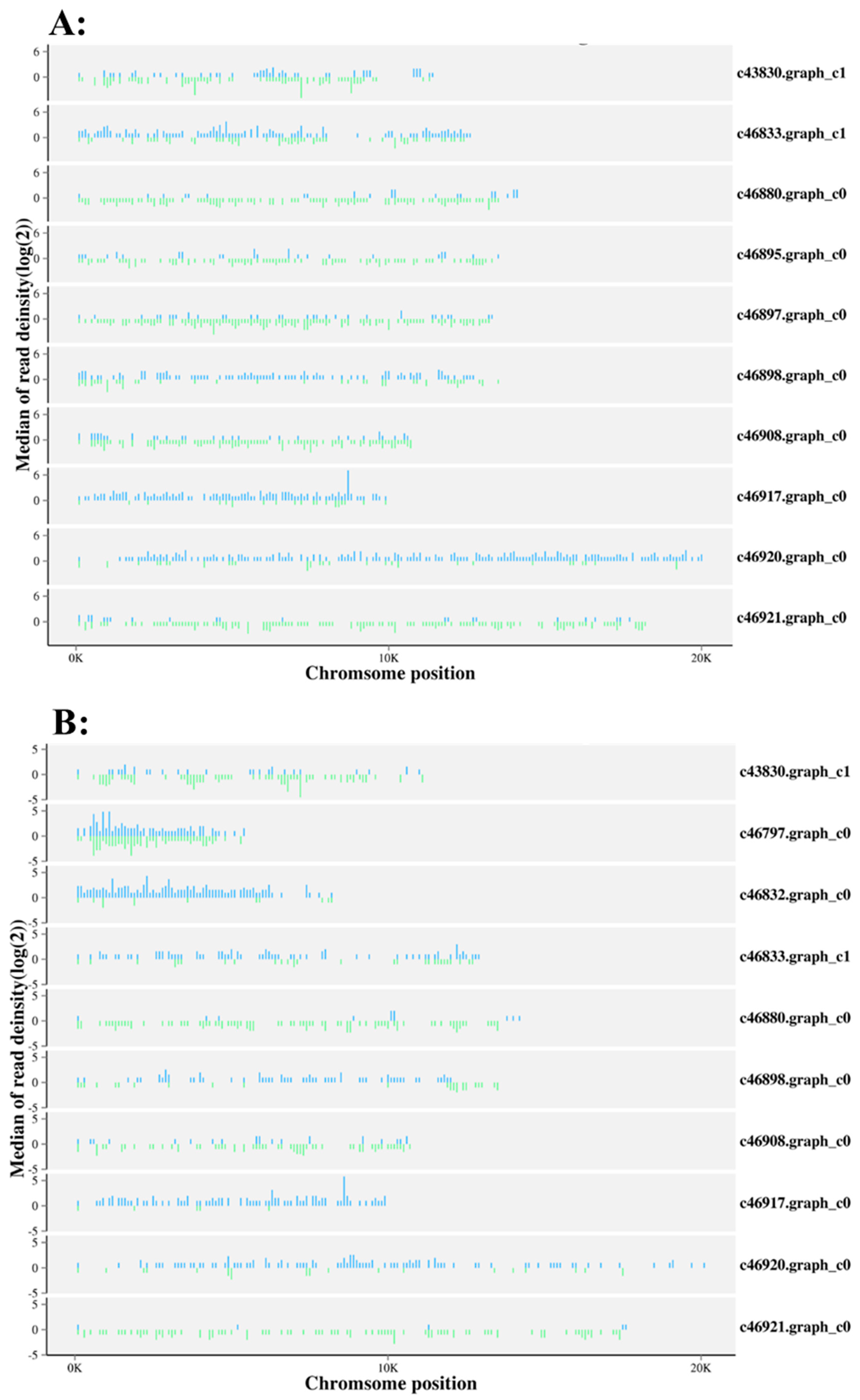
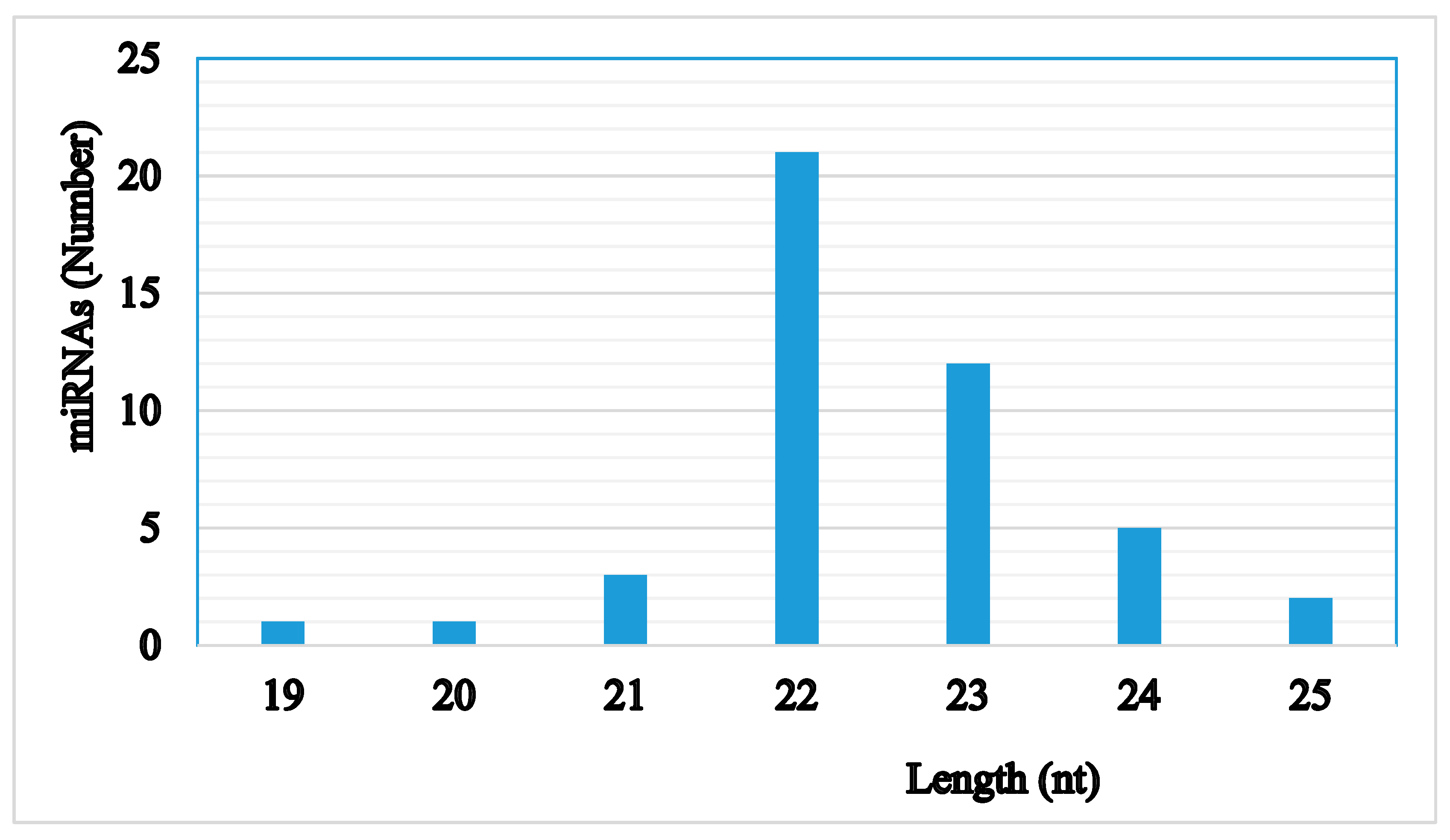
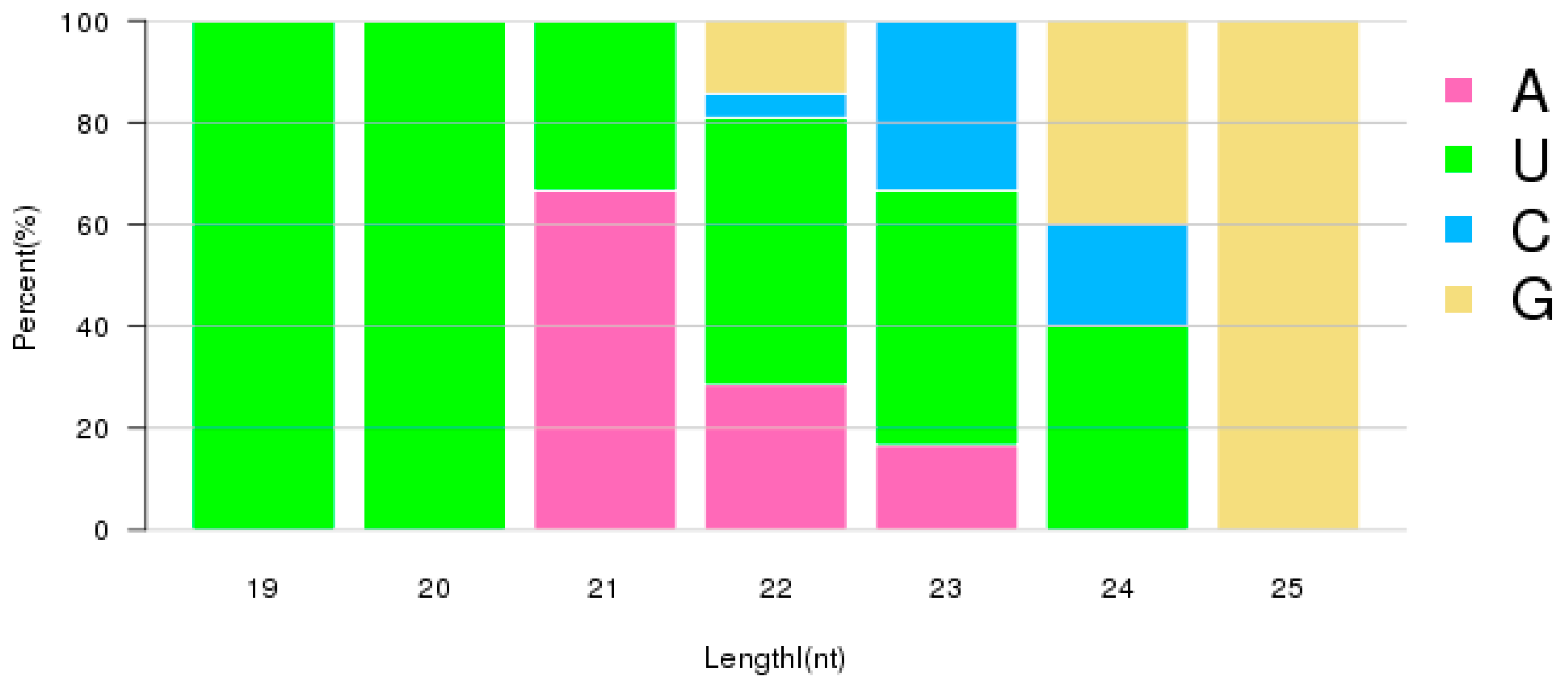
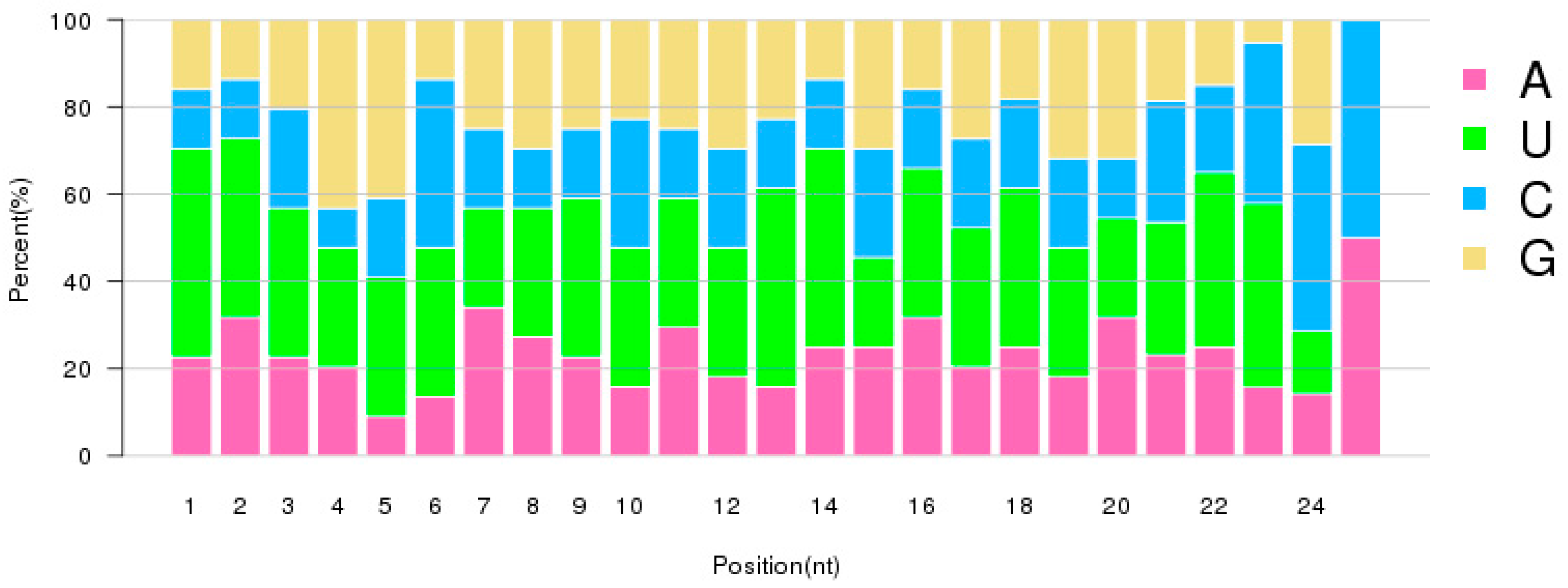
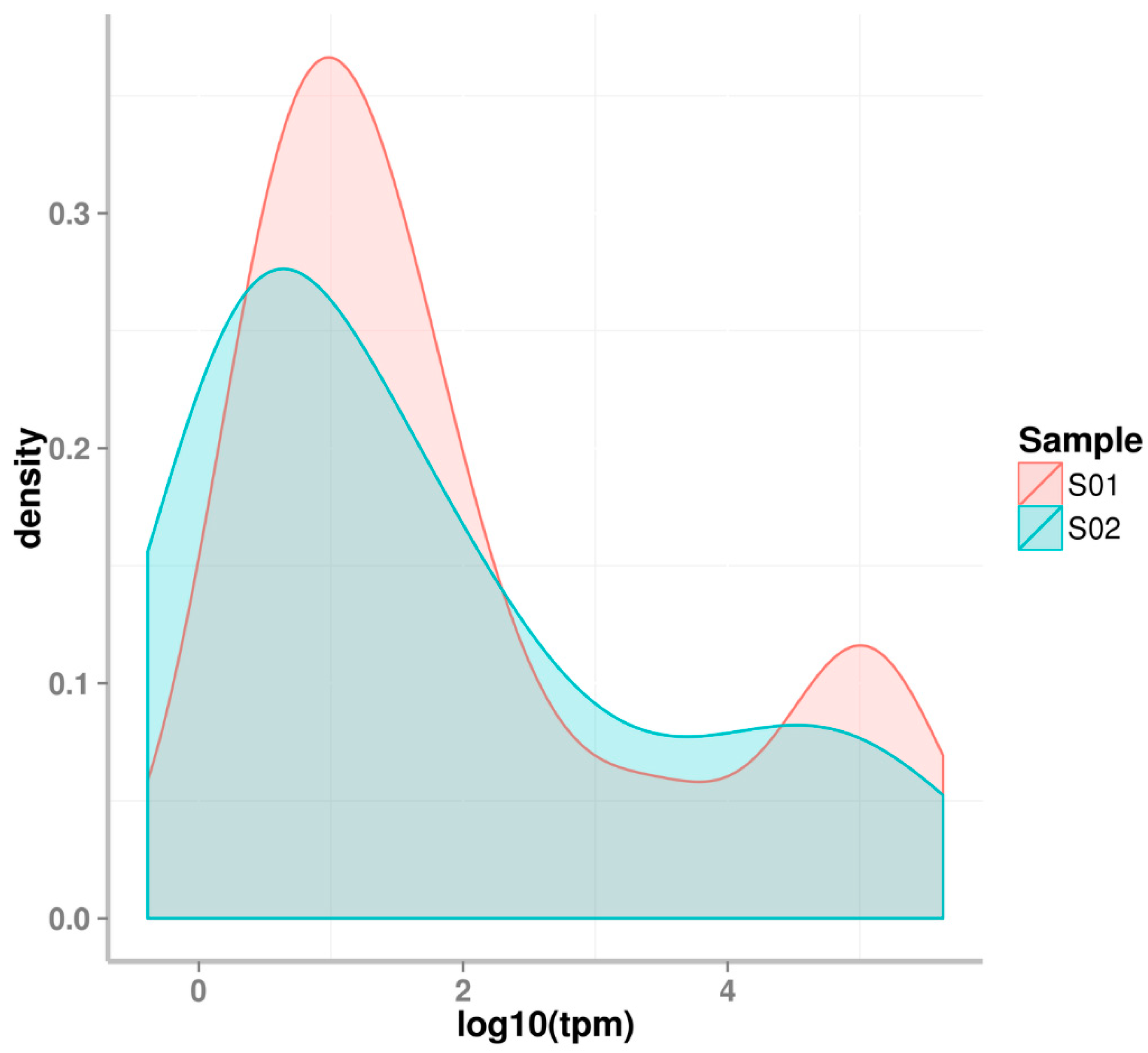
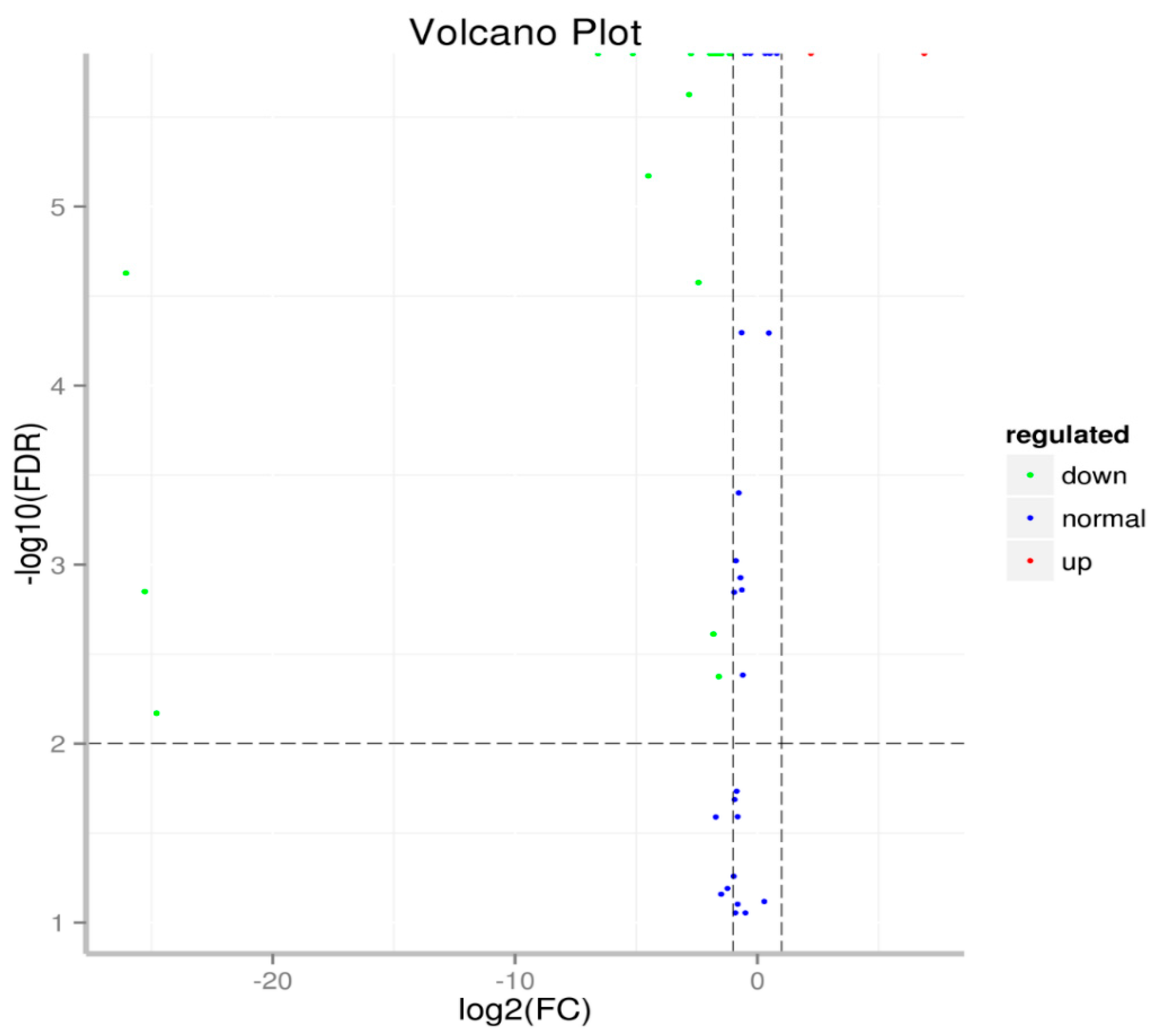


| Samples | Raw Reads | Low Quality | Containing ’N’ Reads | Length <18 | Length >30 | Clean Reads | Q30 (%) |
|---|---|---|---|---|---|---|---|
| Eggs | 19,952,271 | 0 | 46 | 604,254 | 910,019 | 18,437,952 | 98.12 |
| J2 | 17,526,380 | 0 | 171 | 1,611,967 | 524,776 | 15,389,466 | 98.13 |
| Types | Eggs | J2 | ||
|---|---|---|---|---|
| Number | Percentage (%) | Number | Percentage (%) | |
| rRNA | 3,840,307 | 20.83 | 2,573,694 | 16.72 |
| scRNA | 0 | 0.00 | 0 | 0.00 |
| snRNA | 0 | 0.00 | 0 | 0.00 |
| snoRNA | 3415 | 0.02 | 1359 | 0.01 |
| tRNA | 102,301 | 0.55 | 109,083 | 0.71 |
| Repbase | 2744 | 0.01 | 2417 | 0.02 |
| Unannotated | 14,489,185 | 78.59 | 12,702,913 | 82.54 |
| Total | 18,437,952 | 100.00 | 15,389,466 | 100.00 |
| miRNA | Eggs | J2 larvae | miRNA | Eggs | J2 larvae |
|---|---|---|---|---|---|
| miR22 | 8.71 | 2.88 | miRN1 | 9.29 | 0.41 |
| Let7 | 101.6 | 140.88 | miRN10 | 2.9 | 2.06 |
| miR1 | 244,810.84 | 426,558.94 | miRN11 | 8.71 | 4.53 |
| miR10227 | 320.48 | 109.16 | miRN12 | 2.9 | 1.65 |
| miR124 | 70,559.95 | 88,381.37 | miRN13 | 43.54 | 25.54 |
| miR239b | 70,511.18 | 736.93 | miRN14 | 42.38 | 27.19 |
| miR2-3p | 85,466.17 | 26,269.27 | miRN15 | 2.9 | 1.24 |
| miR252 | 1541.42 | 7125.89 | miRN16 | 8.71 | 4.94 |
| miR279 | 188,858.69 | 48,799.92 | miRN17 | 4.06 | 2.06 |
| miR3004 | 2.32 | 0.82 | miRN18 | 10.45 | 5.77 |
| miR4000 | 4.06 | 4.94 | miRN19 | 1.16 | 137.17 |
| miR4174 | 4.06 | 1.24 | miRN2 | 13.35 | 2.47 |
| miR4182 | 22.06 | 3.3 | miRN20 | 216.55 | 77.44 |
| miR429 | 23.22 | 11.95 | miRN21 | 14.51 | 2.06 |
| miR4738 | 6.97 | 0 | miRN3 | 4.06 | 0 |
| miR57-5p | 4555.18 | 2055.09 | miRN4 | 8.71 | 2.47 |
| miR7029 | 80.12 | 51.08 | miRN5 | 2.9 | 0 |
| miR7904 | 21,868.45 | 10,275.88 | miRN6 | 2.32 | 1.24 |
| miR7954 | 30.19 | 8.24 | miRN7 | 33.09 | 21.83 |
| miR8411 | 29.03 | 0.82 | miRN8 | 39.48 | 24.3 |
| miR87 | 86,106.54 | 70,471.29 | miRN9 | 199.14 | 140.05 |
| miR8917 | 29.03 | 15.65 | |||
| miR92 | 221,662.23 | 317,693.78 | |||
| miR993-3p | 2726.37 | 796.25 |
| miRNA | S01-Eggs | S02-J2 | p-Value | Log2FC | Regulated |
|---|---|---|---|---|---|
| miR7954 | 30.2 | 8.2 | <0.0001 | −1.874 | down |
| miR57-5p | 4555.2 | 2055.1 | <0.0001 | −1.148 | down |
| miR7904 | 21,868.4 | 10,275.9 | <0.0001 | −1.09 | down |
| miR4182 | 22.1 | 3.3 | <0.0001 | −2.743 | down |
| miR2-3p | 85,466.2 | 26,269.3 | <0.0001 | −1.702 | down |
| miR279 | 188,858.7 | 48,799.9 | <0.0001 | −1.952 | down |
| miR8411 | 29 | 0.8 | <0.0001 | −5.139 | down |
| miRN19 | 1.2 | 137.2 | <0.0001 | 6.8843 | up |
| miR239b | 70,511.2 | 736.9 | <0.0001 | −6.58 | down |
| miR993-3p | 2726.4 | 796.3 | <0.0001 | −1.776 | down |
| miR10227 | 320.5 | 109.2 | <0.0001 | −1.554 | down |
| miR252 | 1541.4 | 7125.9 | <0.0001 | 2.2088 | up |
| miRN20 | 216.6 | 77.4 | <0.0001 | −1.484 | down |
| miRN21 | 14.5 | 2.1 | 0.000001 | −2.817 | down |
| miRN1 | 9.3 | 0.4 | 0.000003 | −4.495 | down |
| miR4738 | 7 | 0 | 0.00001 | −26.05 | down |
| miRN2 | 13.4 | 2.5 | 0.00001 | −2.434 | down |
| miRN3 | 4.1 | 0 | 0.0009 | −25.28 | down |
| miRN4 | 8.7 | 2.5 | 0.0017 | −1.817 | down |
| miR22 | 8.7 | 2.9 | 0.0031 | −1.595 | down |
| miRN5 | 2.9 | 0 | 0.0051 | −24.79 | down |
| Database | Annotated Number | 300 ≤ Length < 1000 * | Length ≥1000 * |
|---|---|---|---|
| COG | 141 | 46 | 30 |
| GO | 180 | 64 | 60 |
| KEGG | 137 | 47 | 45 |
| KOG | 203 | 74 | 69 |
| Pfam | 260 | 95 | 81 |
| Swissprot | 184 | 66 | 68 |
| eggNOG | 268 | 84 | 90 |
| nr | 286 | 99 | 88 |
| All | 344 | 117 | 92 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Nichols, R.L.; Qiu, L.; Sun, R.; Zhang, B.; Pan, X. Small RNA Sequencing Reveals Regulatory Roles of MicroRNAs in the Development of Meloidogyne incognita. Int. J. Mol. Sci. 2019, 20, 5466. https://doi.org/10.3390/ijms20215466
Liu H, Nichols RL, Qiu L, Sun R, Zhang B, Pan X. Small RNA Sequencing Reveals Regulatory Roles of MicroRNAs in the Development of Meloidogyne incognita. International Journal of Molecular Sciences. 2019; 20(21):5466. https://doi.org/10.3390/ijms20215466
Chicago/Turabian StyleLiu, Huawei, Robert L. Nichols, Li Qiu, Runrun Sun, Baohong Zhang, and Xiaoping Pan. 2019. "Small RNA Sequencing Reveals Regulatory Roles of MicroRNAs in the Development of Meloidogyne incognita" International Journal of Molecular Sciences 20, no. 21: 5466. https://doi.org/10.3390/ijms20215466
APA StyleLiu, H., Nichols, R. L., Qiu, L., Sun, R., Zhang, B., & Pan, X. (2019). Small RNA Sequencing Reveals Regulatory Roles of MicroRNAs in the Development of Meloidogyne incognita. International Journal of Molecular Sciences, 20(21), 5466. https://doi.org/10.3390/ijms20215466






