Abstract
Due to the present scenario of climate change, plants have to evolve strategies to survive and perform under a plethora of biotic and abiotic stresses, which restrict plant productivity. Maintenance of plant protein functional conformation and preventing non-native proteins from aggregation, which leads to metabolic disruption, are of prime importance. Plant heat shock proteins (HSPs), as chaperones, play a pivotal role in conferring biotic and abiotic stress tolerance. Moreover, HSP also enhances membrane stability and detoxifies the reactive oxygen species (ROS) by positively regulating the antioxidant enzymes system. Additionally, it uses ROS as a signal to molecules to induce HSP production. HSP also enhances plant immunity by the accumulation and stability of pathogenesis-related (PR) proteins under various biotic stresses. Thus, to unravel the entire plant defense system, the role of HSPs are discussed with a special focus on plant response to biotic and abiotic stresses, which will be helpful in the development of stress tolerance in plant crops.
1. Introduction
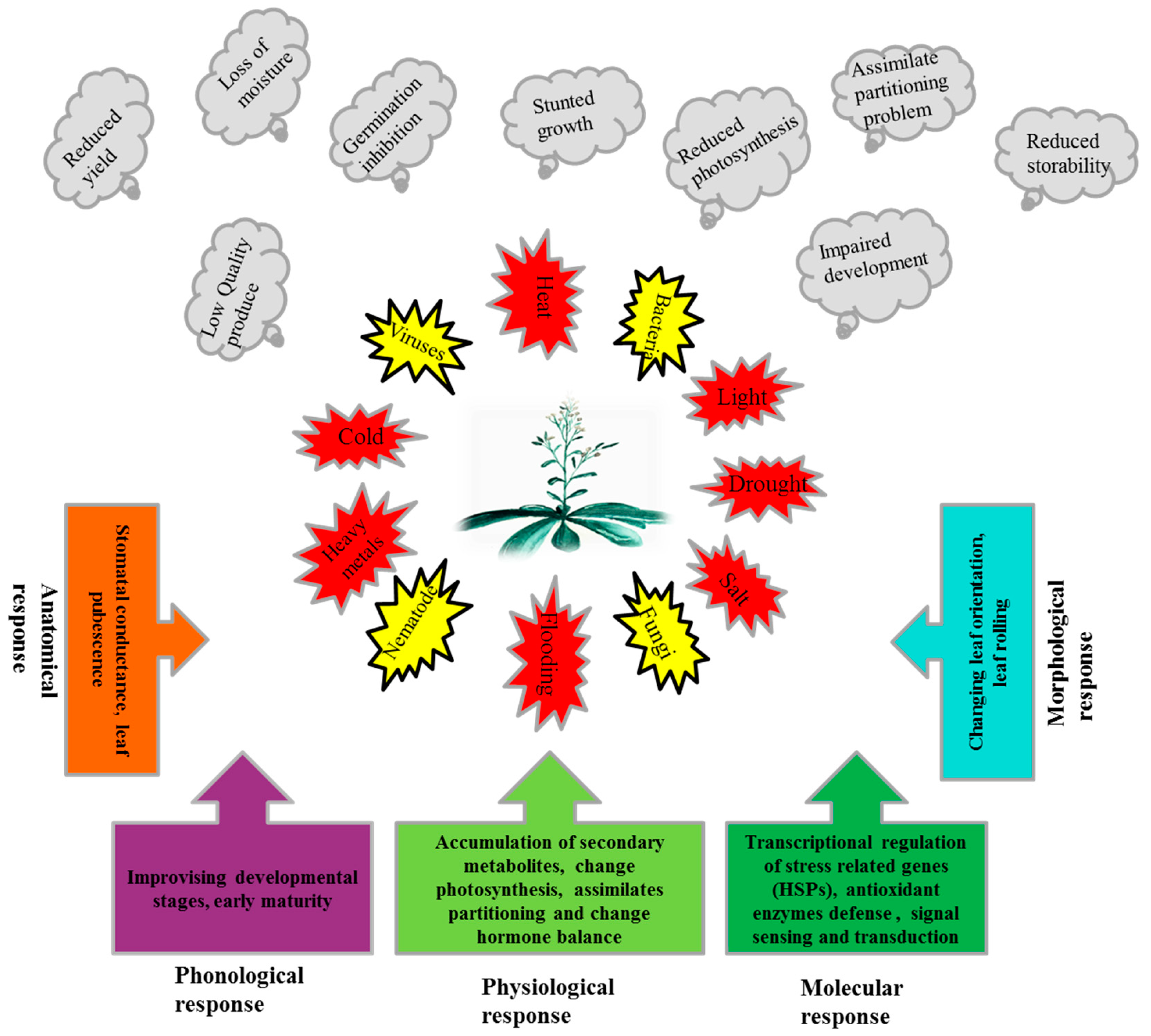
Plants are sessile organisms and are subjected to various threats, both biotic and abiotic. These stresses, individually or in combination, result in huge losses in terms of growth, development, and yield and sometimes threaten the survival of the plant [1]. Plants continuously confront harsh environments like high/low temperatures, drought, salts, heavy metals, light, flooding and physical wounding [2,3,4,5]. Biotic stresses like pathogens (Viruses, Bacteria, Fungi) and pests such as nematodes, insects, and rodents also restrict plant productivity [6,7,8,9,10]. Negative effects of these stresses on the plant germination [11], are stunted growth [12,13], sunburn and scorching of leaves [14], loss of photosynthetic pigment, decreased production of photo-assimilates, and depletion of carbohydrate reserves which results in starvation [15,16,17,18]. Abiotic stresses also negatively affect the reproductive characteristics of plants by enhancing male sterility [19] and increasing premature flower and fruit drop [20] which results in significant low yield and quality. It has been reported that the increase in temperature by 1 °C results in a 4–10% yield decrease [21]. As a consequence of these stresses, reactive oxygen species (ROS) are produced which lead to oxidative stress and, ultimately, results in cell death. ROS could be singlet oxygen (1O2), superoxide radical (O2•−), hydrogen peroxide (H2O2) and hydroxyl radical (OH −), which are produced in cell organelles such as mitochondria, peroxisomes and chloroplasts in oxidative stress situations and react with all types of macromolecules like pigments, proteins, lipids and DNA [22,23].
Plants respond morphologically to elevated temperature and light stress by changing the leaf orientation [13], anatomically by altering stomatal conductance and increased leaf pubescence [24,25], and phenologically by shifting and improvising the developmental stages to escape the abiotic stress condition [26]. Plants also change the metabolic processes and physiology to retain root hydraulic conductance [27], accumulation of the compatible osmolytes, such as sugars, sugar alcohols, proline and phenolic compounds under saline and water-logged conditions, as well as high temperature and water deficit conditions [28]. Moreover, plants manage to maintain photosynthetic machinery [29] by changing their assimilate partitioning a shift occurs from symplastic to apo-plastic [30]. During the onset of the stress situations, plants also improvise the hormonal balance of abscisic acid (ABA), ethylene, and salicylic acid (SA) as a signaling molecule in the systemic acquired resistance. Similarly, jasmonic acid (JA) and other steroids enhance stress tolerance and resistance [31]. Furthermore, secondary metabolites, such as isopropanioid, carotenoid, flavonoid, anthocyanin, lignin, and isoprenoids [30,32], also are produced and accumulated.
Besides these adaptations, plants also have sophisticated adaptive systems at the cellular and molecular levels. During the onset of stress, plants reduce the synthesis of normal protein production, and transcribe and translate heat shock proteins (HSPs). Added to transcriptional regulations, plants also have some sophisticated post-transcriptional modifications which help the plant to cope with these stresses, such as alternative splicing and micro RNA (miRNA). Alternative splicing, which generates multiple copies from a single gene, helps the plants to mitigate abiotic stresses [33]. One of the important plant post-transcriptional modification strategies is the miRNA, which binds to the mRNA at any point to repress translation or direct cleavage of the mRNA. Some of the miRNAs also are involved with abiotic stress tolerance [34].
HSPs have been identified for a long time in cellular biology as proteins, which concentration dramatically increases when cells are grown at higher temperatures. Now, it is established that these are proteins that help newly synthesized proteins to fold, or to protect proteins that might mis-fold and thereby lose their potential functional conformation during a stress event, such as biotic and/or abiotic stress [35]. These stress-responsive biomolecules act as molecular chaperones which perform under stress situations [36]. Generally, proper protein folding, unfolding and transport, in conjunction with their localization in the cell, and, subsequently, disposal and degradation of the non-native proteins, are the main functions of HSPs [37,38,39]. HSPs are important in the plant life cycle as its role clearly extends beyond the protection from biotic and abiotic stresses. Although HSPs (with the exception of ubiquitin) were first characterized due to their response to high temperatures, now many HSPs are found at normal, non-stressed cells, are produced at particular stages of the cell cycle, or during development in the absence of stress [40]. HSPs beside stress-responsive biomolecules, also are involved in plant growth and development under normal conditions, like the flowers, seeds and fruits set, and the development [41], tuberization [42,43,44,45] and nutrient uptake [46]. HSPs are found in different compartments of the cells, such as cytoplasm, and nucleus, and cell organelles like mitochondria, chloroplasts and endoplasmic reticulum [47,48].
It is evident, now, that HSP expression is controlled by transcription factors known as the heat shock factor (HSF). Seen in plants, among the HSF classes, HSFA positively regulates plant tolerance to anoxia, heat, osmotic and oxidative stresses [49]. Found in tomato plants, HSFA1 is considered a master regulator of signal perception, transduction and controlling the expression of stress-responsive genes, including HSPs [4,50], thus, increased expression of HSPs and other stress responsive genes in conjunction with HSFs play an important and significant role in modifying physiological and biochemical processes, which leads to the development of tolerance to stresses [51,52].
Earlier, HSPs were believed to produce under heat stress, as the name indicates, but now it is established that these biomolecules are produced in response to various biotic and abiotic stresses. HSPs show response to biotic and abiotic stress situations by up- or down-regulation, but, sensing signals and transduction, particularly in biotic stress, still needs to be explored [53]. HSPs are grouped into different classes based on their molecular weight in kilo Dalton (kDa), such as HSP100, HSP90, HSP70, HSP60, and small HSPs, respectively [50,51,52,54,55]. These chaperone families are found in a wide variety of organisms and are involved in maintaining cell homeostasis, transportation of newly synthesized proteins across cell organelles, and folding—preventing mis-folded, denatured and aggregated proteins caused by stress conditions [37,56,57]. Industrialization and urbanization have resulted in climate change and global warming, which pose serious challenges for plant growth, development, yield, and quality and sometimes threaten the plants existence. Considering such a scenario, plant biologists and researchers are trying to investigate, identify and confirm certain traits and characteristics that are related to stress tolerance. Similarly, scientists also are employing the recent approaches of omics techniques in the development of transgenic plants through the incorporation and manipulation of the stress-related genes. Regarding this, HSPs are of wide function, both in development as well as in stress tolerance. We focus on, and compile, various scientific developments that took place in the recent past in respect to role of the HSP families in different crop plants against various biotic and abiotic stresses, which will be helpful in future research regarding the development of stress-tolerant varieties.
2. Occurrence of HSP in Plants
The heat shock response is not unique in plants. It was first discovered in the early 1960s by an Italian scientist, F. Ritossa, in Drosophila melanogaster, while working on high-temperature stress [58,59]. HSPs were studied in Saccharomyces cerevisiae, by McAlister et al. (1980) [60], Escherichia coli, by Yamamori et al. (1982) [61], and plants (Glycine max), Lin et al. (1984) [62]. A comparison of the response in different organisms has shown that HSPs are conserved highly across organisms [63]. The evolutionary conservation of the heat shock response shows that the production of HSPs is a fundamental and essential process in all organisms [51].
Plants, due to their sessile nature, have evolved different efficient strategies to ensure their generations in the challenging competitive environment. The numbers of stress related genes, including HSPs, are greater in plants than other organisms due to whole genome duplication and gene retention in their evolutionary past [64]. Additionally, plants contain chloroplasts which have their own genome and de novo protein synthesis machinery [65]. A strategy of the plant to cope with stressful situations (Figure 1), is the synthesis of normal protein reduced while the synthesis of stress-related proteins i.e., HSPs, is enhanced.
Figure 1.
Exposures of plants to different biotic and abiotic stresses, adverse effects of these stresses on plants and response mechanisms of plants to these stresses.
Many HSPs have been reported in a wide range of organisms from prokaryotes to eukaryotes [63,66]. These HSPs across the organisms are conserved highly, with little difference except for HSP33, which differs in plants from that in bacteria [67]. The genes which encode different HSPs are found in different cell compartments, such as the nucleus, mitochondria, chloroplast, endoplasmic reticulum and cytosol [68]. Similarly, the accumulation of these HSPs in different parts of the cell also depends on the intensity of the stress. Nuclear HSPs, for instance, are accumulating in the cytosol at the lower and higher temperatures of 27 °C and 43 °C, respectively, while the same aggregate in chloroplast is at 37 °C [47]. Different HSPs are found and differentially expressed in different species, even in different genotypes but in the same species, as investigated by Korotaeva et al., (2001) and Nieto-Sotelo et al., (2002) [69,70] in small HSPs where five sHSPs showed response to a higher temperature (42 °C) in maize but only one expressed in wheat and rice. Likewise, HSP68 expressed in mitochondria under stress situations in potatoes, tomatoes, and soybeans [71]. Some HSPs showed a tissue-specific response to stress situations; HSP101 was expressed more in reproductive parts like tassels, ear, and endosperm, than in vegetative parts like leaves and roots in maize [72]. Some HSPs responded differently to the varying length of stresses. As Heckathorn et al., (1989) [73] reported, in HSP45, a nuclear protein accumulated in the chloroplast at a 3 h exposure to heat stress, which returned to its native state after removal of stress. Similarly, HSPs triggered differently with different development stages. HSP45, for example, showed a response in the whole plant to a stress situation, while HSP64 and -72 only showed expression in the reproductive parts i.e., pollens [74,75]. It can be deduced that these are the key regulators which show different responses to varying levels of stress in different parts of the plants.
3. Classification and Nomenclature of HSPs
Heat shock proteins are conserved in almost all organisms from bacteria, to fungi, plants and animals, including human beings. HSPs are classified and named based on the molecular weight in kilo Dalton (kDa), which ranges from 8–200 kDa [66]. Based on molecular weight, HSPs generally are classified into the following sub-families: HSP100, HSP90, HSP70, HSP60, and small HSPs [66,67,76,77,78]. The names for HSPs in bacteria are different from plant HSPs, but the classification remains the same on the basis of molecular weight [51] (Table 1). Characteristic features of plant HSPs are nuclear binding domain-I, -II and a middle domain which is preceded by an N- terminal region and followed by a C -terminal extension [79].

Table 1.
Nomenclature of HSPs in prokaryotes and eukaryotes.
4. Regulation of HSP
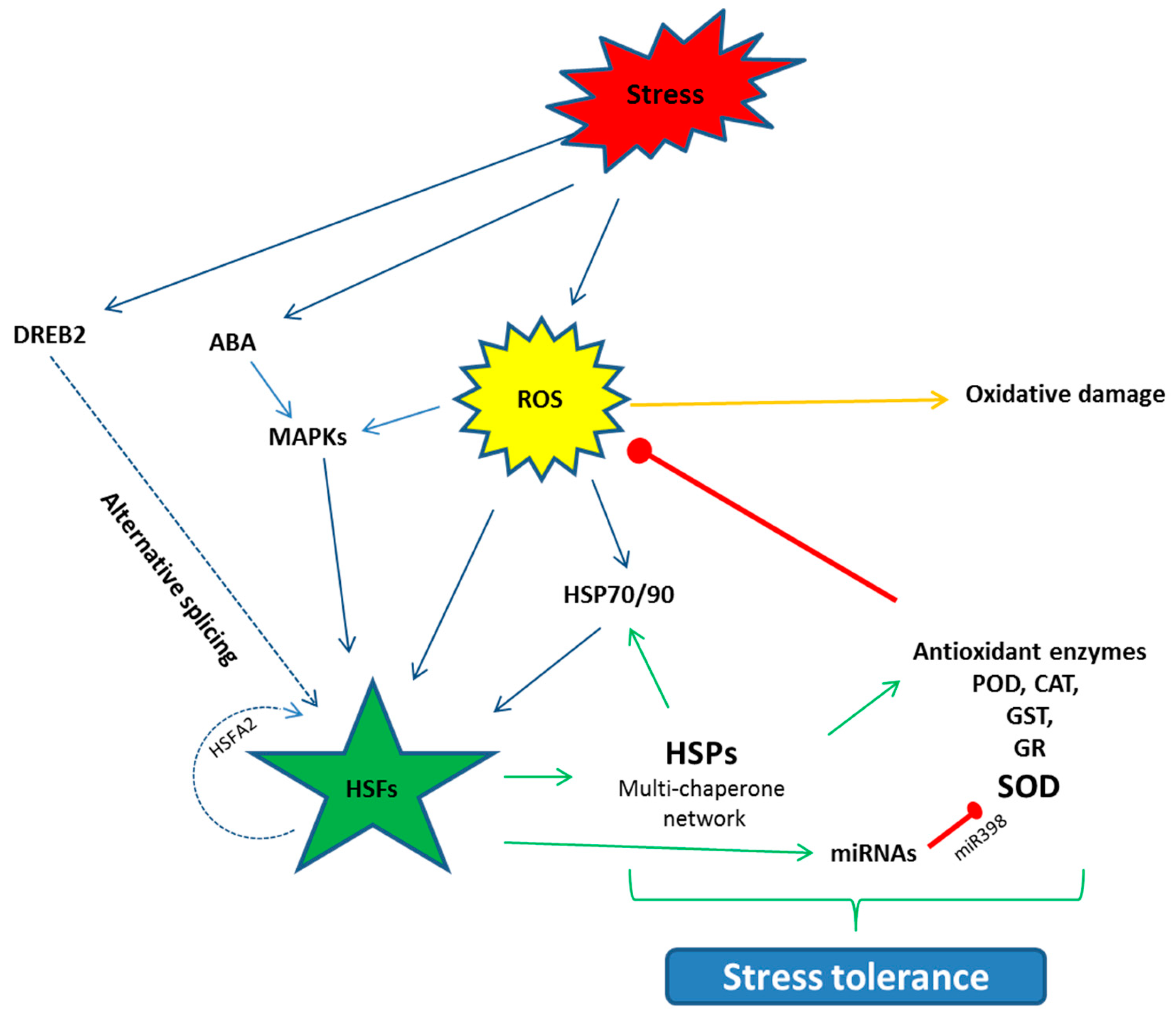
When plants are exposed to stress, the synthesis of normal proteins is decreased while the expression of special genes are up-regulated and, as a result, the synthesis of HSPs is triggered. To respond to stresses, the transcriptional regulation of HSPs is called the heat shock response (HSR) [77]. This HSR is regulated by HSFs in the promoter region, which bind to cis-acting elements known as HSE (Heat shock elements) [80,81]. HSFs are classified as three types: HSFA, B, and C; the functions of these classes vary from each other. Among the HSFs, HSFA regulates the HSPs cycle, which is found in the cytosol in a monomeric state. The activity of the HSFA, under normal conditions, is regulated negatively by HSP90s and are checked in the form of phospho-proteins [82]. During the onset of stress, this repression is reversed and HSP90 dissociates and changes into a functional trimer state. This HSFA homo-trimer then binds to the HSE in the promoter region [83], transcription occurs and HSPs are synthesized [84]. Among the HSFA, HSFA1 acts as the master regulator in tomatoes [50]. HSFA2 is structurally and functionally the same as HSFA1, but only is expressed in stressed plants. Under stress situations, HSFA2 makes a super activator hetero-oligomer structure with HSFA1, which is more efficient than the individual HSFs, which not only regulate the down-stream stress related HSP genes, but also the protective enzyme genes such as GST, GR, POX and APX [49,52]. Some studies also report that HSP gene expression positively regulates the protective enzyme activities. Seen in Arabidopsis, over expression of HSP17.8 enhanced the SOD activity and, in tobacco, HSP16.9 increased the activities of POD, CAT and SOD [85]. Post-transcriptional modification, such as alternative splicing, also regulate the HSFs. HSFA2 under heat stress, for instance, binds to its own promoter region and activates its own transcription in a positive auto-regulatory loop. Similarly, HSFA are regulated by DREB2 under stress, which in turn regulates the stress related genes in many plants [33] (Figure 2). Similarly, miRNAs also play a vital role in the stress response by down-regulation of stress-related genes. Some of the miRNAs are reported to have positive regulation in drought, cold, salinity, hormones and nutrient starvation stresses such as miR159, miR319, miR395, miR402 [34]. Conversely, in Arabidopsis short term heat stress, miR398 negatively regulates the expression of CSD1, CSD2 and CCS, which yield the SOD [85].
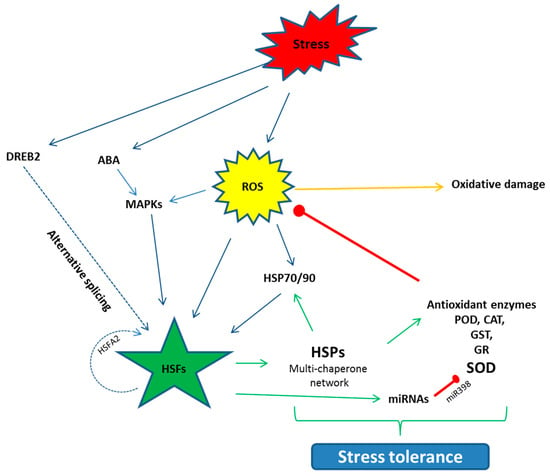
Figure 2.
Schematic diagram of the activation of the HSFs and their interaction with the other pathways to counter stress situations. HSFs are activated directly or indirectly through the event of alternative splicing. HSFs further regulate the down-stream HSPs, antioxidant enzyme genes and miRNAs, which help the plants to develop stress tolerance. Arrows denote the positive while red bars stand for negative interaction. ROS (Reactive oxygen species), ABA (Abscisic acid), MAPK (Mitogen-activated Protein Kinase), DREB (Dehydration responsive element binding protein), POD (Peroxidase), CAT (Catalase), GST (Glutathione S transferase), GR (Glutathione reductase), SOD (Superoxide dismutase).
5. Role of HSPs in Plant Defense
Plants are subjected to various biotic and abiotic stresses, singly or in combination, which adversely affect plant growth, development, and survival [86]. To cope with these stresses, plants have evolved various defense strategies i.e., physical [13], anatomical [25], and physiological [27,87]. Plants also respond to stress situations at the molecular level by altering gene expression, synthesis of stress-related biomolecules and proteins including heat stress proteins, to enable plants to ensure their generation in challenging situations.
5.1. Biotic Stress Tolerance
Plant growth, development, yield, and quality are affected adversely by several biotic factors such as pathogenic bacteria, fungi, viruses, and nematodes. Biotic factors, directly deprive their host plants of their nutrients, which result in reduced plant vigor, growth, productivity and sometimes leads to death of the host plants. Biotic stresses are major cause of pre- and post-harvest losses. Animals have an immune system, which helps them to adapt to biotic stresses such as new diseases and memorized the past infections, while plants lack such a system. Although plants lack this adaptive immune system, they have evolved several sophisticated strategies to counteract these biotic stresses [88]. These defense mechanisms are stored in the plant’s genome at the genetic level, which encode thousands of stress resistance genes. One of the adaptive systems plants employ in response to biotic stress is through regulation of HSPs (Table 2). HSP response to biotic stresses depend on the nature of the causal organisms and plant genotypes, either susceptible or resistant, and the developmental stage [6].

Table 2.
Summary of the role of plant HSP under biotic stresses.
5.1.1. Fungi
The phyto-pathogenic fungi not only cause devastating epidemics, but also cause serious yield losses which challenge global food security. During invasion, these pathogens employ different enzymes, toxins and secrete effector proteins. Different invading fungi have different types of interaction with the host, it could be biotrophic, necrotrophic or hemi-biotrophic [89]. To respond, plants have evolved different defense strategies, signal perception, transduction and activation of the immune system, including stress-related HSPs.
Tomato 3 species exposed to high and low temperatures along with infection by Phytophthora infestans, results in the up-regulation of HSP70 genes and the increased synthesis of HSP70 proteins. An increase in the temperature increases the mRNA and HSP70 genes, however, the increase of HSP70 proteins only occurs in susceptible tomato varieties [90]. Plant defense is affected directly by HSP-hijacked transport of the pathogens to the chloroplast where it forms a large structure that weakens the plant defense system, as reported in Pseudomonas syringae type III effector Hop II [91]. Small HSPs showed differential responses to infection by pathogenic fungi in rice, which were growth stage-specific [92], in relation to Magnaporthe grisea infection. Four of the HSPs (HSP16, 17, 18.1 and 18.2) were up-regulated, while four HSPs (HSP16.6, 17.8, 18.8 and 22) were down-regulated. Knockdown of some of the HSPs has affected the severity of the fungal infection in some cases. Van et al., (2010) [93] studied the Fusarium oxysporum infection in relation to HSP20 and some PR proteins. When HSP20 was silenced, the more severe infection was noted, although the PR proteins were there. A similar trend was noted by silencing the HSP17.6 in tomato Rhizopus nigricans post-harvest fruit decay [94]. Ahmad et al., (2015) [95] reported powdery mildew infection development with suppression of HSP16.9 in genotype SEP0105. Some researchers have established that HSPs are more effective against the pathogen in a complex as compared to individual HSPs. Arabidopsis HSPs along with PR proteins were more effective against Rhizoctonia solani. HSP20 expression along with PR proteins was more than 10 folds in a pathogen resistant genotype AG-8 but was normal in pathogen susceptible genotype AG2-1. Prominent performers out of these HSP20 genes were HSP17.4 and -17.6, overexpressed and silenced, respectively, have the same response for both genotypes [96,97]. One report showed the possible degrading or knockdown mechanism of HSP by the pathogen while developing an infection in the host, as evident in the proteomic study of apple fungus Venturia inaequalis, which displayed knockdown of the HSP21 during infection [98]. ROS triggered the induction and accumulation of HSP72 and HSP75 in two tomato genotypes against abiotic stresses and fungus Oidium neolycopersici [99].
HSPs also are involved in positive interaction with other defense-related proteins to develop resistance against biotic stresses as Yogendra et al., (2015) [100] reported in potato against powdery mildew. HSP17.8 and WRKY simultaneously were regulated transcriptionally in the resistant potato genotypes where HSP chaperone activity kept the defense-related proteins active during abiotic stress. A recent report on powdery mildew infection in sunflowers, susceptible and resistant genotypes in relation to HSP70 demonstrated that HSP70 and other pathogenesis and defense-related proteins were more expressed and accumulated in the resistant genotypes than in the susceptible ones [101].
5.1.2. Bacteria
Plant pathogenic bacteria are important biotic factors that limit plant productivity. Many plant pathogenic bacteria employ a type III secretion system to cause an infection through injection of different effector proteins into host plant cells. Plants respond to many phyto-pathogenic bacteria by pathogen-associated molecular patterns (PAMPs) as a basal defense response [102]. Plants also have evolved effector-triggered immunity (ETI) where resistance (R) protein can recognize the pathogen effector proteins and effective defense is developed against the invading pathogen, particularly the bacteria [6]. HSPs play an important role against virulent bacterial strains, as studied in tobacco against the Ralstonia solanacearum infection, where HSP17 was induced and accumulated in the virulent strain as compared to the avirulent strain [103]. The HSP17 gene, which corresponds to the HSP20 protein, had PR1 and PR4 expression increased soon after infection, even in the avirulent strain of the pathogen. HSP20 knockdown in the presence of the PR protein resulted in the virulence of non-pathogenic R. solanacearum. The same pattern was reported in HSP20′s involvement in resistance against compatible and incompatible Xanthomonas campestris in peppers and oranges, which showed HSP20 responded in basal resistance against the biotic stress [104]. Seen in Arabidopsis, six of the small HSPs were reported as down-regulated to Pseudomonas syringae [105] while, later on, it was explored that this down-regulation was due to salicylic acid, as the pathogen also mediated the SA pathway to cause infection [106]. HSP90, in contrast, has a positive interaction with Ralstonia solani in tobacco. HSP90, PAR1, and SGT1 were silenced; only the PAR1 silenced plants showed infection. HSP90- and SGT1-silenced plants resulted in lower infection, although PAR1 accumulation was found in the silenced plants, which showed a positive interaction of HSP90 with the infection [107].
5.1.3. Viruses
Viruses, upon entry into the plant cell, use the plant cell machinery to perpetuate and spread into the neighboring cells and quickly throughout the plant [9]. Cucumber necrosis virus (CNV) infection in the Nicotiana benthamiana resulted in higher HSP transcripts, which resulted in more accumulation of HSPs. This high concentration of HSP70 positively regulated CNV genomics RNA, coat protein and viron accumulation and infection [108]. Likewise, in Tomato yellow leaf curl virus (TYLCV), nuclear material was lowered after the knockdown of HSP70 genes [109]. The expression of HSP is not only specific to pathogen strains but also depends on the time after inoculation.
Occurring in Arabidopsis, regarding five viral strains at an interval of 1-day post-inoculation, HSP17.4 and HSP 17.6 were induced in only two strains after one day and showed a delayed response to the other three strains after three days, when studying profile expression by a microarray technique [110]. Similar results also were reported by Senthil et al., (2005) [114], who studied the heterologous microarray of potatoes in response to a virulent strain of Sonchus Yellow Net Virus (SYNV) and a virulent strain of Impatiens Necrotic Spot Virus (INSV) in model plant tobacco. HSP18 and HSP20 were up-regulated at four days after inoculation, while were lower at the fifth day post-inoculation to INSV; however, they were not induced by the SYNV strain. Viruses in some cases target the subcellular localization and expression pattern of HSP to develop an infection in the host cells. Li et al., (2015) [126] studied HSP20 in relation to Rice Strip Virus (RSV) where an invading pathogen infected the HSP20 cellular location with a large virus protein RdPd. This negative interaction possibly was responsible for virus spread and infection in the host cell. To contrast, HSP70 had a positive interaction with RSV and Tobacco Mosaic Virus (TMV), where heat treatment also increased the viral infection. HSP70, when silenced, also lowered the viral infection [127]. Some viruses need specific protein in the host cell for assembly and to establish infection in the host cell; for instance, Red Clover Necrotic Mosaic Virus (RCNMV) needs 480 KDa functional proteins to replicate in the host cell with the help of HSP70 and HSP90. This protein synthesis was checked by HSP70 knockdown and so was the infection, although a large complex of viral RNA was accumulated in the host cell but was non-functional [113]. A similar pattern was reported by Chen et al., (2014) [112] where HSP70 accumulation was noticed under viral infection and heat stress in tobacco. When cytoplasmic HSP70 was silenced, no viral infection was noticed.
One interesting report pointed out the role of Tomato Yellow Leaf Curl Virus (TYLCV) in controlling PCD by deactivation of tomato HSFA2. This inactivation resulted in HSP90 silencing, which mitigated the PCD and kept the plant healthy for its replication and infection at a later stage. Furthermore, the Root knot nematode coat protein transport was associated with HSP70 from the cytoplasm into the nucleus of a tomato plant. Silencing of HSP70 in the tomato plant controlled the viral infection. Conversely, HSP90 knockdown promoted viral infection [10,111]. A recent study on Potato Virus Y (PVY) in potato thermo-tolerant and sensitive genotypes under heat stress, HSPs was induced in both genotypes. Regarding thermo-tolerant genotypes, the expression of PR genes was also high, resulting in less viral infection [117]. Resistance against viral infection in Cytosinpeptidemycin application for the control of Rice Black Streaked Dwarf Virus (RBSDV), along with other defense-related genes, antioxidant enzymes and HSP also were up-regulated to control the virus infection [115].
5.1.4. Nematodes
Generally, nematodes species such as the root-knot nematode (Meloidogyne spp.), root-lesion nematode (Pratylenchus spp.) and cyst nematodes (Heterodera and Globodera spp.) are more detrimental to agricultural crops [8]. HSPs are involved in resistance to phyto-nematodes. Li et al., (2015) [7] analyzed RNA-Seq data of Gossypium hirsutum after Rotylenchulus reniformis infection at a three day interval up to 12 days. Twenty-three HSPs were induced in susceptible genotypes and 41 HSPs in resistant genotypes. Expression of HSP to different nematodes is genotype-specific, as studied by Lopes–Caitar et al., (2013) [119] in different soybean genotypes. Six HSP20 showed differential expression to Meloidogyne javenica infection. HSP22, -17.9a, -17.9b and -17.4 showed up-regulation in nematode-susceptible genotypes, while HSP17.6 and -22.4 showed a differential expression in both the susceptible and resistant genotypes. Fuganti et al., (2010) [120] identified gene Gm13G176000, which had a differential expression in soybean nematode-resistant and susceptible genotypes, while screening soybean resistance to nematode Meloidogyne javanica using a microsatellite marker. This later revealed that this gene corresponds to GmHSP17.6, which was induced highly in nematode infection. Studies also showed that some HSPs work in correlation with RghI protein and other antioxidant-related enzymes to develop resistance against nematodes. Kandoth et al., (2011) [121], while working with the Soybean cyst nematode (SCN) (Heterodera spp.), found RghI was differentially expressed, which induced the HSP20 accumulation in both the resistant and sensitive genotypes. Position of cis-acting elements in the promoter region of HSPs could be activated in biotic stresses, depending on the distance from the site of transcription. Meloidogyne incognita infection in the sunflower, HSP17.7, in transgenic tobacco showed that HSE were activated within 83 base pairs (bp) and HSP17.7 were up-regulated beyond 83 bp [122]. This trend was further confirmed by Barcala et al., (2008) [123] in HSP17.6 and HSP18.6, where they showed transcription within 108 and 49 bps, respectively. This also confirmed that, not only the position of HSE, but also other transcription factors like TATA Box and CAAT Box also interacted and influenced the expression of HSP20 in biotic stresses. HSP90 knockdown in tomatoes showed resistance to tomato root knot nematodes through SGT1 and Mi-1-mediated resistance [124]. HSP90, on the other hand, promoted the nematode infection. Lourenço et al., (2015) [125] silenced HSP90 and iso-citrate lyase (ICL) in tobacco, which resulted in 46–77% reduction in eggs of the root knot nematode (RKN) Meloidogyne incognita, as compared to the wild type which showed nematodes can reproduce only in the presence of HSP90 and ICL.
5.2. Abiotic Stress Tolerance
Abiotic stresses are extreme environmental conditions like extreme temperatures, water deficit, and ion imbalance due to heavy metals and salinity, which pose a serious threat to plants survival, yield and quality. Global warming and climate change, due to industrialization, has further worsened the situation. Greenhouse gases, particularly the concentration of CO2, is increasing constantly in the atmosphere, which is estimated to reach 520 ppm from 410 by the year 2100 [36]. Twenty per cent of the world cultivated land and almost 50% of irrigated land is affected by salinity, and yield loss up to 50% due to drought is projected by the year 2050 [128]. The world population by the year 2050 is approximated to be 9 billion [129], so, in such a situation, biotic and abiotic stress-tolerant cultivars need to be developed using transgenic and omic techniques. The role of HSPs has been studied by various scientists under different abiotic stresses (Table 3).

Table 3.
Summary of studies of plant HSP and abiotic stresses.
5.2.1. Temperature Stress
Extreme temperatures are the potential and important environmental stresses that affect plant survival in many ways. Temperatures stresses are discussed in the following sub-sections.
High-Temperature Stress
Accompanying climate change, high temperatures have threatened all organisms, but plants, being directly exposed and unable to change their position, are the most affected [130]. High temperature affects the macro-molecules, like proteins, by misfolding, aggregation and denatured enzymes. It also affects membrane fluidity and its disruption results in the accumulation of ROS and the development of secondary stress [131,132]. Multilevel interactions exist between HSPs and ROS. Plants are wise enough to use ROS as a signal molecule to produce HSPs and other stress-related proteins [85,133].
Several studies indicated that many high molecular weight HSPs showed response under high-temperature stress, such as HSP118, -114, -110, -108, -104, -103, -101, -100 and -97, respectively, [72,134]. Among the HSP100 class, significant HTR was shown by HSP101 [65] and also was involved in thermo-tolerance in Arabidopsis [135]. It was confirmed further in maize that HSP101 was involved in thermo-tolerance [70]. Merret et al., (2017) and Mcloughlin et al., (2016) [136,137] studied this trend a step further in Arabidopsis; they confirmed the role of HSP101 in thermotolerance, but also established that this played a role in recovery after heat shock. Besides the HSP101 role as a chaperone, HSP100 also was involved in development [138]. Low molecular weight HSPs i.e., -18.1 and -17.9, accumulated in the pea while it was treated for four hours at 42 °C. The response and expression of the HSPs also were development stage and different tissue specific. Maize subjected to 40 °C, HSPs (-101, -70 and -17.6) were induced. Above 36 °C, fertilization was reduced, although HSPs were induced in female reproductive parts but, when studied, mature pollens were more sensitive to heat stress [139]. To contrast, HSP70s were expressed more in tomato pollens [140]. Arabidopsis HSP70 expressed more in mitochondria under high temperature stress [141]. Chloroplast HSP70.1, 70.2 and mitochondrial HSP22 also were involved in seed development aside from its role as a chaperone [142,143]. HSP90 also showed increased expression under heat stress situations. HSP90.1 has been reported in rice and Arabidopsis [144,145] and all classes of HSP90 (A, B, and C) in soybeans [146]. Under normal conditions, HSP90 negatively regulated HSF and kept the regulation of all HSP checked [147]. HSP70s and HSP60s chaperonin families are the most studied of the chaperones under heat stress, which maintained protein proper folding using ATPs [38]. Cytosolic HSP70 was involved in heat stress tolerance in Arabidopsis [148]. HSP70s have been studied under high temperature stress in a variety of plant crops; such as witch-grass and alfalfa [149,150]; vegetables like pepper, tomato, cabbage, potato; ornamental plants like chrysanthemum [151,152,153,154,155,156,157]; grain such as wheat [158]; and tea [159]. Xu et al., (2010) [160] observed the expression pattern of chloroplast HSP60, not only in normal conditions but, also, under high-temperature and drought situations. It was responsible for Rubisco assembly, protection and also chloroplast development. Low molecular weight HSPs such as the HSP10, HSP20 and HSP40 families were up-regulated under high-temperature stress situations in various plant crops [5,152,161,162,163,164,165]. Some small HSPs were also genotype-specific and were up-regulated in resistant cultivars, such as foxtail millet, while some small HSPs were down-regulated in sensitive genotypes [53]. Some co-chaperones were involved in thermo-tolerance as HSP40. Correlation of small HSPs with HSP100, HSP70 and HSP60 suggested their role as holders in disaggregation and protein folding [157].
Low-Temperature Stress
Cold stress affects plant enzymes, membrane plasticity, changes physiology and metabolism, sometimes causes water starvation and desiccation which creates a stress condition for the plant that adversely affects plant growth, development and yield. Low temperature also is associated with protein disfunction and denaturing, which induce the accumulation of HSPs [166,167,168]. Many HSPs responded to cold stress and were up-regulated in Arabidopsis, tobacco, maize, rapeseed, chicory, poplar, wheat and barley [167,169,170,171,172,173,174,175,176]. Under low-temperature stress situations, HSPs were induced and translocated into various cell organelles to protect them from cold stress [169]. Bae et al., (2003) [169] investigated this in Arabidopsis treated with cold stress at 40 °C for 6 h. HSP70 were up-regulated and their traffic from the cytoplasm to the nucleus was observed. A similar event was observed in the pea mitochondria when treated at 4 °C for 36 h [177]. Some HSPs accumulated tissue specifically upon low-temperature exposure, as in poplar where HSPs were accumulated in leaves [178]. Regarding rice, low-temperature stress and a gradual decrease in the temperature from 15 °C to 0 °C, with an interval of 5 °C, up-regulated HSP95 and -75 and HSP70 accumulated in the chloroplast, as this was the vulnerable part of the plant to low temperature [179,180]. Some of the HSPs, like HSP90 in wheat and HSP60 and HSP21 in sunflowers down-regulated to cold stress [175,181]. A similar trend also was reported by Hlavackova et al., (2013) and Rinalducci et al., (2011) [167,182] where Rubisco stability was associated with down-regulation of HSP60 and -21 in winter wheat.
5.2.2. Drought Stress
Drought is the crucial and threatening abiotic factor that limits the productivity of many crops in the current changing climatic conditions. This stress, in combination with other abiotic stresses such as high light and temperature stress, negatively affects plant morphological, physiological and molecular characteristics, which leads to lowered photosynthesis, hormonal imbalance, mineral nutrient starvation and an ultimate oxidative stress [230]. Removal of water disrupts the normal structure of the lipid bilayer plasma membrane. This results in the displacement of membrane proteins, denaturation of membrane-based enzymes and, as a result, membrane permeability, physiology and metabolism are adversely affected [231,232]. Dehydration stress also affects the quantity and quality of normal plant proteins and, as a result, stress related proteins including HSPs are induced. HSP70 was up-regulated in drought stress in the seedling of upland rice [207]. Similarly, transgenic Arabidopsis and sugarcane also showed HSP up-regulation and demonstrated drought tolerance [204,211]. The expression pattern of HSPs is also genotype-specific. Burke et al., (1985) [212] studied combined drought and heat stress in irrigated and non-irrigated cotton, where more HSPs accumulated in non-irrigated cotton. Maize heat tolerant and sensitive cultivars were studied under high temperature and dehydration situations, where HSP accumulation was more in drought stress conditions [233]. The same was demonstrated by Benesova et al., (2012) [39], where HSP70 and HSP26 were induced in drought-stressed maize. A study on chickpea HSP70 reported that HSPs were first down-regulated in the early stage of growth in drought-tolerant cultivars. To contrast, HSPs were abundant in drought-sensitive cultivars, which indicated that HSPs responded to drought not only in the specific genotypes but, also, during the developmental stage. Similarly, small HSPs expressed highly in drought-tolerant cultivars as compared to those that were sensitive in chickpea [210]. The same trend also was observed in poplar and Kentucky bluegrass [160,213]. HSP17.7 showed drought tolerance in transgenic rice, and other HSPs also were involved in the acclimation of bryophytes to drought stress [209,234]. Proteomics studies revealed that nuclear and HSPs in the extracellular matrix were both up-regulated to drought stress [235,236,237,238].
5.2.3. Salinity Stress
Increased level of salt in cultivable land is also a limiting factor for agricultural production. Twenty percent of cultivable land, and nearly half of the world irrigated land, is affected by salinity [128]. Studies show that many HSPs are induced and up-regulated in saline stress situations like HSP70 in rice seedlings [197], wheat [195], and poplar HSP70-9,-12 and -33 [203]. Furthermore, HSP40 in rice [165] and poplar, HSP100-21 and -75), HSP90-9 and -12), HSP60-31, -33, -38 and -49), HSP40-113 and -117, and HSP21 were also up-regulated under salt stress [203]. Seen in wheat hybrid Jinan 177 and its salt-resistant hybrid, protein profiling showed HSPs and chaperones were induced highly under salt stress [196]. HSPs were studied in relation to programmed cell death (PCD) in a rice root higher salt situation, where mitochondrial HSP70 were the up-regulated proteins that possibly were involved in PCD regulation [198]. Soybean proteomic studies showed a differential HSPs expression of HSP90, chloroplast HSP70, HSP60 and HSP20 under salt stress [201]. Different HSPs in Arabidopsis like HSP 90 [146,239], HSP100, Clp (B1, B2), Clp (D1, D2) and small HSPs in rice [199,240] showed tolerance to high salinity stress. The role of HSPs in response to salinity stress is also genotype-specific, as recorded in soybean, where HSPs were induced more in salt resistant cultivars [202].
5.2.4. Light Stress
Plants, being autotrophs, require light for photosynthesis. Excess light damages the photosynthetic apparatus and plants undergo a phenomenon known as photorespiration. During this process, toxic chemicals rather than sugars, along with ROS, are produced. These toxic chemicals in chloroplast could damage the photosystem II permanently by excessive absorption of light [241]. Rossel et al., (2002) [215] reported many HSPs were up-regulated upon high light stress in Arabidopsis. A similar over-accumulation of nuclear HSP70 was observed in Chlamydomonas [216]. The thylakoid proteome analysis of Arabidopsis was studied with respect to high light saturation involving isoforms of chloroplast HSP70 along with the accumulation of other osmolytes like anthocyanins and ascorbates [214]. Seen in the marine ecosystem where low light created a stress, HSP70, ClpB1, Sti, and HSP60 were up-regulated [242]. Under HLS, small HSP23 was seen to be involved in the post-transcriptional regulation in Chenopodium rubrum cell suspension [69].
5.2.5. Chemical Pollutant Stress
Plant productivity is restricted by chemical pollutants in the soil media, such as heavy metals. These pollutants affect plant growth either by displacement of essential cations from specific binding sites or generation of oxidative stress by the generation of ROS. One direct way of disruption was, upon uptake into the cell, a direct reaction took place with proteins due to an affinity for Thionyl-, Histidyl- and Carboxyl- groups [243,244].
Many HSPs were induced by heavy metal stress. HSP70s were differentially expressed and accumulated in the roots of tomatoes [217]. Similarly, the HSP70 sub-family, DnaK (Bip), was up-regulated in rice seedlings [218]. Arabidopsis exposure to cadmium stress induced many HSPs [222]. Similarly, increased expression was reported for HSP80 and HSP17.9 in rice [219], HSP 90s in Lotus corniculatus [245], HSP17.7 in carrots [226] and HSP26 in soybeans [220] under cadmium, lead and arsenic stresses. Using a comparative proteomic analysis of poplar under cadmium stress, a differential expression pattern of HSP was noted. Similarly, in soybeans, two-folds higher accumulation of HSP was recorded in Cd-accumulating genotypes, while there was less HSP70 expression in lower Cd-accumulating varieties, which showed that HSP expression was also genotype-specific [246]. When flax was cultured on heavy metal treated media, many heavy metal binding proteins, including HSP70 accumulation, were enhanced, while HSP83 showed a down-regulation [168]. HSP90.3 enhanced Cadmium stress tolerance by lowering germination potential in Arabidopsis, mediating the antioxidant enzymes [247].
5.2.6. Flooding Stress
Waterlogging/flooding is also an environmental limiting factor that hinders plant growth and development. A gradual decrease of redox potential and oxygen in the soil are the ill effects of flooding [248]. Studies show that HSPs are involved in plant resistance against flooding stress by up-regulation and higher gene expression, which is organ-specific. As noticed by Chen et al., (2014) [228] in the soybean plasma membrane where HSP70 accumulated more than 10 folds, this occurrence was more in cotyledon than the roots of the soybean. Discussed in another proteomic study by the same group of researchers, HSP60 was differentially regulated in soybeans [229]. To contrast, HSPs were induced in flooding stress, but were not mandatory for resistance in flooding stress and were genotype specific. Regarding resistant and susceptible cultivars of rice to anoxia and hypoxia conditions, HSPs were more up-regulated in the sensitive cultivars than resistant genotypes [230]. Proteomics study of flooding stress in relation to PCD in maize revealed that HSP70s were more up-regulated [231]. The same pattern of flooding tolerance was studied in rice protoplast where ectopic mtHSP70 expression protected H2O2 induced PCD [230]. Similarly, Arabidopsis showed that anoxia tolerance was enhanced via HSFA2-mediated production of HSP70 and HSP101 [224], which indicated the involvement and multifarious role of HSPs in flooding situations.
5.2.7. Oxidative/Combined Stress
Since plants are exposed to many stresses simultaneously, such as light, this creates high temperature stress that leads to dehydration. Such situations lead to oxidative or secondary stress and plants have to adjust their signaling pathways and metabolisms to ensure their growth and development [132,197,249,250]. Oxidative stress generates ROS which, in high concentrations, are harmful to cellular structures. HSPs respond to multiple stress situations and enable the plants to cope with the challenging environment. HSP70 expression was higher in tobacco to heat stress but was even higher to the combined stress of heat and drought [251]. Ectopic expression of genes from soybeans in Arabidopsis GmHSP90 showed tolerance to heat, salinity and osmotic stresses, although response in salinity was not as high as to combined stresses [146]. A similar pattern was observed with small HSPs in rice to multiple stresses [252,253]. Overexpression of HSP17.6 in Arabidopsis enhanced tolerance to salinity combined with dehydration, but no response was noted to high temperature stress only [254]. Single or combined stresses led to the production of ROS and oxidative stress which, if not checked timely, are very detrimental to plants [248] (Figure 3). Under oxidative stress, overexpression of organelle and cytosolic HSP90 enhanced tolerance in Arabidopsis. Similar results were reported by Nishizawa-Yokoi et al., (2010) [255], where HSP90 regulated HSFA2, which enhanced tolerance to oxidative stress. Queitsch et al., (2000) [135] reported oxidative stress accumulated HSP100/Clp B, ClpC2 and ClpD1 in rice. HSPs protected vital cellular parts under oxidative stress, as demonstrated by Downs et al., (1999) [256], where small HSPs protected the photosystem II from oxidative stress and photo-inhibition. Different organelle HSPs also responded to oxidative stress. mtHSP22 accumulation was enhanced in tomatoes under oxidative stress [257]. Small HSPs responded to oxidative stress, as HSP16.4 and HSP17 accumulated in multiple stress situations in Arabidopsis and carrots, respectively [16,223].
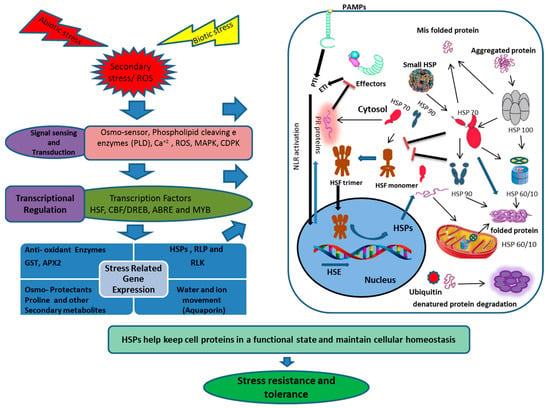
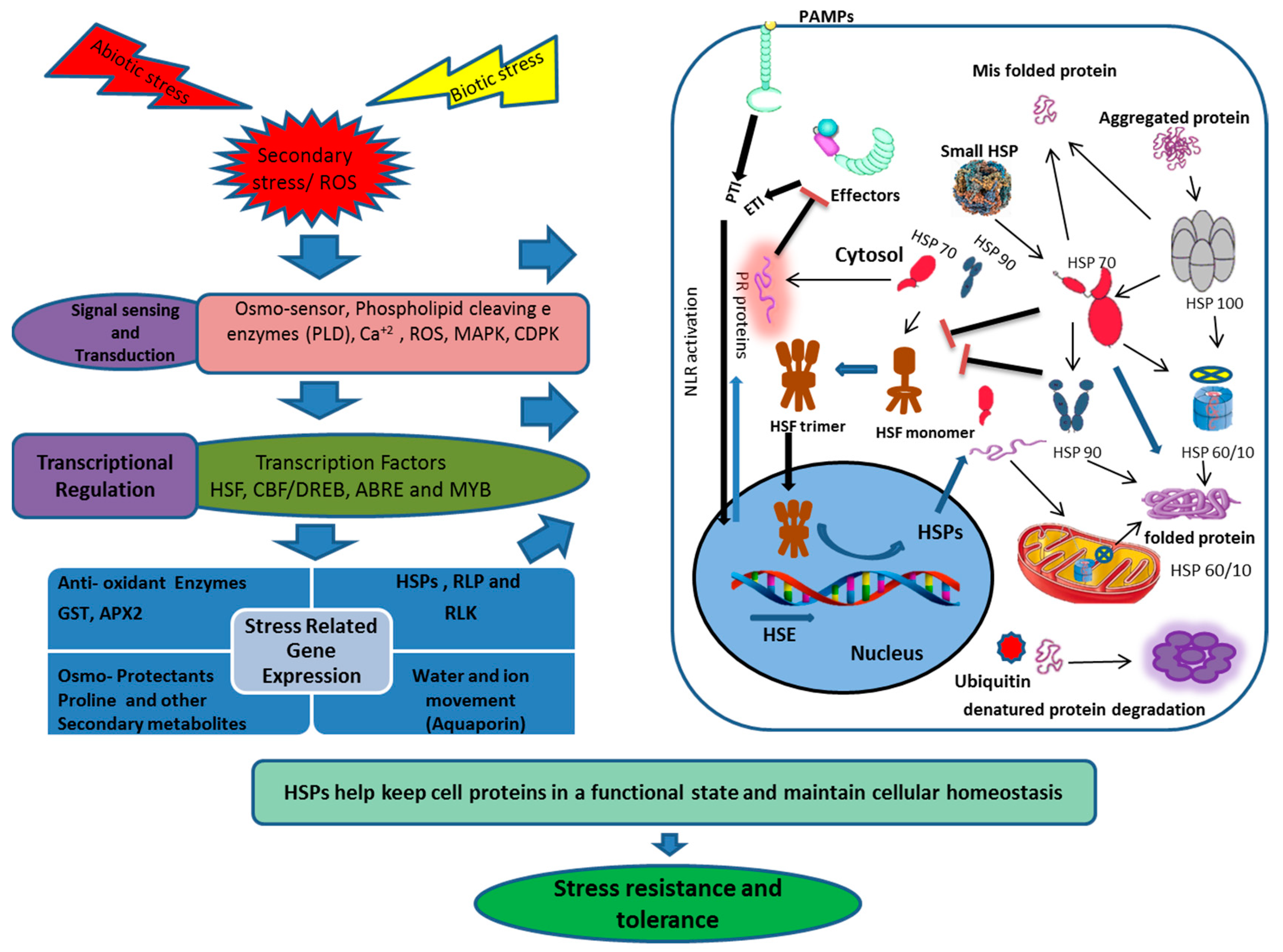
Figure 3.
Schematic presentation of the HSP transcriptional regulation, transport and disposal, under biotic and abiotic stresses. The diagram integrates both positive (Arrows) and negative (Bars) regulatory mechanisms. Biotic and abiotic stresses provoke the HSFs through calcium accumulation, recognition of invading pathogen effector proteins, ROS or misfolding and aggregation of cell proteins, which results in activation of HSP and other stress responsive proteins. ROS (Reactive oxygen species), PLD (Phospholipase D), MAPK (Mitogen-activated protein kinase), CDPK (Calcium-dependent protein kinase), HSF (Heat shock factor), CBF (C-repeat binding factor), DREB (Dehydration response element binding protein), ABRE (Abscisic acid-responsive element), MYB (Myeloblastosis), HSP (Heat shock protein), RLKs (Receptor-like kinases), RLP (Receptor-like proteins), PAMP (Pathogenesis-associated molecular pattern), PTI (Pattern-triggered immunity), ETI (Effector-triggered immunity) NLR (Node-like receptor protein), GST (Glutathione-s-transferase), APX2(Ascorbate peroxidase 2).
6. Conclusions and Future Prospects
Plants are exposed to several biotic and abiotic stresses, which not only limit the performance of the plants in term of productivity, but also their quality and storability. Plants have evolved various morphological, anatomical, physiological, phonological and molecular level strategies to deal with stress situations (Figure 1). Plants respond to stress at the molecular level by transcriptional regulation of stress-related proteins, including HSPs. HSPs are classified into different classes, based on approximate molecular weight, as HSP100, -90, -70, -60 and small HSPs. HSPs prevent protein aggregation and maintain a non-native protein’s functional conformation and cell homeostasis under stress situations. The majority of HSPs are up-regulated under various biotic and abiotic stresses, while a few are down-regulated (Table 1). Moreover, HSPs as chaperones also play a role in membrane stability [228], using ROS as a signal molecule and scavenging by positively regulating the antioxidant enzymes [85,115,133], along with plant growth and development under normal conditions [43,46,258,259]. Small HSP (Ubiquitin) also is responsible for denatured protein degradation and disposal. Stress response is a complex process; HSPs play a pivotal role in stress response and can be employed in transgenic plant development. Studies on HSPs are limited to model plants under controlled laboratory conditions; many studies in other plant crops are limited to gene expression due to lack of suitable mutants. HSPs have a wide range of members, and every member plays a significant role across different networks. The response of HSP is genotype and tissue specific. so the signal perception and cascade need to be explored fully in control and stress situations. The response of HSPs to biotic stresses is evident, for example, but signal sensing and regulation are not yet fully elucidated.
Someday, a combination of advanced technologies, such as microarray and omics techniques, at different developmental stages, genotypes and under different biotic and abiotic stresses, alone or in combination, will lead us to better understand the role of HSPs and the associated signaling pathways. More studies are needed, with a focus on different important crop plants under natural field conditions, with respect to the HSP network. Additionally, future research should be extended to explore other regulatory mechanisms such as alternative splicing, miRNAs and their interaction and cross talk with the complex HSFs, HSPs, phyto-hormones and protective enzymes on plant growth, development and metabolism, under normal and stress conditions. This will provide avenues for the development of stress-resistant and tolerant crops through biotechnological approaches and molecular breeding.
Author Contributions
Conceptualization, S.u.H. and Z.-H.G., Writing-Original Draft Preparation, S.u.H., Investigation, A.K. and M.A., Software and Visualization, W.-X.G. and H.-X.Z., Writing-Review & Editing, S.u.H., and A.M.K., Resources, Z.-H.G., Supervision, Z.-H.G., Project Administration, Z.-H.G. and A.-M.W., Funding Acquisition, Z.-H.G. and A.-M.W.
Funding
This work was supported through funding from National Key R&D Program of China (No. 2016YFD0101900) and the National Natural Science Foundation of China (No. U1603102 and No. 31272163).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mittler, R. Abiotic Stress, the Field Environment and Stress Combination. Trends Plant Sci. 2006, 11, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Swindell, W.R.; Huebner, M.; Weber, A.P. Transcriptional Profiling of Arabidopsis Heat Shock Proteins and Transcription Factors Reveals Extensive Overlap Between Heat and Non-Heat Stress Response Pathways. BMC Genom. 2007, 8, 125. [Google Scholar] [CrossRef] [PubMed]
- Al-Whaibi, M.H. Plant Heat-Shock Proteins: A Mini Review. J. King Saud Univ. Sci. 2011, 23, 139–150. [Google Scholar] [CrossRef]
- Guo, M.; Liu, J.; Ma, X.; Luo, D.; Gong, Z.; Lu, M. The Plant Heat Stress Transcription Factors (HSFs): Structure, Regulation, and Function in Response to Abiotic Stresses. Front. Plant Sci. 2016, 7, 114. [Google Scholar] [CrossRef]
- Xu, Y.; Zhan, C.; Huang, B. Heat Shock Proteins in Association with Heat Tolerance in Grasses. Int. J. Proteom. 2011, 2011, 529648. [Google Scholar] [CrossRef]
- Dodds, P.; Rathjen, J. Plant Immunity: Towards an Integrated View of Plant-Pathogen Interactions. Nat. Rev. Genet. 2010, 11, 539. [Google Scholar] [CrossRef]
- Li, R.; Rashotte, A.M.; Singh, N.K.; Lawrence, K.S.; Weaver, D.B.; Locy, R.D. Transcriptome Analysis of Cotton (Gossypium hirsutum L.) Genotypes that are Susceptible, Resistant, and Hypersensitive to Reniform Nematode (Rotylenchulus Reniformis). PLoS ONE 2015, 10, e0143261. [Google Scholar] [CrossRef]
- Jones, J.T.; Haegeman, A.; Danchin, E.G.J.; Gaur, H.S.; Helder, J.; Jones, M.G.K.; Kikuchi, T.; Manzanilla-Lopez, R.; Palomares-Rius, J.E.; Wesemael, W.M.L.; et al. Top 10 Plant-Parasitic Nematodes in Molecular Plant Pathology. Mol. Plant Pathol. 2013, 14, 946–961. [Google Scholar] [CrossRef]
- Rybicki, E.P. A Top Ten List for Economically Important Plant Viruses. Arch. Virol. 2015, 160, 17–20. [Google Scholar] [CrossRef]
- Gorovits, R.; Moshe, A.; Amrani, L.; Kleinberger, R.; Anfoka, G.; Czosnek, H. The Six Tomato Yellow Leaf Curl Virus Genes Expressed Individually in Tomato Induce Different Levels of Plant Stress Response Attenuation. Cell Stress Chaperones 2017, 22, 345–355. [Google Scholar] [CrossRef]
- Cheng, L.; Zou, Y.; Ding, S.; Zhang, J.; Yu, X.; Cao, J.; Lu, G. Polyamine Accumulation in Transgenic Tomato Enhances the Tolerance to High Temperature Stress. J. Integr. Plant Biol. 2009, 51, 489–499. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Pathak, A.D.; Gupta, P.S.; Shrivastava, A.K.; Srivastava, A.K. Hydrogen Peroxide-Scavenging Enzymes Impart Tolerance to High Temperature Induced Oxidative Stress in Sugarcane. J. Environ. Biol. 2012, 33, 657. [Google Scholar] [PubMed]
- Wahid, A.; Gelani, S.; Ashraf, M.; Foolad, M.R. Heat Tolerance in Plants: An Overview. Environ. Exp. Bot. 2007, 61, 199–223. [Google Scholar] [CrossRef]
- Rodriguez, M.; Canales, E.; Borras-Hidalgo, O. Molecular Aspects of Abiotic Stress in Plants. Biotecnol. Apl. 2005, 22, 1–10. [Google Scholar]
- Tan, W.; Wei Meng, Q.; Brestic, M.; Olsovska, K.; Yang, X. Photosynthesis is Improved by Exogenous Calcium in Heat-Stressed Tobacco Plants. J. Plant Physiol. 2011, 168, 2063–2071. [Google Scholar] [CrossRef]
- Jiang, C.; Xu, J.; Zhang, H.A.O.; Zhang, X.; Shi, J.; Li, M.I.N.; Ming, F. A Cytosolic Class I Small Heat Shock Protein, RcHSP17. 8, of Rosa Chinensis Confers Resistance to a Variety of Stresses to Escherichia coli, Yeast and Arabidopsis thaliana. Plant Cell Environ. 2009, 32, 1046–1059. [Google Scholar] [CrossRef]
- Demirevska-Kepova, K.; Holzer, R.; Simova-Stoilova, L.; Feller, U. Heat Stress Effects on Ribulose-1, 5-Bisphosphate Carboxylase/Oxygenase, Rubisco Binding Protein and Rubisco Activase in Wheat Leaves. Biol. Plant 2005, 49, 521–525. [Google Scholar] [CrossRef]
- Djanaguiraman, M.; Annie Sheeba, J.; Durga Devi, D.; Bangarusamy, U. Cotton Leaf Senescence can be Delayed by Nitrophenolate Spray Through Enhanced Antioxidant Defence System. J. Agron. Crop. Sci. 2009, 195, 213–224. [Google Scholar] [CrossRef]
- Young, L.W.; Wilen, R.W.; Bonham-Smith, P.C. High Temperature Stress of Brassica napus During Flowering Reduces Micro-And Megagametophyte Fertility, Induces Fruit Abortion, and Disrupts Seed Production. J. Exp. Bot. 2004, 55, 485–495. [Google Scholar] [CrossRef]
- Tubiello, F.N.; Soussana, J.F.; Howden, S.M. Crop and Pasture Response to Climate Change. Proc. Natl. Acad. Sci. USA 2007, 104, 19686–19690. [Google Scholar] [CrossRef]
- Wang, X.; Cai, J.; Liu, F.; Jin, M.; Yu, H.; Jiang, D.; Wollenweber, B.; Dai, T.; Cao, W. Pre-Anthesis High Temperature Acclimation Alleviates the Negative Effects of Post-Anthesis Heat Stress on Stem Stored Carbohydrates Remobilization and Grain Starch Accumulation in Wheat. J. Cereal Sci. 2012, 55, 331–336. [Google Scholar] [CrossRef]
- Karuppanapandian, T.; Wang, H.W.; Prabakaran, N.; Jeyalakshmi, K.; Kwon, M.; Manoharan, K.; Kim, W. 2, 4-Dichlorophenoxyacetic Acid-Induced Leaf Senescence in Mung Bean (Vigna radiata L. Wilczek) and Senescence Inhibition by Co-Treatment with Silver Nanoparticles. Plant Physiol. Biochem. 2011, 49, 168–177. [Google Scholar] [CrossRef] [PubMed]
- Moller, I.M.; Jensen, P.E.; Hansson, A. Oxidative Modifications to Cellular Components in Plants. Annu. Rev. Plant Biol. 2007, 58, 459–481. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Jia, W.; Yang, J.; Ismail, A.M. Role of ABA in Integrating Plant Responses to Drought and Salt Stresses. Field Crop. Res. 2006, 97, 111–119. [Google Scholar] [CrossRef]
- Banon, S.; Fernandez, J.A.; Franco, J.A.; Torrecillas, A.; Alarcon, J.J.; Sanchez-Blanco, M.J. Effects of Water Stress and Night Temperature Preconditioning on Water Relations and Morphological and Anatomical Changes of Lotus Creticus Plants. Sci. Hortic. (Amst) 2004, 101, 333–342. [Google Scholar] [CrossRef]
- Sato, S.; Kamiyama, M.; Iwata, T.; Makita, N.; Furukawa, H.; Ikeda, H. Moderate Increase of Mean Daily Temperature Adversely Affects Fruit Set of Lycopersicon esculentum by Disrupting Specific Physiological Processes in Male Reproductive Development. Ann. Bot. 2006, 97, 731–738. [Google Scholar] [CrossRef]
- Morales, D.; Rodriguez, P.; Dell’Amico, J.; Nicolas, E.; Torrecillas, A.; Sanchez-Blanco, M.J. High-Temperature Preconditioning and Thermal Shock Imposition Affects Water Relations, Gas Exchange and Root Hydraulic Conductivity in Tomato. Biol. Plant 2003, 47, 203. [Google Scholar] [CrossRef]
- Wahid, A.; Close, T.J. Expression of Dehydrins Under Heat Stress and Their Relationship with Water Relations of Sugarcane Leaves. Biol. Plant 2007, 51, 104–109. [Google Scholar] [CrossRef]
- Salvucci, M.E.; Crafts-Brandner, S.J. Inhibition of Photosynthesis by Heat Stress: The Activation State of Rubisco as a Limiting Factor in Photosynthesis. Physiol. Plant 2004, 120, 179–186. [Google Scholar] [CrossRef]
- Wahid, A.; Ghazanfar, A. Possible Involvement of Some Secondary Metabolites in Salt Tolerance of Sugarcane. J. Plant Physiol. 2006, 163, 723–730. [Google Scholar] [CrossRef]
- Wang, L.J.; Li, S.H. Salicylic Acid-Induced Heat or Cold Tolerance in Relation to Ca2+ Homeostasis and Antioxidant Systems in Young Grape Plants. Plant Sci. 2006, 170, 685–694. [Google Scholar] [CrossRef]
- Sharkey, T.D. Effects of Moderate Heat Stress on Photosynthesis: Importance of Thylakoid Reactions, Rubisco Deactivation, Reactive Oxygen Species, and Thermotolerance Provided by Isoprene. Plant Cell Environ. 2005, 28, 269–277. [Google Scholar] [CrossRef]
- Laloum, T.; Martin, G.; Duque, P. Alternative Splicing Control of Abiotic Stress Responses. Trends Plant Sci. 2018, 23, 140–150. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Pan, X.; Cobb, G.P.; Anderson, T.A. Plant Microrna: A Small Regulatory Molecule with Big Impact. Dev. Biol. 2006, 289, 3–16. [Google Scholar] [CrossRef]
- Weinmann, H.; Ottow, E. 7.09-Recent Development in Novel Anticancer Therapies. In Comprehensive Medicinal Chemistry II; Taylor, J.B., Triggle, D.J., Eds.; Elsevier: Oxford, UK, 2007; pp. 221–251. ISBN 978-0-08-045044-5. [Google Scholar]
- Ahuja, I.; De Vos, R.C.H.; Bones, A.M.; Hall, R.D. Plant Molecular Stress Responses Face Climate Change. Trends Plant Sci. 2010, 15, 664–674. [Google Scholar] [CrossRef]
- Balchin, D.; Hayer-Hartl, M.; Hartl, F.U. In Vivo Aspects of Protein Folding and Quality Control. Science 2016, 353, acc4353. [Google Scholar] [CrossRef]
- Hartl, F.U.; Bracher, A.; Hayer-Hartl, M. Molecular c Haperones in Protein Folding and Proteostasis. Nature 2011, 475, 324–332. [Google Scholar] [CrossRef]
- Benesova, M.; Hola, D.; Fischer, L.; Jedelsky, P.L.; Hnilicka, F.; Wilhelmova, N.; Rothova, O.; Kocova, M.; Prochazkova, D.; Honnerova, J. The Physiology and Proteomics of Drought Tolerance in Maize: Early Stomatal Closure as a Cause of Lower Tolerance to Short-Term Dehydration. PLoS ONE 2012, 7, e38017. [Google Scholar] [CrossRef]
- Vierling, E. The Roles of Heat Shock Proteins in Plants. Annu. Rev. Plant Biol. 1991, 42, 579–620. [Google Scholar] [CrossRef]
- Eck, E.R.H.B.; Ettig, S.E.F.; Nake, C.L.K.; Artig, K.A.H.; Hattarai, T.R.B. Specifi c and Unspecifi c Responses of Plants to Cold and Drought Stress. J. Biosci. 2007, 32, 501–510. [Google Scholar] [CrossRef]
- Lehesranta, S.J.; Davies, H.V.; Shepherd, L.V.T.; Koistinen, K.M.; Massat, N.; Nunan, N.; McNicol, J.W.; Karenlampi, S.O. Proteomic Analysis of the Potato Tuber Life Cycle. Proteomics 2006, 6, 6042–6052. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, L.; Narula, K.; Basu, S.; Shekhar, S.; Ghosh, S.; Datta, A.; Chakraborty, N.; Chakraborty, S. Comparative Proteomics Reveals a Role for Seed Storage Protein AmA1 in Cellular Growth, Development, and Nutrient Accumulation. J. Proteome Res. 2013, 12, 4904–4930. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, L.; Chakraborty, S.; Jaiswal, D.K.; Gupta, S.; Datta, A.; Chakraborty, N. Comparative Proteomics of Tuber Induction, Development and Maturation Reveal the Complexity of Tuberization Process in Potato (Solanum tuberosum L.). J. Proteome Res. 2008, 7, 3803–3817. [Google Scholar] [CrossRef] [PubMed]
- Ahn, Y.J.; Zimmerman, J.L. Introduction of the Carrot HSP17.7 into Potato (Solanum tuberosum L.) Enhances Cellular Membrane Stability and Tuberization in Vitro. Plant Cell Environ. 2006, 29, 95–104. [Google Scholar] [CrossRef]
- Shekhar, S.; Mishra, D.; Gayali, S.; Buragohain, A.K.; Chakraborty, S.; Chakraborty, N. Comparison of Proteomic and Metabolomic Profiles of Two Contrasting Ecotypes of Sweetpotato (Ipomoea batata L.). J. Proteom. 2016, 143, 306–317. [Google Scholar] [CrossRef]
- Waters, E.R.; Lee, G.J.; Vierling, E. Evolution, Structure and Function of the Small Heat Shock Proteins in Plants. J. Exp. Bot. 1996, 47, 325–338. [Google Scholar] [CrossRef]
- Boston, R.S.; Viitanen, P.V.; Vierling, E. Molecular Chaperones and Protein Folding in Plants. In Post-Transcriptional Control of Gene Expression in Plants; Springer: Berlin, Germany, 1996; pp. 191–222. [Google Scholar]
- Zhuang, L.; Cao, W.; Wang, J.; Yu, J.; Yang, Z.; Huang, B. Characterization and Functional Analysis of FaHsfC1b from Festuca arundinacea Conferring Heat Tolerance in Arabidopsis. Int. J. Mol. Sci. 2018, 19, 2702. [Google Scholar] [CrossRef]
- Mishra, S.K.; Tripp, J.; Winkelhaus, S.; Tschiersch, B.; Theres, K.; Nover, L.; Scharf, K.D. In the Complex Family of Heat Stress Transcription Factors, HsfA1 Has a Unique Role as Master Regulator of Thermotolerance in Tomato. Genes Dev. 2002, 16, 1555–1567. [Google Scholar] [CrossRef]
- Kotak, S.; Larkindale, J.; Lee, U.; Von Koskull-Doring, P.; Vierling, E.; Scharf, K.D. Complexity of the Heat Stress Response in Plants. Curr. Opin. Plant Biol. 2007, 10, 310–316. [Google Scholar] [CrossRef]
- Scharf, K.D.; Berberich, T.; Ebersberger, I.; Nover, L. The Plant Heat Stress Transcription Factor (HSF) Family: Structure, Function and Evolution. Biochim. Biophys. Acta Gene Regul. Mech. 2012, 1819, 104–119. [Google Scholar] [CrossRef]
- Singh, R.K.; Jaishankar, J.; Muthamilarasan, M.; Shweta, S.; Dangi, A.; Prasad, M. Genome-Wide Analysis of Heat Shock Proteins in C 4 Model, Foxtail Millet Identifies Potential Candidates for Crop Improvement Under Abiotic Stress. Sci. Rep. 2016, 6, 32641. [Google Scholar] [CrossRef] [PubMed]
- Shaw, A.K.; Bhardwaj, P.K.; Ghosh, S.; Roy, S.; Saha, S.; Sherpa, A.R.; Saha, S.K.; Hossain, Z. β-Aminobutyric Acid Mediated Drought Stress Alleviation in Maize (Zea mays L.). Environ. Sci. Pollut. Res. 2016, 23, 2437–2453. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Vinocur, B.; Shoseyov, O.; Altman, A. Role of Plant Heat-Shock Proteins and Molecular Chaperones in the Abiotic Stress Response. Trends Plant Sci. 2004, 9, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Ratajczak, E.; Zietkiewicz, S.; Liberek, K. Distinct Activities of Escherichia Coli Small Heat Shock Proteins IbpA and IbpB Promote Efficient Protein Disaggregation. J. Mol. Biol. 2009, 386, 178–189. [Google Scholar] [CrossRef]
- Tyedmers, J.; Mogk, A.; Bukau, B. Cellular Strategies for Controlling Protein Aggregation. Nat. Rev. Mol. Cell Biol. 2010, 11, 777–788. [Google Scholar] [CrossRef]
- Ritosa, F. A New Puffing Pattern Induced by Heat Shock and DNP in Drosophi1ae. Cell. Mol. Life Sci. 1962, 12, 571–573. [Google Scholar] [CrossRef]
- Tissieres, A.; Mitchell, H.K.; Tracy, U.M. Protein Synthesis in Salivary Glands of Drosophila melanogaster: Relation to Chromosome Puffs. J. Mol. Biol. 1974, 84, 389–398. [Google Scholar] [CrossRef]
- McAlister, L.; Finkelstein, D.B. Heat Shock Proteins and Thermal Resistance in Yeast. Biochem. Biophys. Res. Commun. 1980, 93, 819–824. [Google Scholar] [CrossRef]
- Yamamori, T.; Yura, T. Genetic Control of Heat-Shock Protein Synthesis and its Bearing on Growth and Thermal Resistance in Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 1982, 79, 860–864. [Google Scholar] [CrossRef]
- Lin, C.Y.; Roberts, J.K.; Key, J.L. Acquisition of Thermotolerance in Soybean Seedlings: Synthesis and Accumulation of Heat Shock Proteins and Their Cellular Localization. Plant Physiol. 1984, 74, 152–160. [Google Scholar] [CrossRef]
- Bharti, K.; Nover, L. Heat Stress Response in Plants: A Complex Game with Chaperones and more than Twenty Heat Stress Transcription Factors. J. Biosci. 2004, 29, 471–487. [Google Scholar]
- Sterck, L.; Rombauts, S.; Vandepoele, K.; Rouze, P.; Van De Peer, Y. How Many Genes are There in Plants (… and Why are They There)? Curr. Opin. Plant Biol. 2007, 10, 199–203. [Google Scholar] [CrossRef] [PubMed]
- Lee, U.; Rioflorido, I.; Hong, S.; Larkindale, J.; Waters, E.R.; Vierling, E. The Arabidopsis ClpB/Hsp100 Family of Proteins: Chaperones for Stress and Chloroplast Development. Plant J. 2007, 49, 115–127. [Google Scholar] [CrossRef] [PubMed]
- Schoffl, F.; Prandl, R.; Reindl, A. Molecular Responses to Heat Stress; Scientific Research: Wuhan, China, 1999; pp. 81–98. ISBN 1570595631. [Google Scholar]
- Schlesinger, M.J. Heat Shock Proteins. J. Biol. Chem. 1990, 265, 12111–12114. [Google Scholar] [PubMed]
- Liu, D.; Zhang, X.; Cheng, Y.; Takano, T.; Liu, S. RHSP90 Gene Expression in Response to Several Environmental Stresses in Rice (Oryza sativa L.). Plant Physiol. Biochem. 2006, 44, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Korotaeva, N.E.; Antipina, A.I.; Grabelnykh, O.I.; Varakina, N.N.; Borovskii, G.B.; Voinikov, V.K. Mitochondrial Low-Molecular-Weight Heat-Shock Proteins and the Tolerance of Cereal Mitochondria to Hyperthermia. Russ. J. Plant Physiol. 2001, 48, 798–803. [Google Scholar] [CrossRef]
- Nieto-Sotelo, J.; Martinez, L.M.; Ponce, G.; Cassab, G.I.; Alagon, A.; Meeley, R.B.; Ribaut, J.M.; Yang, R. Maize HSP101 Plays Important Roles in Both Induced and Basal Thermotolerance and Primary Root Growth. Plant Cell 2002, 14, 1621–1633. [Google Scholar] [CrossRef]
- Neumann, D.; Emmermann, M.; Thierfelder, J.M.; Zur Nieden, U.; Clericus, M.; Braun, H.P.; Nover, L.; Schmitz, U.K. HSP68—A DnaK-Like Heat-Stress Protein of Plant Mitochondria. Planta 1993, 190, 32–43. [Google Scholar] [CrossRef]
- Young, T.E.; Ling, J.; Geisler-Lee, C.J.; Tanguay, R.L.; Caldwell, C.; Gallie, D.R. Developmental and Thermal Regulation of the Maize Heat Shock Protein, HSP101. Plant Physiol. 2001, 127, 777–791. [Google Scholar] [CrossRef]
- Heckathorn, S.A.; Downs, C.A.; Coleman, J.S. Nuclear-Encoded Chloroplast Proteins Accumulate in the Cytosol During Severe Heat Stress. Int. J. Plant Sci. 1998, 159, 39–45. [Google Scholar] [CrossRef]
- Frova, C.; Taramino, G.; Binelli, G. Heat-Shock Proteins During Pollen Development in Maize. Dev. Genet. 1989, 10, 324–332. [Google Scholar] [CrossRef]
- Ristic, Z.; Williams, G.; Yang, G.; Martin, B.; Fullerton, S. Dehydration, Damage to Cellular Membranes, and Heat-Shock Proteins in Maize Hybrids From Different Climates. J. Plant Physiol. 1996, 149, 424–432. [Google Scholar] [CrossRef]
- Kotak, S.; Vierling, E.; Baumlein, H.; von Koskull-Doring, P. A novel transcriptional cascade regulating expression of heat stress proteins during seed development of Arabidopsis. Plant Cell 2007, 19, 182–195. [Google Scholar] [CrossRef] [PubMed]
- Shamovsky, I.; Nudler, E. New Insights into the Mechanism of Heat Shock Response Activation. Cell. Mol. Life Sci. 2008, 65, 855–861. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Liu, J.H.; Lu, J.P.; Zhai, Y.F.; Wang, H.; Gong, Z.H.; Wang, S.B.; Lu, M.H. Genome-Wide Analysis of the CaHsp20 Gene Family in Pepper: Comprehensive Sequence and Expression Profile Analysis Under Heat Stress. Front. Plant Sci. 2015, 6, 806. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Zhao, X.; Burkholder, W.F.; Gragerov, A.; Ogata, C.M.; Gottesman, M.E.; Hendrickson, W.A. Structural Analysis of Substrate Binding by the Molecular Chaperone DnaK. Science 1996, 272, 1606–1614. [Google Scholar] [CrossRef] [PubMed]
- Akerfelt, M.; Morimoto, R.I.; Sistonen, L. Heat Shock Factors: Integrators of Cell Stress, Development and Lifespan. Nat. Rev. Mol. Cell Biol. 2010, 11, 545–552. [Google Scholar] [CrossRef] [PubMed]
- Pirkkala, L.; Nykanen, P.; Sistonen, L.E.A. Roles of the Heat Shock Transcription Factors in Regulation of the Heat Shock Response and Beyond. FASEB J. 2001, 15, 1118–1131. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Bharadwaj, S.; O’Carroll, R.; Ovsenek, N. HSP90 Interacts with and Regulates the Activity of Heat Shock Factor 1 in Xenopus oocytes. Mol. Cell. Biol. 1998, 18, 4949–4960. [Google Scholar] [CrossRef]
- Zou, J.; Guo, Y.; Guettouche, T.; Smith, D.F.; Voellmy, R. Repression of Heat Shock Transcription Factor HSF1 Activation by HSP90 (HSP90 Complex) that Forms a Stress-Sensitive Complex with HSF1. Cell 1998, 94, 471–480. [Google Scholar] [CrossRef]
- Calderwood, S.K.; Xie, Y.; Wang, X.; Khaleque, M.A.; Chou, S.D.; Murshid, A.; Prince, T.; Zhang, Y. Signal Transduction Pathways Leading to Heat Shock Transcription. Sign. Transduct. Insights 2010, 2, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Driedonks, N.; Xu, J.; Peters, J.L.; Park, S.; Rieu, I. Multi-Level Interactions Between Heat Shock Factors, Heat Shock Proteins, and the Redox System Regulate Acclimation to Heat. Front. Plant Sci. 2015, 6, 999. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Kim, Y.; Song, L.; Coutu, J.; Coutu, A.; Ciftci-Yilmaz, S.; Lee, H.; Stevenson, B.; Zhu, J.K. Gain-And Loss-Of-Function Mutations in Zat10 Enhance the Tolerance of Plants to Abiotic Stress. FEBS Lett. 2006, 580, 6537–6542. [Google Scholar] [CrossRef] [PubMed]
- Wahid, A. Physiological Implications of Metabolite Biosynthesis for Net Assimilation and Heat-Stress Tolerance of Sugarcane (Saccharum officinarum) Sprouts. J. Plant Res. 2007, 120, 219–228. [Google Scholar] [CrossRef]
- Singla, J.; Krattinger, S.G. Biotic Stress Resistance Genes in Wheat; Elsevier: Amsterdam, The Netherlands, 2016; ISBN 978-0-08-100596-5. [Google Scholar]
- Koeck, M.; Hardham, A.R.; Dodds, P.N. The Role of Effectors of Biotrophic and Hemibiotrophic Fungi in Infection. Cell. Microbiol. 2011, 13, 1849–1857. [Google Scholar] [CrossRef]
- Kubienova, L.; Sedlarova, M.; Viteckova Wunschova, A.; Piterkova, J.; Luhova, L.; Mieslerova, B.; Lebeda, A.; Navratil, M.; Petrivalsky, M. Effect of extreme temperatures on Powdery Mildew development and Hsp70 induction in Tomato and wild Solanum spp. Plant Prot. Sci. 2013, 49, S41–S54. [Google Scholar] [CrossRef]
- Jelenska, J.; Van Hal, J.A.; Greenberg, J.T. Pseudomonas syringae Hijacks Plant Stress Chaperone Machinery for Virulence. Proc. Natl. Acad. Sci. USA 2010, 107, 13177–13182. [Google Scholar] [CrossRef]
- Sarkar, N.K.; Kim, Y.K.; Grover, A. Rice sHsp Genes: Genomic Organization and Expression Profiling Under Stress and Development. BMC Genom. 2009, 10, 393. [Google Scholar] [CrossRef]
- Van Ooijen, G.; Lukasik, E.; Van Den Burg, H.A.; Vossen, J.H.; Cornelissen, B.J.C.; Takken, F.L.W. The Small Heat Shock Protein 20 RSI2 Interacts with and is Required for Stability and Function of Tomato Resistance Protein I-2. Plant J. 2010, 63, 563–572. [Google Scholar] [CrossRef]
- Pan, X.; Zhu, B.; Luo, Y.; Fu, D. Unraveling the Protein Network of Tomato Fruit in Response to Necrotrophic Phytopathogenic Rhizopus nigricans. PLoS ONE 2013, 8, e73034. [Google Scholar] [CrossRef]
- Ahmed, A.A.; Pedersen, C.; Schultz-Larsen, T.; Kwaaitaal, M.; Jorgensen, H.J.L.; Thordal-Christensen, H. Barley Powdery Mildew Effector Candidate CSEP0105 Inhibits Chaperone Activity of Small Heat Shock Protein. Plant Physiol. 2015, 168, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Jiang, N.; Liu, J.; Liu, W.; Wang, G.L. The Role of Effectors and Host Immunity in Plant-Necrotrophic Fungal Interactions. Virulence 2014, 5, 722–732. [Google Scholar] [CrossRef] [PubMed]
- Foley, R.C.; Gleason, C.A.; Anderson, J.P.; Hamann, T.; Singh, K.B. Genetic and Genomic Analysis of Rhizoctonia solani Interactions with Arabidopsis; Evidence of Resistance Mediated Through NADPH Oxidases. PLoS ONE 2013, 8, e56814. [Google Scholar] [CrossRef] [PubMed]
- Husselmann, L.H.H. Analysis of the Early Events in the Interaction between Venturia inaequalis and the Susceptible Golden Delicious Apple (Malus domestica Borkh.). Ph.D. Thesis, University of the Western Cape, Belleville, Cape Town, Western Cape, South Africa, 2014. [Google Scholar]
- Piterkova, J.; Luhova, L.; Mieslerova, B.; Lebeda, A.; Petrivalsky, M. Nitric Oxide and Reactive Oxygen Species Regulate the Accumulation of Heat Shock Proteins in Tomato Leaves in Response to Heat Shock and Pathogen Infection. Plant Sci. 2013, 207, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Yogendra, K.N.; Kumar, A.; Sarkar, K.; Li, Y.; Pushpa, D.; Mosa, K.A.; Duggavathi, R.; Kushalappa, A.C. Transcription Factor StWRKY1 Regulates Phenylpropanoid Metabolites Conferring Late Blight Resistance in Potato. J. Exp. Bot. 2015, 66, 7377–7389. [Google Scholar] [CrossRef]
- Kallamadi, P.R.; Dandu, K.; Kirti, P.B. An Insight into Powdery Mildew–Infected, Susceptible, Resistant and Immune Sunflower Genotypes. Proteomics 2018, 18, 1700418. [Google Scholar] [CrossRef]
- Thomma, B.P.H.J.; Nurnberger, T.; Joosten, M.H.A.J. Of PAMPs and Effectors: The Blurred PTI-ETI Dichotomy. Plant Cell 2011, 23, 4–15. [Google Scholar] [CrossRef]
- Maimbo, M.; Ohnishi, K.; Hikichi, Y.; Yoshioka, H.; Kiba, A. Induction of a Small Heat Shock Protein and its Functional Roles in Nicotiana Plants in the Defense Response Against Ralstonia solanacearum. Plant Physiol. 2007, 145, 1588–1599. [Google Scholar] [CrossRef]
- Garofalo, C.G.; Garavaglia, B.S.; Dunger, G.; Gottig, N.; Orellano, E.G.; Ottado, J. Expression Analysis of Small Heat Shock Proteins During Compatible and Incompatible Plant-Pathogen Interactions. Adv. Stud. Biol. 2009, 5, 197–205. [Google Scholar]
- Bricchi, I.; Bertea, C.M.; Occhipinti, A.; Paponov, I.A.; Maffei, M.E. Dynamics of Membrane Potential Variation and Gene Expression Induced by Spodoptera littoralis, Myzus persicae, and pseudomonas syringae in Arabidopsis. PLoS ONE 2012, 7, e46673. [Google Scholar] [CrossRef]
- Pavlova, E.L.; Rikhvanov, E.G.; Tauson, E.L.; Varakina, N.N.; Gamburg, K.Z.; Rusaleva, T.M.; Borovskii, G.B.; Voinikov, V.K. Effect of Salicylic Acid on the Development of Induced Thermotolerance and Induction of Heat Shock Protein Synthesis in the Arabidopsis Thaliana Cell Culture. Russ. J. Plant Physiol. 2009, 56, 68–73. [Google Scholar] [CrossRef]
- Ito, M.; Ohnishi, K.; Hikichi, Y.; Kiba, A. Molecular Chaperons and Co-Chaperons, Hsp90, RAR1, and SGT1 Negatively Regulate Bacterial wilt Disease Caused by Ralstonia solanacearum in Nicotiana benthamiana. Plant Signal. Behav. 2014, 10, e970410. [Google Scholar] [CrossRef] [PubMed]
- Alam, S.B.; Rochon, D. Cucumber Necrosis Virus Recruits Cellular Heat Shock Protein 70 Homologs at Several Stages of Infection. J. Virol. 2016, 90, 3302–3317. [Google Scholar] [CrossRef] [PubMed]
- Gorovits, R.; Moshe, A.; Ghanim, M.; Czosnek, H. Recruitment of the Host Plant Heat Shock Protein 70 by Tomato Yellow Leaf Curl Virus Coat Protein is Required for Virus Infection. PLoS ONE 2013, 8, e70280. [Google Scholar] [CrossRef]
- Whitham, S.A.; Quan, S.; Chang, H.; Cooper, B.; Estes, B.; Zhu, T.; Wang, X.; Hou, Y. Diverse RNA Viruses Elicit the Expression of Common Sets of Genes in Susceptible Arabidopsis Thaliana Plants. Plant J. 2003, 33, 271–283. [Google Scholar] [CrossRef]
- Gorovits, R.; Czosnek, H. The Involvement of Heat Shock Proteins in the Establishment of Tomato Yellow Leaf Curl Virus Infection. Front. Plant Sci. 2017, 8, 355. [Google Scholar] [CrossRef]
- Chen, Z.; Zhou, T.; Wu, X.; Hong, Y.; Fan, Z.; Li, H. Influence of Cytoplasmic Heat Shock Protein 70 on Viral Infection of Nicotiana benthamiana. Mol. Plant Pathol. 2008, 9, 809–817. [Google Scholar] [CrossRef]
- Mine, A.; Hyodo, K.; Tajima, Y.; Kusumanegara, K.; Taniguchi, T.; Kaido, M.; Mise, K.; Taniguchi, H.; Okuno, T. Differential Roles of Hsp70 and Hsp90 in the Assembly of the Replicase Complex of a Positive-Strand RNA Plant Virus. J. Virol. 2012, 86, 91–104. [Google Scholar] [CrossRef]
- Senthil, G.; Liu, H.; Puram, V.G.; Clark, A.; Stromberg, A.; Goodin, M.M. Specific and Common Changes in Nicotiana benthamiana Gene Expression in Response to Infection by Enveloped Viruses. J. Gen. Virol. 2005, 86, 2615–2625. [Google Scholar] [CrossRef]
- Yu, L.; Wang, W.; Zeng, S.; Chen, Z.; Yang, A.; Shi, J.; Zhao, X.; Song, B. Label-Free Quantitative Proteomics Analysis of Cytosinpeptidemycin Responses in Southern Rice Black-Streaked Dwarf Virus-Infected Rice. Pestic. Biochem. Physiol. 2018, 147, 20–26. [Google Scholar] [CrossRef]
- Jiang, S.; Lu, Y.; Li, K.; Lin, L.; Zheng, H.; Yan, F.; Chen, J. Heat Shock Protein 70 is Necessary for R ice Stripe Virus Infection in Plants. Mol. Plant Pathol. 2014, 15, 907–917. [Google Scholar] [CrossRef] [PubMed]
- Makarova, S.; Makhotenko, A.; Spechenkova, N.; Love, A.J.; Kalinina, N.O.; Taliansky, M. Interactiver Responses of Potato (Solanum tuberosum L.) Plants to Heat Stress and Infection with Potato Virus Y. Front. Microbiol. 2018, 9, 2582. [Google Scholar] [CrossRef] [PubMed]
- Calderwood, S.K. Heat Shock Proteins and Plants; Springer International Publishing: Cham, Switzerland; Boston, MA, USA, 2016; ISBN 9783319463391. [Google Scholar]
- Lopes-Caitar, V.S.; De Carvalho, M.C.C.G.; Darben, L.M.; Kuwahara, M.K.; Nepomuceno, A.L.; Dias, W.P.; Abdelnoor, R.V.; Marcelino-Guimaraes, F.C. Genome-Wide Analysis of the Hsp 20 Gene Family in Soybean: Comprehensive Sequence, Genomic Organization and Expression Profile Analysis Under Abiotic and Biotic Stresses. BMC Genom. 2013, 14, 577. [Google Scholar] [CrossRef] [PubMed]
- Fuganti, R.; Machado, M.D.F.P.D.; Lopes, V.S.; Silva, J.F.V.; Arias, C.A.A.; Marin, S.R.R.; Binneck, E.; Abdelnoor, R.V.; Marcelino, F.C.; Nepomuceno, A.L. Size of A T (n) Insertions in Promoter Region Modulates Gmhsp17.6-L mRNA Transcript Levels. Biomed. Res. Int. 2010, 2010, 847673. [Google Scholar] [CrossRef]
- Kandoth, P.K.; Ithal, N.; Recknor, J.; Maier, T.; Nettleton, D.; Baum, T.J.; Mitchum, M.G. The Soybean Rhg1 Locus for Resistance to the Soybean Cyst Nematode Heterodera Glycines Regulates Expression of a Large Number of Stress-And Defense-Related Genes in Degenerating Feeding Cells. Plant Physiol. 2011, 155, 960–975. [Google Scholar] [CrossRef]
- Escobar, C.; Barcala, M.; Portillo, M.; Almoguera, C.; Jordano, J.; Fenoll, C. Induction of the Hahsp17. 7G4 Promoter by Root-Knot Nematodes: Involvement of Heat-Shock Elements in Promoter Activity in Giant Cells. Mol. Plant Microbe Interact. 2003, 16, 1062–1068. [Google Scholar] [CrossRef]
- Barcala, M.; Garcia, A.; Cubas, P.; Almoguera, C.; Jordano, J.; Fenoll, C.; Escobar, C. Distinct Heat-Shock Element Arrangements that Mediate the Heat Shock, But Not the Late-Embryogenesis Induction of Small Heat-Shock Proteins, Correlate with Promoter Activation in Root-Knot Nematode Feeding Cells. Plant Mol. Biol. 2008, 66, 151–164. [Google Scholar] [CrossRef]
- Bhattarai, K.K.; Li, Q.; Liu, Y.; Dinesh-Kumar, S.P.; Kaloshian, I. The MI-1-Mediated Pest Resistance Requires Hsp90 and Sgt1. Plant Physiol. 2007, 144, 312–323. [Google Scholar] [CrossRef]
- Lourenço-Tessutti, I.; Antonino De Souza, J., Jr.; Martins-De-SA, D.; Viana, A.A.; Carneiro, R.; Togawa, R.; Engler, J.; Batista, J.; Cristina Mattar Silva, M.; Fragoso, R.; et al. Knock-Down of Heat-Shock Protein 90 and Isocitrate Lyase gene expression reduced Root-Knot Nematode reproduction. Phytopathology 2015, 105, 628–637. [Google Scholar] [CrossRef]
- Li, J.; Xiang, C.Y.; Yang, J.; Chen, J.P.; Zhang, H.M. Interaction of HSP20 with a Viral RdRp Changes its Sub-Cellular Localization and Distribution Pattern in Plants. Sci. Rep. 2015, 11, 14016. [Google Scholar] [CrossRef]
- Bolhassani, A.; Agi, E. Heat shock proteins in infection. Clin. Chim. Acta 2019, 498, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Plant Salt Tolerance. Trends Plant Sci. 2001, 6, 66–71. [Google Scholar] [CrossRef]
- Reguera, M.; Peleg, Z.; Blumwald, E. Targeting Metabolic Pathways for Genetic Engineering Abiotic Stress-Tolerance in Crops. Biochim. Biophys. Acta. Gene Regul. Mech. 2012, 1819, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Bita, C.; Gerats, T. Plant Tolerance to High Temperature in a Changing Environment: Scientific Fundamentals and Production of Heat Stress-Tolerant Crops. Front. Plant Sci. 2013, 4, 273. [Google Scholar] [CrossRef] [PubMed]
- Fragkostefanakis, S.; Roeth, S.; Schleiff, E.; Scharf, K. Prospects of Engineering Thermotolerance in Crops Through Modulation of Heat Stress Transcription Factor and Heat Shock Protein Networks. Plant Cell Environ. 2015, 38, 1881–1895. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Finka, A.; Goloubinoff, P. How Do Plants Feel the Heat? Trends Biochem. Sci. 2012, 37, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Lavania, D.; Dhingra, A.; Siddiqui, M.H.; Al-Whaibi, M.H.; Grover, A. Current Status of the Production of High Temperature Tolerant Transgenic Crops for Cultivation in Warmer Climates. Plant Physiol. Biochem. 2015, 86, 100–108. [Google Scholar] [CrossRef]
- Singla, S.L.; Pareek, A.; Grover, A. Plant Hsp100 Family with Special Reference to Rice. J. Biosci. 1998, 23, 337–345. [Google Scholar] [CrossRef]
- Queitsch, C.; Hong, S.W.; Vierling, E.; Lindquist, S. Heat Shock Protein 101 Plays a Crucial Role in Thermotolerance in Arabidopsis. Plant Cell 2000, 12, 479–492. [Google Scholar] [CrossRef]
- Merret, R.; Carpenier, M.C.; Favory, J.J.; Picart, C.; Descombin, J.; Bousquet-Antonelli, C.; Tillard, P.; Lejay, L.; Deragon, J.M.; Charng, Y. Heat-Shock Protein HSP101 Affects the Release of Ribosomal Protein mRNAs for Recovery after Heat Shock. Plant Physiol. 2017, 174, 1216–1225. [Google Scholar] [CrossRef]
- McLoughlin, F.; Basha, E.; Fowler, M.E.; Kim, M.; Bordowitz, J.; Katiyar-Agarwal, S.; Vierling, E. Class I and II Small Heat Shock Proteins Together with HSP101 Protect Protein Translation Factors During Heat Stress. Plant Physiol. 2016, 172, 1221–1236. [Google Scholar] [CrossRef] [PubMed]
- Pyatrikas, D.V.; Rikhvanov, E.G.; Fedoseeva, I.V.; Varakina, N.N.; Rusaleva, T.M.; Tauson, E.L.; Stepanov, A.V.; Borovskii, G.B.; Voinikov, V.K. Mitochondrial Retrograde Regulation of HSP101 Expression in Arabidopsis Thaliana Under Heat Stress and Amiodarone Action. Russ. J. Plant Physiol. 2014, 61, 80–89. [Google Scholar] [CrossRef]
- Dupuis, I.; Dumas, C. Influence of Temperature Stress on in Vitro Fertilization and Heat Shock Protein Synthesis in Maize (Zea mays L.) Reproductive Tissues. Plant Physiol. 1990, 94, 665–670. [Google Scholar] [CrossRef] [PubMed]
- Duck, N.B.; Folk, W.R. Hsp70 Heat Shock Protein Cognate is Expressed and Stored in Developing Tomato Pollen. Plant Mol. Biol. 1994, 26, 1031–1039. [Google Scholar] [CrossRef] [PubMed]
- Sung, D.Y.; Vierling, E.; Guy, C.L. Comprehensive Expression Profile Analysis of the Arabidopsis Hsp70 Gene Family. Plant Physiol. 2001, 126, 789–800. [Google Scholar] [CrossRef] [PubMed]
- Su, P.H.; Li, H. Arabidopsis Stromal 70-kD Heat Shock Proteins are Essential for Plant Development and Important for Thermotolerance of Germinating Seeds. Plant Physiol. 2008, 146, 1231–1241. [Google Scholar] [CrossRef] [PubMed]
- Avelange-Macherel, M.H.; Payet, N.; Lalanne, D.; Neveu, M.; Tolleter, D.; Burstin, J.; Macherel, D. Variability Within a Pea Core Collection of LEAM and HSP22, Two Mitochondrial Seed Proteins Involved in Stress Tolerance. Plant Cell Environ. 2015, 38, 1299–1311. [Google Scholar] [CrossRef]
- Hu, W.; Hu, G.; Han, B. Genome-Wide Survey and Expression Profiling of Heat Shock Proteins and Heat Shock Factors Revealed Overlapped and Stress Specific Response Under Abiotic Stresses in Rice. Plant Sci. 2009, 176, 583–590. [Google Scholar] [CrossRef]
- Prasad, B.D.; Goel, S.; Krishna, P. In Silico Identification of Carboxylate Clamp Type Tetratricopeptide Repeat Proteins in Arabidopsis and Rice as Putative Co-Chaperones of Hsp90/Hsp70. PLoS ONE 2010, 5, e12761. [Google Scholar] [CrossRef]
- Xu, J.; Xue, C.; Xue, D.; Zhao, J.; Gai, J.; Guo, N.; Xing, H. Overexpression of GmHsp90s, a Heat Shock Protein 90 (Hsp90) Gene Family Cloning From Soybean, Decrease Damage of Abiotic Stresses in Arabidopsis Thaliana. PLoS ONE 2013, 8, e69810. [Google Scholar] [CrossRef]
- Yamada, K.; Fukao, Y.; Hayashi, M.; Fukazawa, M.; Suzuki, I.; Nishimura, M. Cytosolic HSP90 Regulates the Heat Shock Response that is Responsible for Heat Acclimation in Arabidopsis Thaliana. J. Biol. Chem. 2007, 282, 37794–37804. [Google Scholar] [CrossRef] [PubMed]
- Jungkunz, I.; Link, K.; Vogel, F.; Voll, L.M.; Sonnewald, S.; Sonnewald, U. AtHsp70-15-Deficient Arabidopsis Plants are Characterized by Reduced Growth, a Constitutive Cytosolic Protein Response and Enhanced Resistance to TuMV. Plant J. 2011, 66, 983–995. [Google Scholar] [CrossRef] [PubMed]
- Song, G.; Yuan, S.; Wen, X.; Xie, Z.; Lou, L.; Hu, B.; Cai, Q.; Xu, B. Transcriptome Analysis of Cd-Treated Switchgrass Root Revealed Novel Transcripts and the Importance of HSF / HSP Network in Switchgrass Cd Tolerance. Plant Cell Rep. 2018, 37, 1485–1497. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Long, R.; Zhang, T.; Wang, Z.; Zhang, F.; Yang, Q.; Kang, J.; Sun, Y. Molecular Cloning and Functional Analysis of the Drought Tolerance Gene MsHSP70 From Alfalfa (Medicago sativa L.). J. Plant Res. 2017, 130, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Liu, J.; Ma, X.; Zhai, Y.; Gong, Z.; Lu, M. Plant Science Genome-Wide Analysis of the Hsp70 Family Genes in Pepper (Capsicum annuum L.) and Functional Identification of CaHsp70-2 Involvement in Heat Stress. Plant Sci. 2016, 252, 246–256. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Cheng, G.; Khan, A.; Wei, A.; Yu, Q.; Yang, S.; Luo, D. CaHSP16.4, a Small Heat Shock Protein Gene in Pepper, is Involved in Heat and Drought Tolerance. Protoplasma 2019, 256, 39–51. [Google Scholar] [CrossRef]
- Usman, M.G.; Rafii, M.Y.; Martini, M.Y.; Yusuff, O.A. Introgression of Heat Shock Protein (Hsp70 and sHsp) Genes into the Malaysian Elite Chilli Variety Kulai (Capsicum annuum L.) Through the Application of Marker-Assisted Backcrossing (MAB). Cell Stress Chaperones 2018, 23, 223–234. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, S.; Lv, J.; Liu, Z.; Wang, Y.; Ma, N.; Meng, Q. SUMO E3 Ligase SlSIZ1 Facilitates Heat Tolerance in Tomato. Plant Cell Physiol. 2017, 59, 58–71. [Google Scholar] [CrossRef]
- Lee, S.S.; Jung, W.Y.; Park, H.J.; Lee, A.; Kwon, S.; Cho, H.S. Genome-Wide Analysis of Alternative Splicing in an Inbred Cabbage (Brassica oleracea L.) line‘HO’in Response to Heat Stress. Curr. Genom. 2018, 19, 12–20. [Google Scholar] [CrossRef]
- Liu, J.; Pang, X.; Cheng, Y.; Yin, Y.; Zhang, Q.; Su, W.; Hu, B. The Hsp70 Gene Family in Solanum tuberosum: Genome-Wide Identification, Phylogeny, and Expression Patterns. Sci. Rep. 2018, 8, 16628. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, M.; Zhang, Q. Proteomic Analysis of the Heat Stress Response in Leaves of Two Contrasting Chrysanthemum Varieties. Plant Omics 2014, 7, 229. [Google Scholar]
- Id, H.W.; Shi, N.; An, X.; Liu, C.; Fu, H.; Cao, L.; Feng, Y. Candidate Genes for Yellow Leaf Color in Common Wheat (Triticum aestivum L.) and Major Related Metabolic Pathways According to Transcriptome Profiling. Int. J. Mol. Sci. 2018, 19, 1594. [Google Scholar] [CrossRef]
- Chen, J.; Gao, T.; Wan, S.; Zhang, Y.; Yang, J.; Yu, Y.; Wang, W. Genome-Wide Identification, Classification and Expression Analysis of the HSP Gene Superfamily in Tea Plant (Camellia sinensis). Int. J. Mol. Sci. 2018, 19, 2633. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Huang, B. Comparative Analysis of Drought Responsive Proteins in Kentucky Bluegrass Cultivars Contrasting in Drought Tolerance. Crop. Sci. 2010, 50, 2543–2552. [Google Scholar] [CrossRef]
- Kumar, N.; Suyal, D.C.; Sharma, I.P.; Verma, A.; Singh, H. Elucidating Stress Proteins in Rice (Oryza sativa L.) Genotype Under Elevated Temperature: A Proteomic Approach to Understand Heat Stress Response. 3 Biotech 2017, 7, 205. [Google Scholar] [CrossRef]
- Liao, J.L.; Zhou, H.W.; Zhang, H.Y.; Zhong, P.A.; Huang, Y.J. Comparative Proteomic Analysis of Differentially Expressed Proteins in the Early Milky Stage of Rice Grains During High Temperature Stress. J. Exp. Bot. 2013, 65, 655–671. [Google Scholar] [CrossRef]
- Lin, C.J.; Li, C.Y.; Lin, S.K.; Yang, F.H.; Huang, J.J.; Liu, Y.H.; Lur, H.S. Influence of High Temperature during Grain Filling on the Accumulation of Storage Proteins and Grain Quality in Rice (Oryza sativa L.). J. Agric. Food Chem. 2010, 58, 10545–10552. [Google Scholar] [CrossRef]
- Majoul, T.; Bancel, E.; Triboi, E.; Ben Hamida, J.; Branlard, G. Proteomic Analysis of the Effect of Heat Stress on Hexaploid Wheat Grain: Characterization of Heat-Responsive Proteins From Non-Prolamins Fraction. Proteomics 2004, 4, 505–513. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, H.; Shao, L.Y.; Yan, X.; Peng, H.; Ouyang, J.X.; Li, S.B. Expression and Function Analysis of a Rice OsHSP40 Gene Under Salt Stress. Genes Genom. 2018, 11, 1–8. [Google Scholar] [CrossRef]
- Hashimoto, M.; Komatsu, S. Proteomic Analysis of Rice Seedlings During Cold Stress. Proteomics 2007, 7, 1293–1302. [Google Scholar] [CrossRef]
- Hlavackova, I.; Vitamvas, P.; Santrueek, J.; Kosova, K.; Zelenkova, S.; Prasil, I.T.; Ovesna, J.; Hynek, R.; Kodicek, M. Proteins Involved in Distinct Phases of Cold Hardening Process in Frost Resistant Winter Barley (Hordeum vulgare L.) Cv Luxor. Int. J. Mol. Sci. 2013, 14, 8000–8024. [Google Scholar] [CrossRef] [PubMed]
- Kosova, K.; Vitamvas, P.; Prasil, I.T.; Renaut, J. Plant Proteome Changes Under Abiotic Stress—Contribution of Proteomics Studies to Understanding Plant Stress Response. J. Proteom. 2011, 74, 1301–1322. [Google Scholar] [CrossRef] [PubMed]
- Bae, M.S.; Cho, E.J.; Choi, E.; Park, O.K. Analysis of the Arabidopsis Nuclear Proteome and its Response to Cold Stress. Plant J. 2003, 36, 652–663. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Zhang, C.; Yang, H.; Yang, Y.; Huang, C.; Tian, Y.; Lu, X. Proteomic Analysis of Cold Stress Responses in Tobacco Seedlings. Afr. J. Biotechnol. 2011, 10, 18991–19004. [Google Scholar]
- Kollipara, K.P.; Saab, I.N.; Wych, R.D.; Lauer, M.J.; Singletary, G.W. Expression Profiling of Reciprocal Maize Hybrids Divergent for Cold Germination and Desiccation Tolerance. Plant Physiol. 2002, 129, 974–992. [Google Scholar] [CrossRef]
- Reddy, R.K.; Chaudhary, S.; Patil, P.; Krishna, P. The 90 kDa Heat Shock Protein (hsp90) is Expressed Throughout Brassica napus Seed Development and Germination. Plant Sci. 1998, 131, 131–137. [Google Scholar] [CrossRef]
- Degand, H.; Faber, A.; Dauchot, N.; Mingeot, D.; Watillon, B.; Van Cutsem, P.; Morsomme, P.; Boutry, M. Proteomic Analysis of Chicory Root Identifies Proteins Typically Involved in Cold Acclimation. Proteomics 2009, 9, 2903–2907. [Google Scholar] [CrossRef]
- Ghosh, D.; Xu, J. Abiotic Stress Responses in Plant Roots: A Proteomics Perspective. Front. Plant Sci. 2014, 5, 6. [Google Scholar] [CrossRef]
- Vitamvas, P.; Prasil, I.T.; Kosova, K.; Planchon, S.; Renaut, J. Analysis of Proteome and Frost Tolerance in Chromosome 5A and 5B Reciprocal Substitution Lines Between Two Winter Wheats During Long-Term Cold Acclimation. Proteomics 2012, 12, 68–85. [Google Scholar] [CrossRef]
- Ono, K.; Hibino, T.; Kohinata, T.; Suzuki, S.; Tanaka, Y.; Nakamura, T.; Takabe, T. Overexpression of DnaK From a Halotolerant Cyanobacterium Aphanothece halophytica Enhances the High-Temperatue Tolerance of Tobacco During Germination and Early Growth. Plant Sci. 2001, 160, 455–461. [Google Scholar] [CrossRef]
- Taylor, N.L.; Heazlewood, J.L.; Day, D.A.; Millar, A.H. Differential Impact of Environmental Stresses on the Pea Mitochondrial Proteome. Mol. Cell. Proteom. 2005, 4, 1122–1133. [Google Scholar] [CrossRef] [PubMed]
- Renaut, J.; Lutts, S.; Hoffmann, L.; Hausman, J. Responses of Poplar to Chilling Temperatures: Proteomic and Physiological Aspects. Plant Biol. 2004, 6, 81–90. [Google Scholar] [PubMed]
- Hahn, M.; Walbot, V. Effects of Cold-Treatment on Protein Synthesis and mRNA Levels in Rice Leaves. Plant Physiol. 1989, 91, 930–938. [Google Scholar] [CrossRef]
- Cui, S.; Huang, F.; Wang, J.; Ma, X.; Cheng, Y.; Liu, J. A Proteomic Analysis of Cold Stress Responses in Rice Seedlings. Proteomics 2005, 5, 3162–3172. [Google Scholar] [CrossRef] [PubMed]
- Balbuena, T.S.; Salas, J.J.; Martinez-Force, E.; Garces, R.; Thelen, J.J. Proteome Analysis of Cold Acclimation in Sunflower. J. Proteome Res. 2011, 10, 2330–2346. [Google Scholar] [CrossRef]
- Rinalducci, S.; Egidi, M.G.; Mahfoozi, S.; Godehkahriz, S.J.; Zolla, L. The Influence of Temperature on Plant Development in a Vernalization-Requiring Winter Wheat: A 2-DE Based Proteomic Investigation. J. Proteom. 2011, 74, 643–659. [Google Scholar] [CrossRef]
- Chauhan, H.; Khurana, N.; Nijhavan, A.; Khurana, J.P.; Khurana, P. The Wheat Chloroplastic Small Heat Shock Protein (sHSP26) is Involved in Seed Maturation and Germination and Imparts Tolerance to Heat Stress. Plant Cell Environ. 2012, 35, 1912–1931. [Google Scholar] [CrossRef]
- Pratt, W.B.; Toft, D.O. Regulation of Signaling Protein Function and Trafficking by the hsp90/hsp70-Based Chaperone Machinery. Exp. Biol. Med. 2003, 228, 111–133. [Google Scholar] [CrossRef]
- Xu, X.; Song, H.; Zhou, Z.; Shi, N.; Ying, Q.; Wang, H. Functional Characterization of AtHsp90.3 in Saccharomyces cerevisiae and Arabidopsis Thaliana Under Heat Stress. Biotechnol. Lett. 2010, 32, 979–987. [Google Scholar] [CrossRef]
- Kim, S.R.; An, G. Rice Chloroplast-Localized Heat Shock Protein 70, OsHsp70CP1, is Essential for Chloroplast Development Under High-Temperature Conditions. J. Plant Physiol. 2013, 170, 854–863. [Google Scholar] [CrossRef]
- Yu, J.; Cheng, Y.; Feng, K.; Ruan, M.; Ye, Q.; Wang, R.; Li, Z.; Zhou, G.; Yao, Z.; Yang, Y. Genome-Wide Identification and Expression Profiling of Tomato Hsp20 Gene Family in Response to Biotic and Abiotic Stresses. Front. Plant Sci. 2016, 7, 1215. [Google Scholar] [CrossRef] [PubMed]
- Derocher, A.E.; Helm, K.W.; Lauzon, L.M.; Vierling, E. Expression of a Conserved Family of Cytoplasmic Low Molecular Weight Heat Shock Proteins during Heat Stress and Recovery. Plant Physiol. 1991, 96, 1038–1047. [Google Scholar] [CrossRef] [PubMed]
- Haq, S.U.L.; Khan, A.; Ali, M.; Gai, W.X.; Zhang, H.X.; Yu, Q.H.; Yang, S.B.; Wei, A.M.; Gong, Z.H. Knockdown of CaHSP60-6 Confers Enhanced Sensitivity to Heat Stress in Pepper (Capsicum annuum L.). Planta 2019. [Google Scholar] [CrossRef] [PubMed]
- Kosova, K.; Vitamvas, P.; Planchon, S.; Renaut, J.; Vankova, R.; Prasil, I.T. Proteome Analysis of Cold Response in Spring and Winter Wheat (Triticum aestivum) Crowns Reveals Similarities in Stress Adaptation and Differences in Regulatory Processes between the Growth Habits. J. Proteome Res. 2013, 12, 4830–4845. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Aoki, N.; Matsumura, H.; Okamura, M.; Ohsugi, R.; Shimono, H. Cooling Water Before Panicle Initiation Increases Chilling-Induced Male Sterility and Disables Chilling-Induced Expression of Genes Encoding OsFKBP65 and Heat Shock Proteins in Rice Spikelets. Plant Cell Environ. 2015, 38, 1255–1274. [Google Scholar] [CrossRef] [PubMed]
- Sabehat, A.; Lurie, S.; Weiss, D. Expression of Small Heat-Shock Proteins at Low Temperatures: A Possible Role in Protecting Against Chilling Injuries. Plant Physiol. 1998, 117, 651–658. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Chen, J.; Kuang, J.; Chen, W.; Lu, W. Expression of sHSP Genes as Affected by Heat Shock and Cold Acclimation in Relation to Chilling Tolerance in Plum Fruit. Postharvest Biol. Technol. 2010, 55, 91–96. [Google Scholar] [CrossRef]
- Rozenzvieg, D.; Elmaci, C.; Samach, A.; Lurie, S.; Porat, R. Isolation of Four Heat Shock Protein cDNAs From Grapefruit Peel Tissue and Characterization of Their Expression in Response to Heat and Chilling Temperature Stresses. Physiol. Plant 2004, 121, 421–428. [Google Scholar] [CrossRef]
- Sobhanian, H.; Aghaei, K.; Komatsu, S. Changes in the Plant Proteome Resulting From Salt Stress: Toward the Creation of Salt-Tolerant Crops? J. Proteom. 2011, 74, 1323–1337. [Google Scholar] [CrossRef]
- Wang, M.; Peng, Z.; Li, C.; Li, F.; Liu, C.; Xia, G. Proteomic Analysis on a High Salt Tolerance Introgression Strain of Triticum aestivum/Thinopyrum ponticum. Proteomics 2008, 8, 1470–1489. [Google Scholar] [CrossRef]
- Ngara, R.; Ndimba, B.K. Understanding the Complex Nature of Salinity and Drought-Stress Response in Cereals Using Proteomics Technologies. Proteomics 2014, 14, 611–621. [Google Scholar] [CrossRef] [PubMed]
- Han, F.; Chen, H.; Li, X.J.; Yang, M.F.; Liu, G.S.; Shen, S.H. A Comparative Proteomic Analysis of Rice Seedlings Under Various High-Temperature stresses. Biochim. Biophys. Acta. Proteins Proteom. 2009, 1794, 1625–1634. [Google Scholar] [CrossRef] [PubMed]
- Muthusamy, S.K.; Dalal, M.; Chinnusamy, V.; Bansal, K.C. Differential Regulation of Genes Coding for Organelle and Cytosolic ClpATPases Under Biotic and Abiotic Stresses in Wheat. Front. Plant Sci. 2016, 7, 929. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.C.; Grover, A. Constitutive Over-Expression of Rice ClpD1 Protein Enhances Tolerance to Salt and Desiccation Stresses in Transgenic Arabidopsis Plants. Plant Sci. 2016, 250, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Fan, P.; Li, Y. Overexpression of Organellar and Cytosolic AtHSP90 in Arabidopsis Thaliana Impairs Plant Tolerance to Oxidative Stress. Plant Mol. Biol. Report. 2009, 27, 342–349. [Google Scholar] [CrossRef]
- Pi, E.; Qu, L.; Hu, J.; Huang, Y.; Qiu, L.; Lu, H.; Jiang, B.; Liu, C.; Peng, T.; Zhao, Y. Mechanisms of Soybean Roots’ Tolerances to Salinity Revealed by Proteomic and Phosphoproteomic Comparisons Between Two Cultivars. Mol. Cell. Proteom. 2016, 15, 266–288. [Google Scholar] [CrossRef]
- Manaa, A.; Ben Ahmed, H.; Valot, B.; Bouchet, J.P.; Aschi-Smiti, S.; Causse, M.; Faurobert, M. Salt and Genotype Impact on Plant Physiology and Root Proteome Variations in Tomato. J. Exp. Bot. 2011, 62, 2797–2813. [Google Scholar] [CrossRef]
- Yer, E.N.; Baloglu, M.C.; Ayan, S. Identification and Expression Profiling of All Hsp Family Member Genes Under Salinity Stress in Different Poplar Clones. Gene 2018, 678, 324–336. [Google Scholar] [CrossRef]
- Song, A.; Zhu, X.; Chen, F.; Gao, H.; Jiang, J.; Chen, S. A Chrysanthemum Heat Shock Protein Confers Tolerance to Abiotic Stress. Int. J. Mol. Sci. 2014, 15, 5063–5078. [Google Scholar] [CrossRef]
- Alvim, F.C.; Carolino, S.M.B.; Cascardo, J.C.M.; Nunes, C.C.; Martinez, C.A.; Otoni, W.C.; Fontes, E.P.B. Enhanced Accumulation of BiP in Transgenic Plants Confers Tolerance to Water Stress. Plant Physiol. 2001, 126, 1042–1054. [Google Scholar] [CrossRef]
- Reddy, P.S.; Kishor, P.B.K.; Seiler, C.; Kuhlmann, M.; Eschen-Lippold, L.; Lee, J.; Reddy, M.K.; Sreenivasulu, N. Unraveling Regulation of the Small Heat Shock Proteins by the Heat Shock Factor HvHsfB2c in Barley: Its Implications in Drought Stress Response and Seed Development. PLoS ONE 2014, 9, e89125. [Google Scholar] [CrossRef]
- Shu, L.; Lou, Q.; Ma, C.; Ding, W.; Zhou, J.; Wu, J.; Feng, F.; Lu, X.; Luo, L.; Xu, G. Genetic, Proteomic and Metabolic Analysis of the Regulation of Energy Storage in Rice Seedlings in Response to Drought. Proteomics 2011, 11, 4122–4138. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, L.; Gupta, S.; Mishra, S.K.; Pandey, G.; Kumar, S.; Chauhan, P.S.; Chakrabarty, D.; Nautiyal, C.S. Elucidation of Complex Nature of PEG Induced Drought-Stress Response in Rice Root Using Comparative Proteomics Approach. Front. Plant Sci. 2016, 7, 1466. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Yokoya, S. Enhanced Tolerance to Drought Stress in Transgenic Rice Plants Overexpressing a Small Heat-Shock Protein, sHSP17. 7. Plant Cell Rep. 2008, 27, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Subba, P.; Barua, P.; Kumar, R.; Datta, A.; Soni, K.K.; Chakraborty, S.; Chakraborty, N. Phosphoproteomic Dynamics of Chickpea (Cicer arietinum L.) Reveals Shared and Distinct Components of Dehydration Response. J. Proteome Res. 2013, 12, 5025–5047. [Google Scholar] [CrossRef]
- Augustine, S.M.; Cherian, A.V.; Syamaladevi, D.P.; Subramonian, N. Erianthus arundinaceus HSP70 (EaHSP70) Acts as a Key Regulator in the Formation of Anisotropic Interdigitation in Sugarcane (Saccharum Spp. Hybrid) in Response to Drought Stress. Plant Cell Physiol. 2015, 56, 2368–2380. [Google Scholar] [CrossRef]
- Burke, J.J.; Hatfield, J.L.; Klein, R.R.; Mullet, J.E. Accumulation of Heat Shock Proteins in Field-Grown Cotton. Plant Physiol. 1985, 78, 394–398. [Google Scholar] [CrossRef]
- Bonhomme, L.; Monclus, R.; Vincent, D.; Carpin, S.; Lomenech, A.; Plomion, C.; Brignolas, F.; Morabito, D. Leaf Proteome Analysis of Eight Populus euramericana Genotypes: Genetic Variation in Drought Response and in Water-Use Efficiency Involves Photosynthesis-Related Proteins. Proteomics 2009, 9, 4121–4142. [Google Scholar] [CrossRef]
- Giacomelli, L.; Rudella, A.; Van Wijk, K.J. High Light Response of the Thylakoid Proteome in Arabidopsis wild Type and the Ascorbate-Deficient Mutant Vtc2-2. A Comparative Proteomics Study. Plant Physiol. 2006, 141, 685–701. [Google Scholar] [CrossRef]
- Rossel, J.B.; Wilson, I.W.; Pogson, B.J. Global Changes in Gene Expression in Response to High Light in Arabidopsis. Plant Physiol. 2002, 130, 1109–1120. [Google Scholar] [CrossRef]
- Kropat, J.; Oster, U.; Rudiger, W.; Beck, C.F. Chlorophyll Precursors are Signals of Chloroplast Origin Involved in Light Induction of Nuclear Heat-Shock Genes. Proc. Natl. Acad. Sci. USA 1997, 94, 14168–14172. [Google Scholar] [CrossRef]
- Rodriguez-Celma, J.; Rellan-Alvarez, R.; Abadia, A.; Abadia, J.; Lopez-Millan, A.F. Changes Induced by Two Levels of Cadmium Toxicity in the 2-DE Protein Profile of Tomato Roots. J. Proteom. 2010, 73, 1694–1706. [Google Scholar] [CrossRef]
- Ahsan, N.; Lee, S.H.; Lee, D.G.; Lee, H.; Lee, S.W.; Bahk, J.D.; Lee, B.H. Physiological and Protein Profiles Alternation of Germinating Rice Seedlings Exposed to Acute Cadmium Toxicity. C. R. Biol. 2007, 330, 735–746. [Google Scholar] [CrossRef]
- Yang, Y.; Li, X.; Yang, S.; Zhou, Y.; Dong, C.; Ren, J.; Sun, X.; Yang, Y. Comparative Physiological and Proteomic Analysis Reveals the Leaf Response to Cadmium-Induced Stress in Poplar (Populus yunnanensis). PLoS ONE 2015, 10, e0137396. [Google Scholar] [CrossRef]
- Czarnecka, E.; Nagao, R.T.; Key, J.L.; Gurley, W.B. Characterization of Gmhsp26-A, a Stress Gene Encoding a Divergent Heat Shock Protein of Soybean: Heavy-Metal-Induced Inhibition of Intron Processing. Mol. Cell. Biol. 1988, 8, 1113–1122. [Google Scholar] [CrossRef]
- Lee, J.; Ahn, Y.J. Heterologous Expression of a Carrot Small Heat Shock Protein Increased Escherichia Coli Viability Under Lead and Arsenic Stresses. HortScience 2013, 48, 1323–1326. [Google Scholar] [CrossRef]
- Sarry, J.; Kuhn, L.; Ducruix, C.; Lafaye, A.; Junot, C.; Hugouvieux, V.; Jourdain, A.; Bastien, O.; Fievet, J.B.; Vailhen, D. The Early Responses of Arabidopsis Thaliana Cells to Cadmium Exposure Explored by Protein and Metabolite Profiling Analyses. Proteomics 2006, 6, 2180–2198. [Google Scholar] [CrossRef]
- Navascues, J.; Perez-Rontome, C.; Sanchez, D.H.; Staudinger, C.; Wienkoop, S.; Rellan-Alvarez, R.; Becana, M. Oxidative Stress is a Consequence, Not a Cause, of Aluminum Toxicity in the Forage Legume Lotus corniculatus. New Phytol. 2012, 193, 625–636. [Google Scholar] [CrossRef]
- Banti, V.; Mafessoni, F.; Loreti, E.; Alpi, A.; Perata, P. The Heat-Inducible Transcription Factor HsfA2 Enhances Anoxia Tolerance in Arabidopsis. Plant Physiol. 2010, 152, 1471–1483. [Google Scholar] [CrossRef]
- Huther, C.M.; Martinazzo, E.G.; Rombaldi, C.V.; Bacarin, M.A. Effects of Flooding Stress in Micro-Tom’tomato Plants Transformed with Different Levels of Mitochondrial sHSP23. 6. Braz. J. Biol. 2017, 77, 43–51. [Google Scholar] [CrossRef]
- Qi, Y.; Wang, H.; Zou, Y.; Liu, C.; Liu, Y.; Wang, Y.; Zhang, W. Over-Expression of Mitochondrial Heat Shock Protein 70 Suppresses Programmed Cell Death in Rice. Febs Lett. 2011, 585, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, X.; Wang, H.; Bao, Y.; Zhang, W. Examination of the Leaf Proteome During Flooding Stress and the Induction of Programmed Cell Death in Maize. Proteome Sci. 2014, 12, 33. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Makino, T.; Yasue, H. Proteomic and Biochemical Analyses of the Cotyledon and Root of Flooding-Stressed Soybean Plants. PLoS ONE 2013, 8, e65301. [Google Scholar] [CrossRef]
- Komatsu, S.; Yamamoto, A.; Nakamura, T.; Nouri, M.Z.; Nanjo, Y.; Nishizawa, K.; Furukawa, K. Comprehensive Analysis of Mitochondria in Roots and Hypocotyls of Soybean Under Flooding Stress Using Proteomics and Metabolomics Techniques. J. Proteome Res. 2011, 10, 3993–4004. [Google Scholar] [CrossRef]
- Salehi-Lisar, S.Y.; Bakhshayeshan-Agdam, H. Drought Stress in Plants: Causes, Consequences, and Tolerance, Physiology and Biochemistry; Springer International Publishing: Cham, Switzerland; Berlin, Germany, 2016; pp. 1–16. ISBN 978-3-319-28899-4. [Google Scholar]
- Chaves, M.M.; Maroco, J.P.; Pereira, J.S. Understanding Plant Responses to Drought—From Genes to the Whole Plant. Funct. Plant Biol. 2003, 30, 239–264. [Google Scholar] [CrossRef]
- Ramanjulu, S.; Bartels, D. Drought-And Desiccation-Induced Modulation of Gene Expression in Plants. Plant Cell Environ. 2002, 25, 141–151. [Google Scholar] [CrossRef]
- Ristic, Z.; Gifford, D.J.; Cass, D.D. Heat Shock Proteins in Two Lines of Zea mays L. That Differ in Drought and Heat Resistance. Plant Physiol. 1991, 97, 1430–1434. [Google Scholar] [CrossRef]
- Cruz De Carvalho, R.; Bernardes Da Silva, A.; Soares, R.; Almeida, A.M.; Coelho, A.V.; Marques Da Silva, J.; Branquinho, C. Differential Proteomics of Dehydration and Rehydration in Bryophytes: Evidence Towards a Common Desiccation Tolerance Mechanism. Plant Cell Environ. 2014, 37, 1499–1515. [Google Scholar] [CrossRef]
- Pandey, A.; Chakraborty, S.; Datta, A.; Chakraborty, N. Proteomics Approach to Identify Dehydration Responsive Nuclear Proteins From Chickpea (Cicer arietinum L.). Mol. Cell. Proteom. 2008, 7, 88–107. [Google Scholar] [CrossRef]
- Pandey, A.; Rajamani, U.; Verma, J.; Subba, P.; Chakraborty, N.; Datta, A.; Chakraborty, S.; Chakraborty, N. Identification of Extracellular Matrix Proteins of Rice (Oryza sativa L.) Involved in Dehydration-Responsive Network: A Proteomic Approach. J. Proteome Res. 2010, 9, 3443–3464. [Google Scholar] [CrossRef]
- Bhushan, D.; Jaiswal, D.K.; Ray, D.; Basu, D.; Datta, A.; Chakraborty, S.; Chakraborty, N. Dehydration-Responsive Reversible and Irreversible Changes in the Extracellular Matrix: Comparative Proteomics of Chickpea Genotypes with Contrasting Tolerance. J. Proteome Res. 2011, 10, 2027–2046. [Google Scholar] [CrossRef]
- Choudhary, M.K.; Basu, D.; Datta, A.; Chakraborty, N.; Chakraborty, S. Dehydration-Responsive Nuclear Proteome of Rice (Oryza sativa L.) Illustrates Protein Network, Novel Regulators of Cellular Adaptation, and Evolutionary Perspective. Mol. Cell. Proteom. 2009, 8, 1579–1598. [Google Scholar] [CrossRef]
- Song, H.; Zhao, R.; Fan, P.; Wang, X.; Chen, X.; Li, Y. Overexpression of AtHsp90. 2, AtHsp90. 5 and AtHsp90. 7 in Arabidopsis Thaliana Enhances Plant Sensitivity to Salt and Drought Stresses. Planta 2009, 229, 955–964. [Google Scholar] [CrossRef]
- Timperio, A.M.; Egidi, M.G.; Zolla, L. Proteomics Applied on Plant Abiotic Stresses: Role of Heat Shock Proteins (HSP). J. Proteom. 2008, 71, 391–411. [Google Scholar] [CrossRef]
- Ogawa, I.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. Time Course Analysis of Gene Regulation Under Cadmium Stress in Rice. Plant Soil 2009, 325, 97. [Google Scholar] [CrossRef]
- Kumar, M.; Padula, M.P.; Davey, P.; Pernice, M.; Jiang, Z.; Sablok, G.; Contreras-Porcia, L.; Ralph, P.J. Proteome Analysis Reveals Extensive Light Stress-Response Reprogramming in the Seagrass Zostera muelleri (Alismatales, Zosteraceae) Metabolism. Front. Plant Sci. 2017, 7, 2023. [Google Scholar] [CrossRef]
- Sharma, S.S.; Dietz, K.J. The Relationship Between Metal Toxicity and Cellular Redox Imbalance. Trends Plant Sci. 2009, 14, 43–50. [Google Scholar] [CrossRef]
- Hossain, Z.; Khatoon A FAU Komatsu, S.; Komatsu, S. Soybean Proteomics for Unraveling Abiotic Stress Response Mechanism. J. Proteome Res. 2013, 12, 4670–4684. [Google Scholar] [CrossRef]
- Hossain, Z.; Hajika, M.; Komatsu, S. Comparative Proteome Analysis of High and Low Cadmium Accumulating Soybeans Under Cadmium Stress. Amino Acids 2012, 43, 2393–2416. [Google Scholar] [CrossRef]
- Song, H.M.; Wang, H.Z.; Xu, X.B. Overexpression of AtHsp90. 3 in Arabidopsis Thaliana Impairs Plant Tolerance to Heavy Metal Stress. Biol. Plant 2012, 56, 197–199. [Google Scholar] [CrossRef]
- Hossain, Z.; Lopez-Climent, M.F.; Arbona, V.; Perez-Clemente, R.M.; Gomez-Cadenas, A. Modulation of the Antioxidant System in Citrus Under Waterlogging and Subsequent Drainage. J. Plant Physiol. 2009, 166, 1391–1404. [Google Scholar] [CrossRef]
- Kilian, J.; Whitehead, D.; Horak, J.; Wanke, D.; Weinl, S.; Batistic, O.; D’angelo, C.; Bornberg-Bauer, E.; Kudla, J.; Harter, K. The AtGenExpress Global Stress Expression Data Set: Protocols, Evaluation and Model Data Analysis of UV-B Light, Drought and Cold Stress Responses. Plant J. 2007, 50, 347–363. [Google Scholar] [CrossRef]
- Scarpeci, T.E.; Zanor, M.I.; Valle, E.M. Investigating the Role of Plant Heat Shock Proteins During Oxidative Stress. Plant Signal. Behav. 2008, 3, 856–857. [Google Scholar] [CrossRef]
- Rizhsky, L.; Liang, H.; Mittler, R. The Combined Effect of Drought Stress and Heat Shock on Gene Expression in Tobacco. Plant Physiol. 2002, 130, 1143–1151. [Google Scholar] [CrossRef]
- Zou, J.; Liu, C.; Liu, A.; Zou, D.; Chen, X. Overexpression of OsHsp17.0 and OsHsp23.7 Enhances Drought and Salt Tolerance in Rice. J. Plant Physiol. 2012, 169, 628–635. [Google Scholar] [CrossRef]
- Wang, A.; Yu, X.; Mao, Y.; Liu, Y.; Liu, G.; Liu, Y.; Niu, X. Overexpression of a Small Heat-Shock-Protein Gene Enhances Tolerance to Abiotic Stresses in Rice. Plant Breed. 2015, 134, 384–393. [Google Scholar] [CrossRef]
- Sun, W.; Bernard, C.; Van De Cotte, B.; Van Montagu, M.; Verbruggen, N. At-HSP17.6A, Encoding a Small Heat-Shock Protein in Arabidopsis, Can Enhance Osmotolerance Upon Overexpression. Plant J. 2001, 27, 407–415. [Google Scholar] [CrossRef]
- Nishizawa-Yokoi, A.; Tainaka, H.; Yoshida, E.; Tamoi, M.; Yabuta, Y.; Shigeoka, S. The 26S Proteasome Function and Hsp90 Activity Involved in the Regulation of HsfA2 Expression in Response to Oxidative Stress. Plant Cell Physiol. 2010, 51, 486–496. [Google Scholar] [CrossRef]
- Downs, C.A.; Ryan, S.L.; Heckathorn, S.A. The Chloroplast Small Heat-Shock Protein: Evidence for a General Role in Protecting Photosystem II Against Oxidative Stress and Photoinhibition. J. Plant Physiol. 1999, 155, 488–496. [Google Scholar] [CrossRef]
- Banzet, N.; Richaud, C.; Deveaux, Y.; Kazmaier, M.; Gagnon, J.; Triantaphylides, C. Accumulation of Small Heat Shock Proteins, Including Mitochondrial HSP22, Induced by Oxidative Stress and Adaptive Response in Tomato Cells. Plant J. 1998, 13, 519–527. [Google Scholar] [CrossRef]
- Siddique, M.; Gernhard, S.; Von Koskull-Doring, P.; Vierling, E.; Scharf, K.D. The Plant sHSP Superfamily: Five New Members in Arabidopsis Thaliana with Unexpected Properties. Cell Stress Chaperones 2008, 13, 183–197. [Google Scholar] [CrossRef] [PubMed]
- Koo, H.J.; Park, S.M.; Kim, K.P.; Suh, M.C.; Lee, M.O.; Lee, S.K.; Xia, X.; Hong, C.B. Small Heat Shock Proteins Can Release Light Dependence of Tobacco Seed During Germination. Plant Physiol. 2015, 167, 1030–1038. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).