Comprehensive Analysis of the Chitinase Gene Family in Cucumber (Cucumis sativus L.): From Gene Identification and Evolution to Expression in Response to Fusarium oxysporum
Abstract
1. Introduction
2. Results
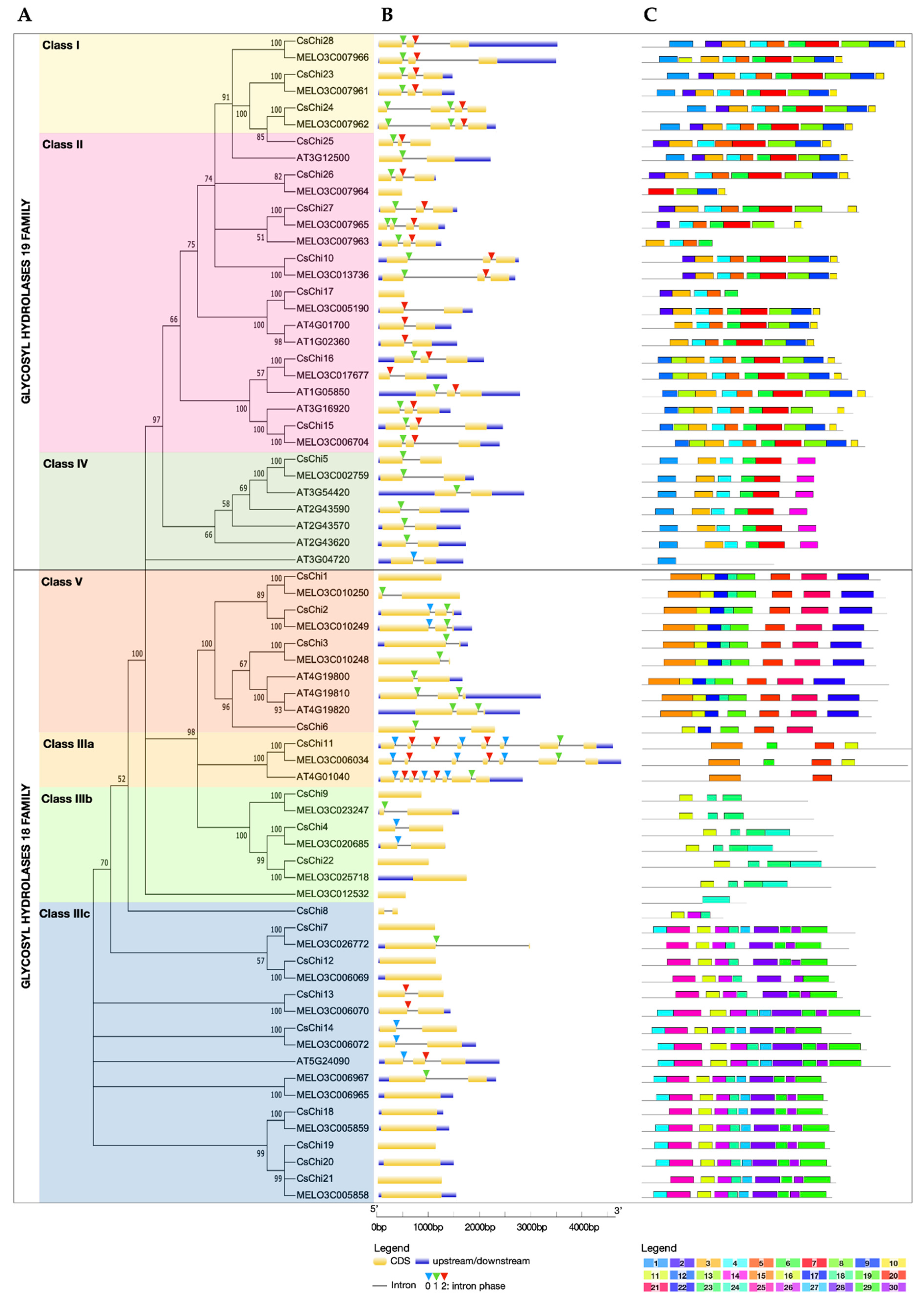
2.1. Genome-Wide Identification and Phylogenetic Analysis of Cucumber Chitinase Genes
2.2. Gene Structure and Conserved Motifs Analyses of Chitinase Genes
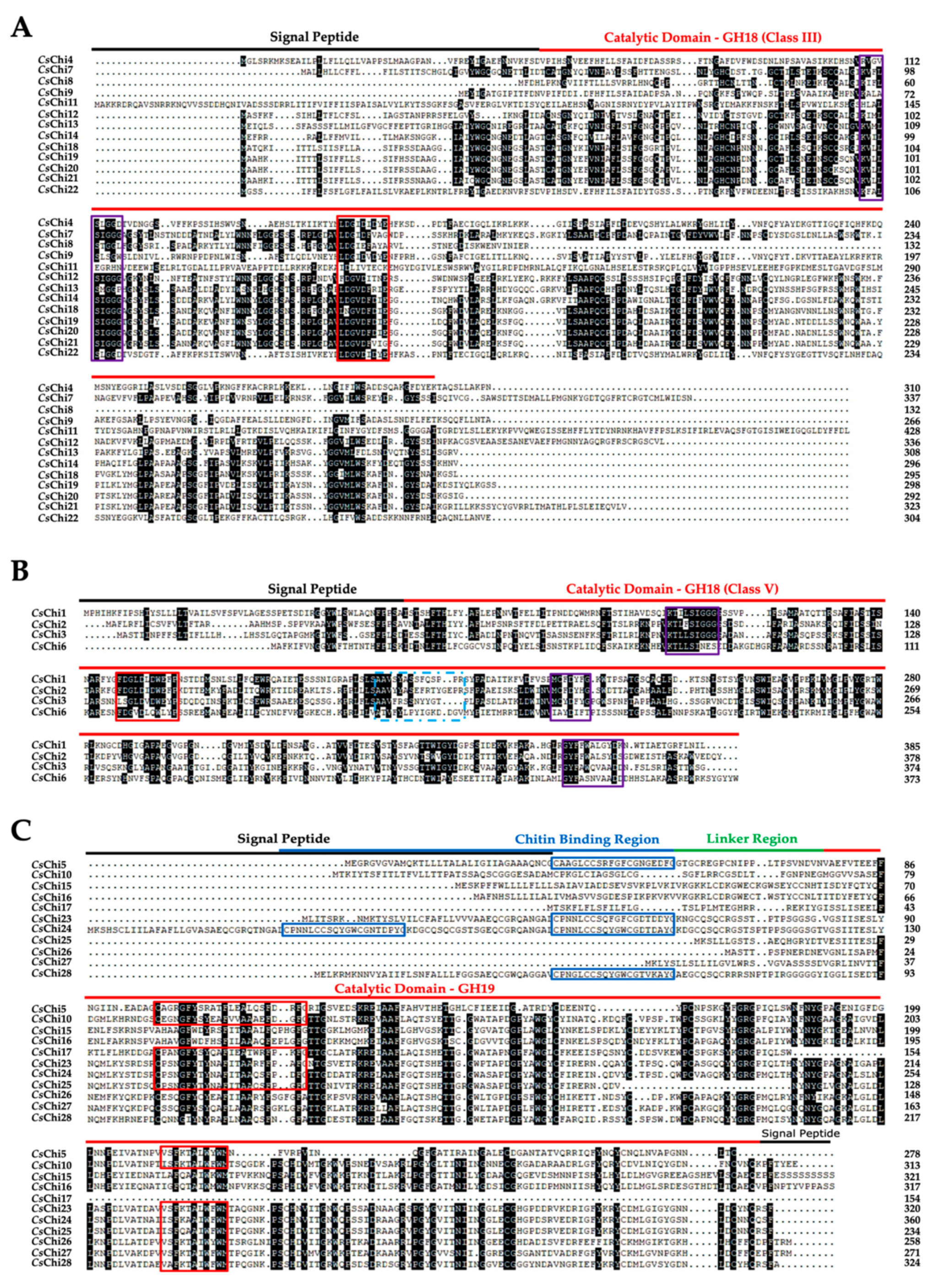
2.3. Conserved Domains and Active Site Analysis of CsChi Genes
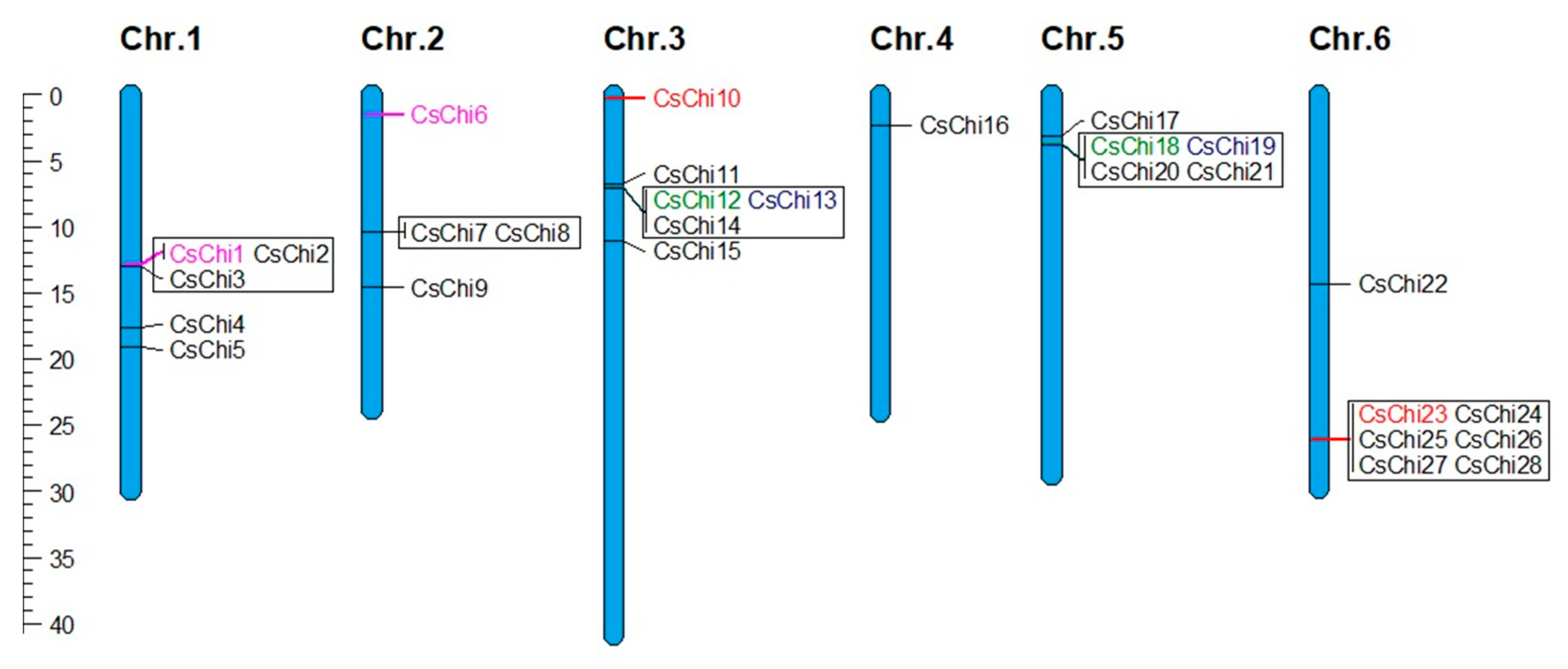
2.4. Chromosomal Distribution and Gene Duplication Analysis of CsChi Genes
2.5. Cis-Regulatory Elements in the Promoter of CsChi Genes
2.6. Spatial Expression Profiles of CsChi Genes
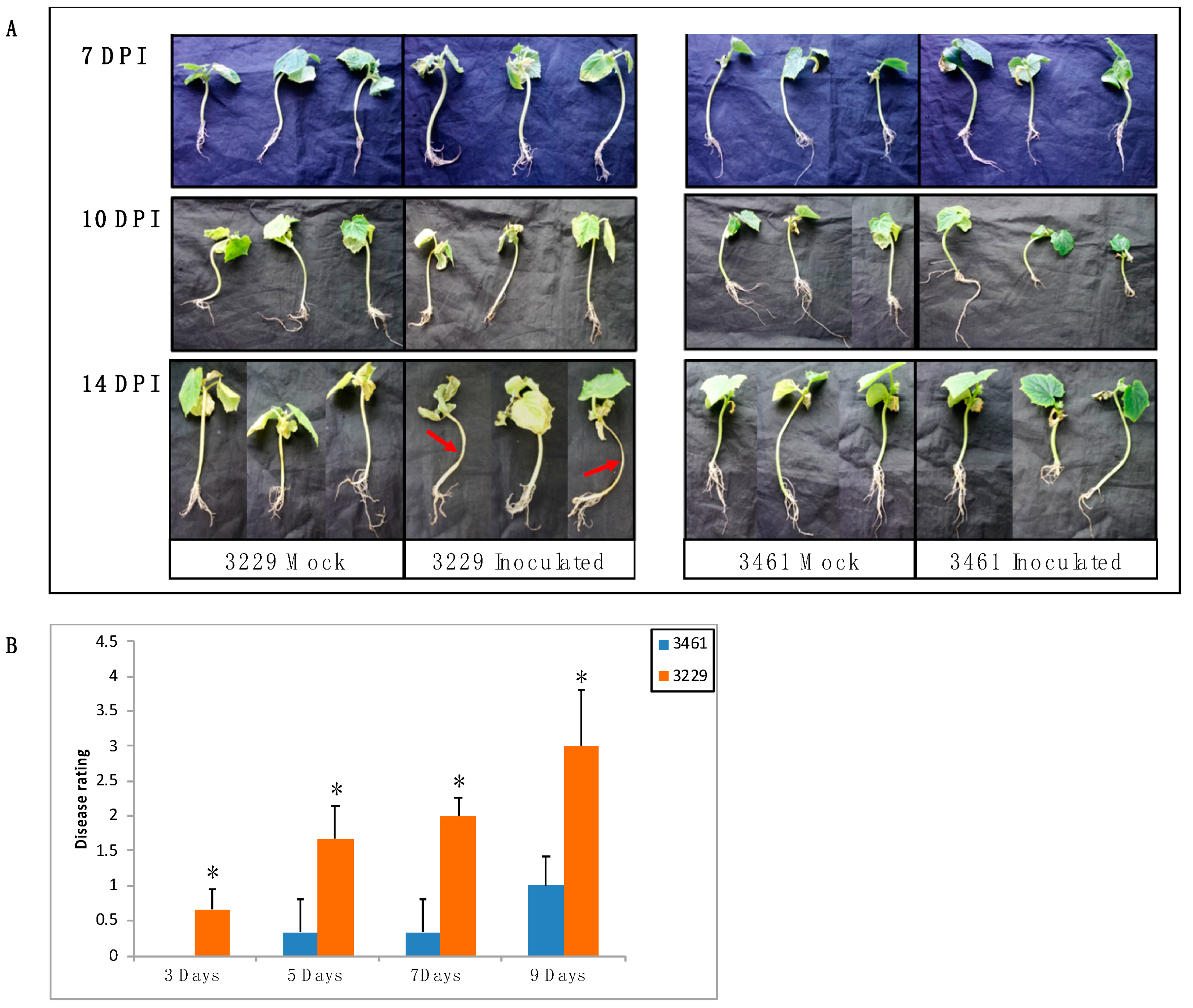
2.7. Expression Analysis of CsChi Genes in Response to F. oxysporum
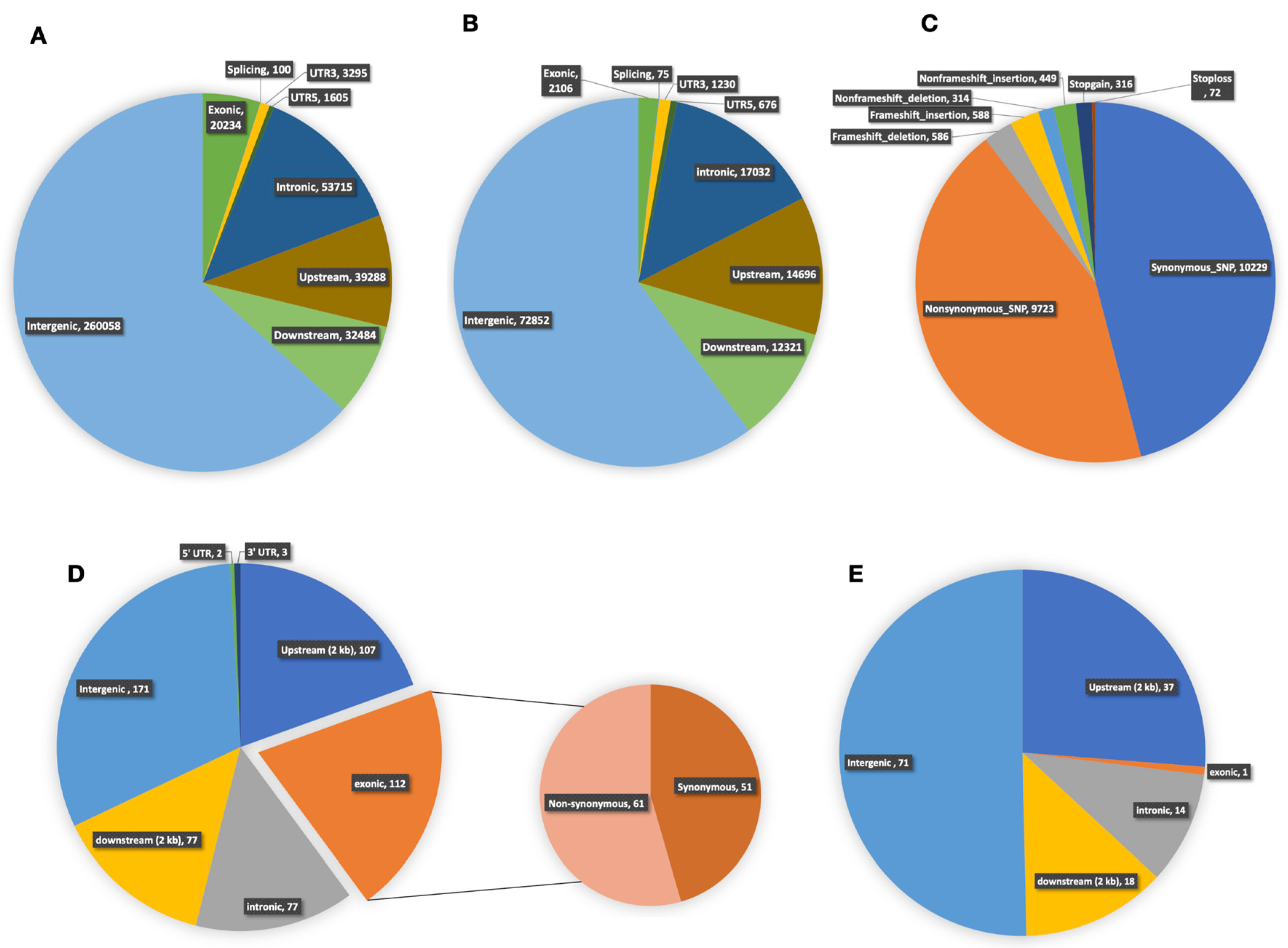
2.8. Whole-Genome Re-Sequencing and Variation Analysis between Lines 3229 and 3461
2.9. DNA Polymorphisms in Region of Chitinase Genes
3. Discussion
4. Materials and Methods
4.1. Sequence Acquisition and Identification of Cucumber Chitinase Genes
4.2. Phylogenetic Tree, Gene Structures and Conserved Motifs Analyses of Chitinase Genes
4.3. Chromosomal Location and Identification of Homologous A. thaliana Members
4.4. Cis-Regulatory Elements Analysis
4.5. Gene Expression Analysis
4.6. Plant Material and Fusarium Infection Assay
4.7. RNA Extraction and Quantitative Real-Time Reverse Transcription PCR (qRT-PCR) Analysis
4.8. DNA Extraction, Whole-Genome Resequencing and Variation Analysis
4.9. Validation of Variations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CDS | Coding sequence |
| CLP | Chitinase-like protein |
| GH | Glycosyl Hydrolases |
| InDel | Insertion and deletion |
| MW | Molecular weight |
| pI | Isoelectric point |
| PR | Pathogenesis-related |
| qRT-PCR | Quantitative real-time reverse transcription PCR |
| SI-CLPs | Stabilin-1 interacting chitinase-like proteins |
| SNP | Single-nucleotide polymorphism |
| UTR | Untranslated region |
References
- Renner, T.; Specht, C.D. Molecular Evolution and Functional Evolution of Class I Chitinases for Plant Carnivory in the Caryophyllales. Mol. Biol. Evol. 2012, 29, 2971–2985. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The Carbohydrate-Active Enzymes Database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed]
- Jain, K.K.; Kumar, A.; Shankar, A.; Pandey, D.; Chaudhary, B.; Sharma, K.K. De Novo Transcriptome Assembly and Protein Profiling of Copper-Induced Lignocellulolytic Fungus Ganoderma Lucidum MDU-7 Reveals Genes Involved in Lignocellulose Degradation and Terpenoid Biosynthetic Pathways. Genomics 2019. [Google Scholar] [CrossRef] [PubMed]
- Tyler, L.; Bragg, J.N.; Wu, J.; Yang, X.; Tuskan, G.A.; Vogel, J.P. Annotation and Comparative Analysis of the Glycoside Hydrolase Genes in Brachypodium Distachyon. BMC Genom. 2010, 11, 600. [Google Scholar] [CrossRef]
- Wang, S.; Ye, X.; Chen, J.; Rao, P. A Novel Chitinase Isolated from Vicia Faba and Its Antifungal Activity. Food Res. Int. 2012, 45, 116–122. [Google Scholar] [CrossRef]
- Wang, S.; Shao, B.; Fu, H.; Rao, P. Isolation of a Thermostable Legume Chitinase and Study on the Antifungal Activity. Appl. Microbiol. Biotechnol. 2009, 85, 313–321. [Google Scholar] [CrossRef]
- Li, C.; Li, X.; Bai, C.; Zhang, Y.; Wang, Z. A Chitinase with Antifungal Activity from Naked Oat (Avena Chinensis) Seeds. J. Food Biochem. 2019, 43, e12713. [Google Scholar] [CrossRef]
- Liu, J.J.; Ekramoddoullah, A.K.M.; Zamani, A. A Class IV Chitinase Is Up-Regulated by Fungal Infection and Abiotic Stresses and Associated with Slow-Canker-Growth Resistance to Cronartium Ribicola in Western White Pine (Pinus Monticola). Phytopathology 2005, 95, 284–291. [Google Scholar] [CrossRef]
- Xiao, Y.H.; Li, X.B.; Yang, X.Y.; Luo, M.; Hou, L.; Guo, S.H.; Luo, X.Y.; Pei, Y. Cloning and Characterization of a Balsam Pear Class I Chitinase Gene (Mcchit1) and Its Ectopic Expression Enhances Fungal Resistance in Transgenic Plants. Biosci. Biotechnol. Biochem. 2007, 71, 1211–1219. [Google Scholar] [CrossRef]
- Maximova, S.N.; Marelli, J.P.; Young, A.; Pishak, S.; Verica, J.A.; Guiltinan, M.J. Over-Expression of a Cacao Class I Chitinase Gene in Theobroma Cacao, L. Enhances Resistance against the Pathogen, Colletotrichum Gloeosporioides. Planta 2006, 224, 740–749. [Google Scholar] [CrossRef]
- Dehestani, A.; Kazemitabar, K.; Ahmadian, G.; Jelodar, N.B.; Salmanian, A.H.; Seyedi, M.; Rahimian, H.; Ghasemi, S. Chitinolytic and Antifungal Activity of a Bacillus Pumilus Chitinase Expressed in Arabidopsis. Biotechnol. Lett. 2010, 32, 539–546. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Liu, H.; Jia, Z.; Fang, Q.; Luo, K. Combined Expression of Antimicrobial Genes (Bbchit1 and LJAMP2) in Transgenic Poplar Enhances Resistance to Fungal Pathogens. Tree Physiol. 2012, 32, 1313–1320. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.; Sharma, K.P.; Gaur, R.K.; Gupta, V.K. Role of Chitinase in Plant Defense. Asian J. Biochem. 2011, 6, 29–37. [Google Scholar] [CrossRef]
- Ganesan, M.; Bhanumathi, P.; Ganesh Kumari, K.; Lakshmi Prabha, A.; Song, P.S.; Jayabalan, N. Transgenic Indian Cotton (Gossypium Hirsutum) Harboring Rice Chitinase Gene (Chi II) Confers Resistance to Two Fungal Pathogens. Am. J. Biochem. Biotechnol. 2009, 5, 63–74. [Google Scholar] [CrossRef]
- Xu, J.; Xu, X.; Tian, L.; Wang, G.; Zhang, X.; Wang, X.; Guo, W. Discovery and Identification of Candidate Genes from the Chitinase Gene Family for Verticillium Dahliae Resistance in Cotton. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Arakane, Y.; Muthukrishnan, S. Insect Chitinase and Chitinase-like Proteins. Cell. Mol. Life Sci. 2010, 67, 201–216. [Google Scholar] [CrossRef]
- Passarinho, P.A.; de Vries, S.C. Arabidopsis Chitinases: A Genomic Survey. Arab. B. 2002, 1, e0023. [Google Scholar] [CrossRef]
- Grover, A. Plant Chitinases: Genetic Diversity and Physiological Roles. CRC. Crit. Rev. Plant. Sci. 2012, 31, 57–73. [Google Scholar] [CrossRef]
- Huang, S.; Li, R.; Zhang, Z.; Li, L.; Gu, X.; Fan, W.; Lucas, W.J.; Wang, X.; Xie, B.; Ni, P.; et al. The Genome of the Cucumber, Cucumis Sativus L. Nat. Genet. 2009, 41, 1275–1281. [Google Scholar] [CrossRef]
- Zhang, D.; Meng, K.X.; Hao, Y.H.; Fan, H.Y.; Cui, N.; Wang, S.S.; Song, T.F. Comparative Proteomic Analysis of Cucumber Roots Infected by Fusarium Oxysporum f. Sp. Cucumerium Owen. Physiol. Mol. Plant. Pathol. 2016, 96, 77–84. [Google Scholar] [CrossRef]
- Hu, J.L.; Lin, X.G.; Wang, J.H.; Shen, W.S.; Wu, S.; Peng, S.P.; Mao, T.T. Arbuscular Mycorrhizal Fungal Inoculation Enhances Suppression of Cucumber Fusarium Wilt in Greenhouse Soils. Pedosphere 2010, 20, 586–593. [Google Scholar] [CrossRef]
- Wang, M.; Ling, N.; Dong, X.; Zhu, Y.; Shen, Q.; Guo, S. Thermographic Visualization of Leaf Response in Cucumber Plants Infected with the Soil-Borne Pathogen Fusarium Oxysporum f. Sp. Cucumerinum. Plant. Physiol. Biochem. 2012, 61, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Martínez, R.; Aguilar, M.I.; Guirado, M.L.; Álvarez, A.; Gómez, J. First Report of Fusarium Wilt of Cucumber Caused by Fusarium Oxysporum in Spain. Plant. Pathol. 2003, 52, 410. [Google Scholar] [CrossRef]
- Hammerschmidt, R. Phytoalexins: What Have We Learned After 60 Years? Annu. Rev. Phytopathol. 1999, 37, 285–306. [Google Scholar] [CrossRef]
- Misra, A.K.; Gupta, V. Trichoderma: Biology, Biodiversity and Biotechnology. J. Eco-Friendly Agric. 2009, 4, 99–117. [Google Scholar]
- Gupta, V.; Misra, A.; Gupta, A.; Pandey, B.; Gaur, R. Rapd-Pcr of Trichoderma Isolates and In Vitro Antagonism Against Fusarium Wilt Pathogens of Psidium Guajaval. J. Plant. Prot. Res. 2010, 50, 256–262. [Google Scholar] [CrossRef]
- Minic, Z. Physiological Roles of Plant Glycoside Hydrolases. Planta 2008, 227, 723–740. [Google Scholar] [CrossRef]
- Sasaki, C.; Varum, K.M.; Itoh, Y.; Tamoi, M.; Fukamizo, T. Rice Chitinases: Sugar Recognition Specificities of the Individual Subsites. Glycobiology 2006, 16, 1242–1250. [Google Scholar] [CrossRef]
- Davis, J.M.; Wu, H.; Cooke, J.E.K.; Reed, J.M.; Luce, K.S.; Michler, C.H. Pathogen Challenge, Salicylic Acid, and Jasmonic Acid Regulate Expression of Chitinase Gene Homologs in Pine. Mol. Plant. Microbe Interact. 2002, 15, 380–387. [Google Scholar] [CrossRef]
- Singh, A.; Isaac Kirubakaran, S.; Sakthivel, N. Heterologous Expression of New Antifungal Chitinase from Wheat. Protein Expr. Purif. 2007, 56, 100–109. [Google Scholar] [CrossRef]
- Edreva, A. Pathogenesis-Related Proteins: Research Progress in the Last 15 Years. Gen. Appl. Plant. Physiol. 2005, 31, 105–124. [Google Scholar]
- Passarinho, P.A.; Van Hengel, A.J.; Fransz, P.F.; De Vries, S.C. Expression Pattern of the Arabidopsis Thaliana AtEP3/AtchitIV Endochitinase Gene. Planta 2001, 212, 556–567. [Google Scholar] [CrossRef] [PubMed]
- Patil, N.S.; Jadhav, J.P. Significance of Penicillium Ochrochloron Chitinase as a Biocontrol Agent against Pest Helicoverpa Armigera. Chemosphere 2015, 128, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, Z.; Yan, P.; Huang, S.; Fei, Z.; Lin, K. RNA-Seq Improves Annotation of Protein-Coding Genes in the Cucumber Genome. BMC Genom. 2011, 12. [Google Scholar] [CrossRef]
- Di Rosa, M.; Distefano, G.; Zorena, K.; Malaguarnera, L. Chitinases and immunity: Ancestral molecules with new functions. Immunobiology 2016, 221, 399–411. [Google Scholar] [CrossRef]
- Tobias, P.A.; Christie, N.; Naidoo, S.; Guest, D.I.; Külheim, C. Identification of the Eucalyptus Grandis Chitinase Gene Family and Expression Characterization under Different Biotic Stress Challenges. Tree Physiol. 2017, 37, 565–582. [Google Scholar] [CrossRef]
- Sigrist, C.J.A.; De Castro, E.; Cerutti, L.; Atrice, B.; Cuche, A.; Hulo, N.; Bridge, A.; Bougueleret, L.; Xenarios, I. New and Continuing Developments at PROSITE. Nucleic Acid Res. 2012, 41. [Google Scholar] [CrossRef]
- Chen, J.; Piao, Y.; Liu, Y.; Li, X.; Piao, Z. Genome-Wide Identification and Expression Analysis of Chitinase Gene Family in Brassica Rapa Reveals Its Role in Clubroot Resistance. Plant. Sci. 2018, 270, 257–267. [Google Scholar] [CrossRef]
- Lescot, M. PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for in Silico Analysis of Promoter Sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Chow, C.N.; Zheng, H.Q.; Wu, N.Y.; Chien, C.H.; Huang, H.D.; Lee, T.Y.; Chiang-Hsieh, Y.F.; Hou, P.F.; Yang, T.Y.; Chang, W.C. PlantPAN 2.0: An Update of Plant Promoter Analysis Navigator for Reconstructing Transcriptional Regulatory Networks in Plants. Nucleic Acids Res. 2016, 44, D1154–D1164. [Google Scholar] [CrossRef]
- Richa, K.; Tiwari, I.M.; Kumari, M.; Devanna, B.N.; Sonah, H.; Kumari, A.; Nagar, R.; Sharma, V.; Botella, J.R.; Sharma, T.R. Functional Characterization of Novel Chitinase Genes Present in the Sheath Blight Resistance QTL: QSBR11-1 in Rice Line Tetep. Front. Plant. Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Sugui, J.A.; Deising, H.B. Isolation of Infection-Specific Sequence Tags Expressed during Early Stages of Maize Anthracnose Disease Development. Mol. Plant. Pathol. Pathol. 2002, 3, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Caldo, R.A.; Nettleton, D.; Peng, J.; Wise, R.P. Stage-Specific Suppression of Basal Defense Discriminates Barley Plants Containing Fast-and Delayed-Acting Mla Powdery Mildew Resistance Alleles. Mol. Plant. Microbe Interact. 2006, 19, 939–947. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, T.B.; Balaji, B.; Breeden, J.; Goodwin, S.B. Resistance of Wheat to Mycosphaerella Graminicola Involves Early and Late Peaks of Gene Expression. Physiol. Mol. Plant. Pathol. 2007, 71, 55–68. [Google Scholar] [CrossRef]
- Takahara, H.; Dolf, A.; Endl, E.; O’Connell, R. Flow Cytometric Purification of Colletotrichum Higginsianum Biotrophic Hyphae from Arabidopsis Leaves for Stage-Specific Transcriptome Analysis. Plant. J. 2009, 59, 672–683. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Park, J.; Choi, H.; Burla, B.; Kretzschmar, T.; Lee, Y.; Martinoia, E. Plant ABC Transporters. Arab. B. 2011, 9, e0153. [Google Scholar] [CrossRef]
- Adrangi, S.; Faramarzi, M.A. From Bacteria to Human: A Journey into the World of Chitinases. Biotechnol. Adv. 2013, 31, 1786–1795. [Google Scholar] [CrossRef]
- Casneuf, T.; De Bodt, S.; Raes, J.; Maere, S.; Van de Peer, Y. Nonrandom Divergence of Gene Expression Following Gene and Genome Duplications in the Flowering Plant Arabidopsis Thaliana. Genome Biol. 2006, 7, R13. [Google Scholar] [CrossRef]
- Ganko, E.W.; Meyers, B.C.; Vision, T.J. Divergence in Expression between Duplicated Genes in Arabidopsis. Mol. Biol. Evol. 2007, 24, 2298–2309. [Google Scholar] [CrossRef]
- Hardt, M.; Laine, R.A. Mutation of Active Site Residues in the Chitin-Binding Domain ChBD ChiA1 from Chitinase A1 of Bacillus Circulans Alters Substrate Specificity: Use of a Green Fluorescent Protein Binding Assay. Arch. Biochem. Biophys. 2004, 426, 286–297. [Google Scholar] [CrossRef]
- Jeffares, D.; Penkett, C.J.; Bahler, P.J. Rapidly Regulated Genes Are Intron Poor. Trends Genet. 2008, 24, 375–378. [Google Scholar] [CrossRef]
- Zhou, F.; Guo, Y.; Qiu, L.J. Genome-Wide Identification and Evolutionary Analysis of Leucine-Rich Repeat Receptor-like Protein Kinase Genes in Soybean. BMC Plant. Biol. 2016, 16, 58. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.W.; Wang, X.N.; Fu, L.S.; Sun, J.; Zheng, W.; Li, Z.F.; College, A. Identification of the Trehalose-6-Phosphate Synthase Gene Family in Winter Wheat and Expression Analysis under Conditions of Freezing Stress. J. Genet. 2015, 94, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Sattelmacher, B. The Apoplast and Its Significance for Plant Mineral Nutrition. New Phytol. 2001, 149, 167–192. [Google Scholar] [CrossRef]
- Samac, D.A.; Hironaka, C.M.; Yallaly, P.E.; Shah, D.M. Isolation and Characterization of the Genes Encoding Basic and Acidic Chitinase in Arabidopsis Thaliana. Plant. Physiol. 1990, 93, 907–914. [Google Scholar] [CrossRef]
- De Gerhardt, L.B.A.; Sachetto-Martins, G.; Contarini, M.G.; Sandroni, M.; de, P.; Ferreira, R.; de Lima, V.M.; Cordeiro, M.C.; de Oliveira, D.E.; Margis-Pinheiro, M. Arabidopsis Thaliana Class IV Chitinase Is Early Induced during the Interaction with Xanthomonas Campestris. FEBS Lett. 1997, 419, 69–75. [Google Scholar] [CrossRef]
- Li, P.; Pei, Y.; Sang, X.; Ling, Y.; Yang, Z.; He, G. Transgenic Indica Rice Expressing a Bitter Melon (Momordica Charantia) Class I Chitinase Gene (McCHIT1) Confers Enhanced Resistance to Magnaporthe Grisea and Rhizoctonia Solani. Eur. J. Plant. Pathol. 2009, 125, 533–543. [Google Scholar] [CrossRef]
- Gao, Y.; Zan, X.; Wu, X.; Yao, L.; Chen, Y.; Jia, S.; Zhao, K. Identification of Fungus-Responsive Cis -Acting Element in the Promoter of Brassica Juncea Chitinase Gene, BjCHI1. Plant. Sci. 2014, 215–216, 190–198. [Google Scholar] [CrossRef]
- Kasprzewska, A. Plant Chitinases—Regulation and Function. Cell. Mol. Biol. Lett. 2003, 8, 809–824. [Google Scholar]
- Taira, T.; Toma, N.; Ichi, M.; Takeuchi, M.; Ishihara, M. Tissue Distribution, Synthesis Stage, and Ethylene Induction of Pineapple (Ananas Comosus) Chitinases. Biosci. Biotechnol. Biochem. 2005, 69, 852–854. [Google Scholar] [CrossRef]
- Yamamoto, S.; Nakano, T.; Suzuki, K.; Shinshi, H. Elicitor-Induced Activation of Transcription via W Box-Related Cis-Acting Elements from a Basic Chitinase Gene by WRKY Transcription Factors in Tobacco. Biochim. Biophys. Acta-Gene Struct. Expr. 2004, 1679, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Rakwal, R.; Yang, G.; Komatsu, S. Chitinase Induced by Jasmonic Acid, Methyl Jasmonate, Ethylene and Protein Phosphatase Inhibitors in Rice. Mol. Biol. Rep. 2004, 31, 113–119. [Google Scholar] [CrossRef] [PubMed]
- van Loon, L.C.; Rep, M.; Pieterse, C.M.J. Significance of Inducible Defense-Related Proteins in Infected Plants. Annu. Rev. Phytopathol. 2006, 44, 135–162. [Google Scholar] [CrossRef] [PubMed]
- Kirsch, C.; Logemann, E.; Lippok, B.; Schmelzer, E.; Hahlbrock, K. A Highly Specific Pathogen-Responsive Promoter Element from the Immediate-Early Activated CMPG1 Gene in Petroselinum Crispum. Plant. J. 2001, 26, 217–227. [Google Scholar] [CrossRef]
- Rushton, P.J. Synthetic Plant Promoters Containing Defined Regulatory Elements Provide Novel Insights into Pathogen- and Wound-Induced Signaling. Plant. Cell Online 2002, 14, 749–762. [Google Scholar] [CrossRef]
- de las Mercedes Dana, M.; Pintor-Toro, J.A.; Cubero, B. Transgenic Tobacco Plants Overexpressing Chitinases of Fungal Origin Show Enhanced Resistance to Biotic and Abiotic Stress Agents. Plant. Physiol. 2006, 142, 722–730. [Google Scholar] [CrossRef]
- Zhong, R.; Kays, S.J.; Schroeder, B.P.; Ye, Z.H. Mutation of a Chitinase-Like Gene Causes Ectopic Deposition of Lignin, Aberrant Cell Shapes, and Overproduction of Ethylene. Plant. Cell Online 2002, 14, 165–179. [Google Scholar] [CrossRef]
- Leubner-Metzger, G.; Meins, F.J. Functions and Regulation of Plant β-1,3-Glucanases (PR-2). In Pathogenesis-Related Proteins in Plants; Datta, S.K., Muthukrishnan, S., Eds.; CRC Press: Boca Raton, FL, USA, 1999; pp. 49–76. [Google Scholar]
- Neuhaus, J.M. Plant chitinases (PR-3, PR-4, PR-8, PR-11). In Pathogenesis-Related Proteins in Plants; Datta, S.K., Muthukrishnan, S., Eds.; CRC Press: Boca Raton, FL, USA, 1999; pp. 77–105. [Google Scholar]
- Wu, C.T.; Bradford, K.J. Class I Chitinase and 1,3-Glucanase are Differentially Regulated by Wounding, Methyl Jasmonate, Ethylene, and Gibberellin in Tomato Seeds and Leaves. Plant. Physiol. 2003, 133, 263–273. [Google Scholar] [CrossRef]
- Datta, K.; Tu, J.; Oliva, N.; Ona, I.; Velazhahan, R.; Mew, T.W.; Muthukrishnan, S.; Datta, S.K. Enhanced Resistance to Sheath Blight by Constitutive Expression of Infection-Related Rice Chitinase in Transgenic Elite Indica Rice Cultivars. Plant. Sci. 2001, 160, 405–414. [Google Scholar] [CrossRef]
- Javed, S.; Ahmad, M.; Ahmad, M.; Abdin, M.; Hamid, R.; Khan, M.; Musarrat, J. Chitinases: An Update. J. Pharm. Bioallied Sci. 2013, 5, 21. [Google Scholar] [CrossRef]
- Xin, M.; Wang, X.; Peng, H.; Yao, Y.; Xie, C.; Han, Y.; Ni, Z.; Sun, Q. Transcriptome Comparison of Susceptible and Resistant Wheat in Response to Powdery Mildew Infection. Genomics, Proteomics Bioinforma. 2012, 10, 94–106. [Google Scholar] [CrossRef] [PubMed]
- Takenaka, Y.; Nakano, S.; Tamoi, M.; Sakuda, S.; Fukamizo, T. Chitinase Gene Expression in Response to Environmental Stresses in Arabidopsis Thaliana: Chitinase Inhibitor Allosamidin Enhances Stress Tolerance. Biosci. Biotechnol. Biochem. 2009, 73, 1066–1071. [Google Scholar] [CrossRef] [PubMed]
- Brunner, F.; Stintzi, A.; Fritig, B.; Legrand, M. Substrate Specificities of Tobacco Chitinases. Plant. J. 1998, 14, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Taira, T.; Ohnuma, T.; Yamagami, T.; Aso, Y.; Ishiguro, M.; Ishihara, M. Antifungal Activity of Rye (Secale Cereale) Seed Chitinases: The Different Binding Manner of Class I and Class II Chitinases to the Fungal Cell Walls. Biosci. Biotechnol. Biochem. 2002, 66, 970–977. [Google Scholar] [CrossRef]
- Taira, T.; Mahoe, Y.; Kawamoto, N.; Onaga, S.; Iwasaki, H.; Ohnuma, T.; Fukamizo, T. Cloning and Characterization of a Small Family 19 Chitinase from Moss (Bryum Coronatum). Glycobiology 2011, 21, 644–654. [Google Scholar] [CrossRef]
- Oyeleye, A.; Normi, Y.M. Chitinase: Diversity, Limitations, and Trends in Engineering for Suitable Applications. Biosci. Rep. 2018, 38, BSR2018032300. [Google Scholar] [CrossRef]
- Yamamoto, T.; Iketani, H.; Ieki, H.; Nishizawa, Y.; Notsuka, K.; Hibi, T.; Hayashi, T.; Matsuta, N. Transgenic Grapevine Plants Expressing a Rice Chitinase with Enhanced Resistance to Fungal Pathogens. Plant. Cell Rep. 2000, 19, 639–646. [Google Scholar] [CrossRef]
- Takahashi, W.; Fujimori, M.; Miura, Y.; Komatsu, T.; Nishizawa, Y.; Hibi, T.; Takamizo, T. Increased Resistance to Crown Rust Disease in Transgenic Italian Ryegrass (Lolium Multiflorum Lam.) Expressing the Rice Chitinase Gene. Plant. Cell Rep. 2005, 23, 811–818. [Google Scholar] [CrossRef]
- Chai, C.; Shankar, R.; Jain, M.; Subudhi, P.K. Genome-Wide Discovery of DNA Polymorphisms by Whole Genome Sequencing Differentiates Weedy and Cultivated Rice. Sci. Rep. 2018, 8, 14218. [Google Scholar] [CrossRef]
- Hudson, T. Wanted: Regulatory SNPs. Nat. Genet. 2003, 33, 439–440. [Google Scholar] [CrossRef]
- Chorley, B.N.; Wang, X.; Campbell, M.R.; Pittman, G.S.; Noureddine, M.A.; Bell, D.A. Discovery and Verification of Functional Single Nucleotide Polymorphisms in Regulatory Genomic Regions: Current and Developing Technologies. Mutat. Res. - Rev. Mutat. Res. 2008, 659, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional Classification of Proteins via Subfamily Domain Architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef] [PubMed]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER Web Server: 2018 Update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef] [PubMed]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam Protein Families Database in 2019. Nucleic Acids Res. 2018. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, A.L.; Attwood, T.K.; Babbitt, P.C.; Blum, M.; Bork, P.; Bridge, A.; Brown, S.D.; Chang, H.Y.; El-Gebali, S.; Fraser, M.I.; et al. InterPro in 2019: Improving Coverage, Classification and Access to Protein Sequence Annotations. Nucleic Acids Res. 2018. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2009; pp. 571–607. [Google Scholar]
- Pierleoni, A.; Martelli, P.L.; Fariselli, P.; Casadio, R. BaCelLo: A Balanced Subcellular Localization Predictor. Bioinformatics 2006, 22. [Google Scholar] [CrossRef]
- Nielsen, H. Predicting Secretory Proteins with SignalP. In Protein Function Prediction. Methods in Molecular Biology; Kihara, D., Ed.; Humana Press: Totowa, NJ, USA, 2017; Volume 1611, pp. 59–73. [Google Scholar]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTALW: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An Upgraded Gene Feature Visualization Server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME Suite: Tools for Motif Discovery and Searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Voorrips, R.E. MapChart: Software for the Graphical Presentation of Linkage Maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Christie, N.; Tobias, P.A.; Naidoo, S.; Külheim, C. The Eucalyptus Grandis NBS-LRR Gene Family: Physical Clustering and Expression Hotspots. Front. Plant. Sci. 2016, 6. [Google Scholar] [CrossRef]
- Darzentas, N. Circoletto: Visualizing Sequence Similarity with Circos. Bioinformatics 2010, 26, 2620–2621. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2013, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Ulitsky, I.; Maron-Katz, A.; Shavit, S.; Sagir, D.; Linhart, C.; Elkon, R.; Tanay, A.; Sharan, R.; Shiloh, Y.; Shamir, R. Expander: From Expression Microarrays to Networks and Functions. Nat. Protoc. 2010, 5, 303–322. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, X.; Liu, B.; Wang, W.; Liu, X.; Chen, C.; Liu, X.; Yang, S.; Ren, H. A GAMYB Homologue CsGAMYB1 Regulates Sex Expression of Cucumber via an Ethylene-Independent Pathway. J. Exp. Bot. 2014, 65, 3201–3213. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing Real-Time PCR Data by the Comparative CT Method. Nat. Protoc. 2008, 3, 1101. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce Framework for Analyzing Next-Generation DNA Sequencing Data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]


| Gene Name | Gene ID | Class | Gene Position | CDS (bp) | Size (aa) | MW (kDa) | pI | Localization | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Start | End (+/−) | Signal P | BaCello | |||||||
| CsChi1 | Csa1G267220 | V | 12946374 | 12947531 (+) | 1158 | 385 | 42.11724 | 4.95 | S.P | S.P |
| CsChi2 | Csa1G267230 | V | 12949093 | 12950626 (−) | 1137 | 378 | 42.499 | 7.73 | S.P | S.P |
| CsChi3 | Csa1G267240 | V | 12954742 | 12956249 (−) | 1125 | 374 | 40.43333 | 8.86 | S.P | S.P |
| CsChi4 | Csa1G496300 | III | 17555204 | 17556387 (+) | 933 | 310 | 34.62323 | 6.14 | C | |
| CsChi5 | Csa1G534750 | IV | 19147451 | 19148640 (+) | 837 | 278 | 30.03272 | 4.79 | S.P | S.P |
| CsChi6 | Csa2G008760 | V | 1514322 | 1516483 (−) | 1125 | 374 | 42.66634 | 6.87 | C | |
| CsChi7 | Csa2G193360 | III | 10356474 | 10357487 (−) | 1014 | 337 | 36.81949 | 5.09 | S.P | S.P |
| CsChi8 | Csa2G193370 | III | 10361066 | 10361589 (−) | 399 | 132 | 14.80392 | 8.84 | S.P | S.P |
| CsChi9 | Csa2G302200 | III | 14586790 | 14587590 (+) | 801 | 266 | 29.79955 | 4.6 | N | |
| CsChi10 | Csa3G002420 | II | 304482 | 307249 (+) | 942 | 313 | 33.64859 | 5.14 | S.P | S.P |
| CsChi11 | Csa3G119680 | III | 6826381 | 6830851 (+) | 1290 | 429 | 48.53536 | 6.91 | C | |
| CsChi12 | Csa3G120470 | III | 6993545 | 6994578 (+) | 1011 | 336 | 37.34421 | 5.85 | S.P | S.P |
| CsChi13 | Csa3G120480 | III | 6996557 | 6997717 (+) | 927 | 308 | 34.25192 | 8.54 | S.P | S.P |
| CsChi14 | Csa3G120500 | III | 7006203 | 7007553 (+) | 891 | 296 | 31.94549 | 8.69 | S.P | S.P |
| CsChi15 | Csa3G166220 | II | 10980690 | 10983104 (−) | 969 | 322 | 35.63844 | 6.2 | S.P | S.P |
| CsChi16 | Csa4G017110 | II | 2283413 | 2285478 (−) | 957 | 318 | 35.41338 | 6.23 | S.P | S.P |
| CsChi17 | Csa5G128240 | II | 3140161 | 3140625 (+) | 405 | 154 | 17.33685 | 8.61 | S.P | S.P |
| CsChi18 | Csa5G139730 | III | 3709013 | 3710042 (+) | 888 | 295 | 31.12939 | 9.53 | S.P | C |
| CsChi19 | Csa5G139740 | III | 3711563 | 3712459 (−) | 897 | 298 | 31.92364 | 4.12 | S.P | S.P |
| CsChi20 | Csa5G139760 | III | 3715029 | 3716187 (−) | 879 | 292 | 30.76542 | 4.38 | S.P | S.P |
| CsChi21 | Csa5G139770 | III | 3718639 | 3719610 (−) | 972 | 323 | 34.47814 | 5.52 | S.P | S.P |
| CsChi22 | Csa6G301040 | III | 14333727 | 14334641 (−) | 915 | 304 | 34.10921 | 5.61 | S.P | S.P |
| CsChi23 | Csa6G507520 | I | 26077903 | 26079303 (−) | 963 | 320 | 34.829 | 8.4 | S.P | S.P |
| CsChi24 | Csa6G508020 | I | 26082160 | 26084273 (−) | 1083 | 360 | 38.73501 | 5.71 | S.P | S.P |
| CsChi25 | Csa6G508520 | II | 26090192 | 26091124 (−) | 705 | 234 | 25.79373 | 5.78 | N | |
| CsChi26 | Csa6G509020 | II | 26095683 | 26096793 (+) | 777 | 258 | 29.19341 | 8.78 | N | |
| CsChi27 | Csa6G509030 | II | 26098005 | 26099490 (+) | 818 | 271 | 29.96452 | 9.44 | S.P | S.P |
| CsChi28 | Csa6G509040 | I | 26100924 | 26104416 (+) | 975 | 324 | 35.8648 | 6.31 | S.P | S.P |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bartholomew, E.S.; Black, K.; Feng, Z.; Liu, W.; Shan, N.; Zhang, X.; Wu, L.; Bailey, L.; Zhu, N.; Qi, C.; et al. Comprehensive Analysis of the Chitinase Gene Family in Cucumber (Cucumis sativus L.): From Gene Identification and Evolution to Expression in Response to Fusarium oxysporum. Int. J. Mol. Sci. 2019, 20, 5309. https://doi.org/10.3390/ijms20215309
Bartholomew ES, Black K, Feng Z, Liu W, Shan N, Zhang X, Wu L, Bailey L, Zhu N, Qi C, et al. Comprehensive Analysis of the Chitinase Gene Family in Cucumber (Cucumis sativus L.): From Gene Identification and Evolution to Expression in Response to Fusarium oxysporum. International Journal of Molecular Sciences. 2019; 20(21):5309. https://doi.org/10.3390/ijms20215309
Chicago/Turabian StyleBartholomew, Ezra S., Kezia Black, Zhongxuan Feng, Wan Liu, Nan Shan, Xiao Zhang, Licai Wu, Latoya Bailey, Ning Zhu, Changhong Qi, and et al. 2019. "Comprehensive Analysis of the Chitinase Gene Family in Cucumber (Cucumis sativus L.): From Gene Identification and Evolution to Expression in Response to Fusarium oxysporum" International Journal of Molecular Sciences 20, no. 21: 5309. https://doi.org/10.3390/ijms20215309
APA StyleBartholomew, E. S., Black, K., Feng, Z., Liu, W., Shan, N., Zhang, X., Wu, L., Bailey, L., Zhu, N., Qi, C., Ren, H., & Liu, X. (2019). Comprehensive Analysis of the Chitinase Gene Family in Cucumber (Cucumis sativus L.): From Gene Identification and Evolution to Expression in Response to Fusarium oxysporum. International Journal of Molecular Sciences, 20(21), 5309. https://doi.org/10.3390/ijms20215309






