Abstract
Cigarette smoking causes lung inflammation and tissue damage. Lung fibroblasts play a major role in tissue repair. Previous studies have reported smoking-associated changes in fibroblast responses and methylation patterns. Our aim was to identify the effect of current smoking on miRNA expression in primary lung fibroblasts. Small RNA sequencing was performed on lung fibroblasts from nine current and six ex-smokers with normal lung function. MiR-335-5p and miR-335-3p were significantly downregulated in lung fibroblasts from current compared to ex-smokers (false discovery rate (FDR) <0.05). Differential miR-335-5p expression was validated with RT-qPCR (p-value = 0.01). The results were validated in lung tissue from current and ex-smokers and in bronchial biopsies from non-diseased smokers and never-smokers (p-value <0.05). The methylation pattern of the miR-335 host gene, determined by methylation-specific qPCR, did not differ between current and ex-smokers. To obtain insights into the genes regulated by miR-335-5p in fibroblasts, we overlapped all proven miR-335-5p targets with our previously published miRNA targetome data in lung fibroblasts. This revealed Rb1, CARF, and SGK3 as likely targets of miR-335-5p in lung fibroblasts. Our study indicates that miR-335-5p downregulation due to current smoking may affect its function in lung fibroblasts by targeting Rb1, CARF and SGK3.
1. Introduction
Cigarette smoke consists of a complex mixture of thousands of toxic chemicals and over 1015 reactive oxygen species [1]. Oxidative stress caused by cigarette smoking can dysregulate cell function, and induce the damage and death of the cellular constituents of the lungs [2]. Smoking is a major cause of complex lung diseases such as chronic obstructive pulmonary disease. It is important to investigate its effect in “normal” lung at the molecular level to gain a better understanding before investigating its effect in complex lung diseases. Several studies have shown smoking-induced changes in gene expression patterns in the lung. Epithelial cells are the first cells that encounter the inhaled smoke. Consequently, aberrant gene expression signatures were reported in epithelial cells when comparing current smokers with never-smokers [3,4]. Gene expression can be regulated by microRNAs (miRNAs). These small non-coding RNAs influence their target gene and/or protein expressions by binding based on sequence homology [5]. Marked changes in miRNA expression signatures have also been reported in the bronchial airway epithelial cells, small airway epithelium, whole blood, and induced sputum supernatant of current smokers compared to never-smokers [6,7,8,9]. The altered expression signatures may be due to a direct effect of smoking, but can also be caused by indirect effects such as smoking-induced aberrant DNA methylation patterns [10]. Most of the observed changes in gene expression and methylation are (slowly) reversible, while some of the effects may be permanent [4,10]. A previous study showed that when smokers quit smoking for three months, 65% of the miRNAs that were differentially expressed between current smokers and never-smokers return to the expression level of never-smokers [7].
Lung fibroblasts are the main guardians of connective tissue homeostasis. Therefore, lung fibroblasts, in close concert with other structural cells such as the epithelium, are considered crucial cells for tissue repair and remodeling of the lungs. Cigarette smoke suppresses lung repair by affecting multiple lung cells, as nicely reviewed by Rennard et al. [11]. Specific work in human lung fibroblasts showed that the fibronectin and elastin production was inhibited upon cigarette smoke extract (CSE) exposure [12,13]. In addition, lung fibroblasts were hampered in their proliferation, contractile function, and migration toward fibronectin upon CSE exposure [12,13,14]. Furthermore, human lung fibroblasts showed characteristics of senescence when treated with CSE [15].
To date, no information on differential miRNA expression in lung fibroblasts from donors with different smoking statuses is available. We hypothesized that the miRNA expression profile in lung fibroblasts is different in current smoking compared to ex-smoking donors, and that these changes in miRNA expression may affect the function of the fibroblasts. The aim of our study was to identify smoking status-related miRNA expression changes in lung fibroblasts and to assess miRNA-related functions that may be affected by current smoking.
2. Results
2.1. Subject Characteristics
Clinical characteristics of lung fibroblast, lung tissue, and bronchial biopsy donors are shown in Table 1. No significant difference was observed between current smokers, ex-smokers, and never-smokers in age and forced expiratory volume in one second/forced vital capacity ratio (FEV1/FVC). Moreover, the number of pack-years did not differ between current smokers and ex-smokers. Furthermore, the characteristics of subjects from whom we have obtained lung fibroblasts and those from whom we obtained lung tissue were not significantly different.

Table 1.
Patient characteristics of the donors of lung fibroblasts, lung tissue, and bronchial biopsy.
2.2. Differential miRNA Expression in Lung Fibroblasts of Current and Ex-Smokers
The miRNA expression profiles of lung fibroblasts of nine ex-smoking and six current smoking subjects were determined using small RNA sequencing. Total read counts and percentages of reads mapping to miRBase Release 21 are shown in Table S1. The top 10 most abundant miRNAs in both current smokers and ex-smokers covered 65% of all reads (Figure 1a). MiR-21-5p was the most abundant miRNA in both subgroups.
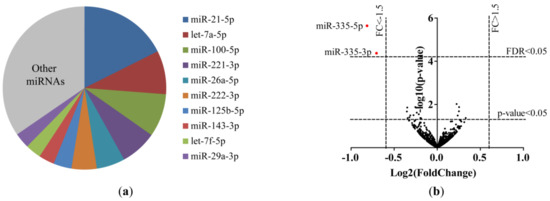
Figure 1.
MicroRNAs (miRNAs) in primary lung fibroblasts of ex-smokers and current smokers. (a) Top 10 most abundant miRNAs in primary lung fibroblasts of ex-smokers and current smokers. (b) Volcano plot of the 1339 miRNAs included in the analyses of the small RNA-sequencing data from lung fibroblasts. The lowest horizontal line represents the nominal p-value cut-off of 0.05. The upper horizontal line represents the false discovery rate (FDR) of 0.05. The two vertical lines represent the negative (left) and positive (right) fold change of 1.5. Differentially expressed miRNAs are indicated with a red dot. MiR-335-5p (FC = –1.8, FDR p-value = 0.0030) and miR-335-3p (FC = –1.6, FDR p-value = 0.0285) were lower expressed in current smokers compared to ex-smokers.
A total of 18 miRNAs (five upregulated and 13 downregulated) differed between current smokers and ex-smokers at a nominal p-value of <0.05 (Table S2). At a false discovery rate (FDR) cut-off <0.05, miR-335-5p and miR-335-3p were significantly differentially expressed with lower expression levels in current smokers compared to ex-smokers (fold change (FC) = –1.8, FDR p-value = 0.003 and FC = –1.6, FDR p-value = 0.0285, respectively; Figure 1b, Figure 2a, Figure S1a). Differential expression of miR-335-5p (p-value = 0.01, Figure 2b), but not miR-335-3p (Figure S1b), was validated using RT-qPCR in the same samples.
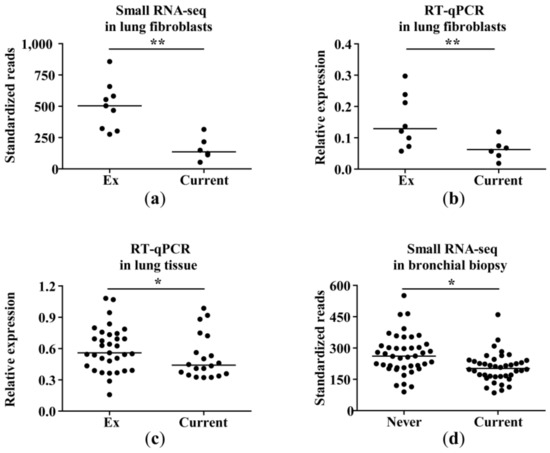
Figure 2.
Differentially expressed miR-335-5p in current smokers. (a) Standardized reads of miR-335-5p in lung fibroblasts of ex-smokers and current smokers, derived from small RNA sequencing data, as presented in Figure 1. ** FDR p-value = 0.0030. (b) Validation of miR-335-5p differential expression in the same lung fibroblasts samples using RT-qPCR. The data are presented as relative expression to RNU48 (2−ΔCp). One ex-smoker sample is missing due to a failure in experimental procedures. ** p-value = 0.0100. (c) MiR-335-5p RT-qPCR analysis in lung tissues of ex-smokers and current smokers. The data are presented as relative expression to RNU48 and RNU44 (2−ΔCp). * p-value = 0.048. (d) MiR-335-5p standardized read counts from small RNA sequencing data of bronchial biopsy samples. * p-value = 0.018.
2.3. Validation of miR-335-5p Differential Expression in Lung Tissue and Bronchial Biopsies
We validated the differential miR-335-5p expression in lung tissue from 20 current smokers compared to 33 ex-smokers (p-value <0.05, Figure 2c). In lung tissue samples of never-smokers, miR-335-5p expression was not significantly different from current smokers (Figure S2). In bronchial biopsies of healthy subjects with normal lung function, we observed a significantly lower expression of miR-335-5p in current smokers compared to never-smokers (p-value <0.05, FC = –1.2, Figure 2d).
To assess whether there is a direct smoke effect on miR-335-5p expression, we treated lung fibroblasts from four ex-smokers with 2.5% and 5% CSE. CSE treatment resulted in slightly decreased miR-335-5p levels in all (2.5% CSE) or three out of four fibroblast samples (5% CSE). This experiment supported our findings (Figure 3) pointing toward a CSE-dependent decrease in miR-335-5p levels.
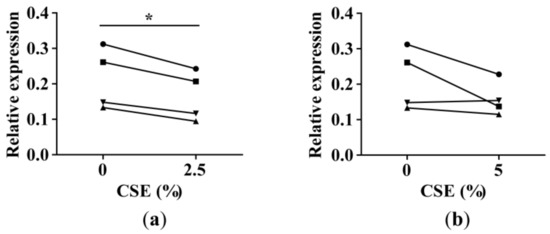
Figure 3.
MiR-335-5p expression in cigarette smoke extract (CSE)-treated lung fibroblasts. The CSE treatment started at passage 5 and lasted until the cells had reached a minimum of three cell divisions, i.e., lung fibroblasts were treated with CSE for 21 days. MiR-335-5p expression in lung fibroblasts of four ex-smokers (each individual is indicated with a symbol) treated with (a) 2.5% and (b) 5% CSE (RT-qPCR). The data are presented as relative expression to RNU48 (2-ΔCp). * p-value <0.05 using paired T-test.
2.4. No Regional Hypermethylation in miR-335 Host Gene in Lung Tissue of Current Smokers
In a previous study of hepatocellular carcinoma, decreased miR-335-5p expression was shown to be associated with the aberrant hypermethylation of a specific CpG island in an enhancer region of the miR-335 host gene [16]. To examine whether the decreased miR-335-5p expression in current smokers is due to hypermethylation, we did a methylation-specific qPCR (MSP) on the lung tissue samples used for measuring the miR-335-5p expression in Figure 2c. The location of miR-335-5p, the analyzed CpG island, and the primers used for the methylation specific qPCR are shown in Figure 4a. We found no differences in the proportion of methylated DNA between current smokers and ex-smokers (Figure 4b). The proportion of methylated DNA was also not correlated with miR-335-5p expression in lung tissue (not shown). Furthermore, the methylation status did not show any obvious change after CSE treatment in lung fibroblasts from ex-smokers (Figure S3a,b).
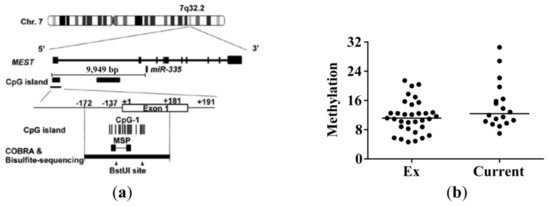
Figure 4.
Methylation status in the CpG island of the enhancer region of miR-335-5p in lung tissue. (a) Location of the primers for methylation-specific qPCR (MSP) and miR-335 (figure adapted from Dohi et al. [16]). (b) Methylation status in specific CpG island was determined in lung tissue from 33 ex-smokers and 18 current smokers. The methylation status was determined as follows: 2^(mean Cp value methylated-specific primers—mean Cp value unmethylated-specific primers). A clear difference was observed between the in vitro methylated and the unmethylated control DNA sample (not shown).
2.5. Predicted and Experimentally Proven Targets of miR-335-5p in the MiRNA Targetome of Lung Fibroblasts
We previously performed argonaute 2-immunoprecipitation (Ago2-IP) to identify the genes that are actively regulated by miRNAs in primary lung fibroblasts from two control subjects. These genes are collectively called the miRNA targetome [17]. To identify miR-335-5p target genes relevant in lung fibroblasts, we first assessed the enrichment of predicted miR-335-5p target genes in our previously published miRNA targetome of lung fibroblasts. In both controls, we observed 16 predicted miR-335-5p target genes in the top 1500 Ago2-IP enriched probes; however, this was not a significant enrichment compared to the proportion of miR-335-5p targets in all expressed genes (Table S3).
Next, we identified 40 experimentally proven target genes of miR-335-5p based on published luciferase reporter assays [18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55] (Table S4). Of these genes, RB transcriptional corepressor 1 (Rb1) [18,19], calcium responsive transcription factor (CARF) [20], and serum/glucocorticoid regulated kinase family member 3 (SGK3) [21] were present in the miRNA targetome of lung fibroblasts. In the RNA sequencing dataset of bronchial biopsies [56], we observed an increase of SGK3 (FC = 1.1, p-value <0.05) in smokers compared to never-smokers, whereas Rb1 and CARF were not significantly different.
3. Discussion
In this study, we found miR-335-5p levels to be lower in the parenchymal lung fibroblasts of current smokers compared to those of ex-smokers, and this was validated in lung tissue. Moreover, we also observed a lower miR-335-5p in bronchial biopsies from healthy current smokers compared to never-smokers. A smoking-related decrease in miR-335-5p was supported by decreased miR-335-5p levels upon the CSE treatment of fibroblasts. The lower expression level of this miRNA in the fibroblasts and lung tissue of current smokers was not associated with hypermethylation of the previously reported CpG island. Next, we found that three previously published miR-335-5p target genes, i.e., Rb1, CARF, and SGK3, were present in the miRNA targetome of lung fibroblasts.
The differential expression of miR-335-5p in lung fibroblasts suggests a potential role of this miRNA in smoking-induced changes in fibroblast function. However, the exact role of miR-335-5p in lung fibroblasts is as yet unknown. In bone-marrow derived human mesenchymal stem cells, the overexpression of miR-335-5p had an inhibitory effect on cell proliferation, migration, and differentiation [34]. This suggests that lower miR-335-5p levels as observed in current smokers and upon CSE exposure in this study could result in enhanced proliferation, as well as in other cell types, such as fibroblasts. However, other studies in lung fibroblasts showed the opposite, i.e., short and long-term CSE exposure reduced the proliferation and migration [13,14]. As the predicted targets of miR-335-5p were not significantly enriched, we searched for the experimentally proven targets. Three of these experimentally proven targets, i.e., Rb1, CARF, and SGK3, were enriched in the Ago2-IP fraction in lung fibroblasts. The presence of these genes in the miRNA targetome shows active targeting by miRNAs, and this might involve targeting by miR-335-5p in lung fibroblasts. As we found a decreased expression of miR-335-5p in lung fibroblast from current smokers, we speculated that these genes might be upregulated in current smokers. SGK3 is a serine/threonine protein kinase, and to our knowledge, the function of this gene in lung fibroblasts is still unknown. Rb1 was the most prominently IP-enriched target gene of miR-335-5p. This protein-coding gene can negatively regulate the cell cycle by interacting with E2F Transcription Factor 1 (E2F1), which is required for the activation of genes involved in the S phase of the cell cycle [57]. A previous study showed that nicotine increased Rb1 expression in non-small cell lung cancer cell lines, and the knockdown of Rb1 inhibited cell proliferation [58]. In contrast to this finding, cigarette smoking has been shown to inhibit the proliferation of lung fibroblasts [13]. The second IP-enriched miR-335-5p target was CARF, which is a transcriptional activator. CARF was shown to induce the transcription of brain-derived neurotrophic factor (BDNF) exon III in rat neurons [59]. BDNF is also expressed in lung fibroblasts, and it was previously reported that BDNF increased the cell proliferation of lung fibroblasts [60]. However, it is unknown whether CARF also induces BDNF transcription in lung fibroblasts. Additional experiments are required to investigate the role of miR-335-5p and the function of the identified target genes in lung fibroblasts.
Furthermore, miR-335-5p has been reported to be involved in different cancer types, either as a tumor suppressor or as an oncomiR [61]. The downregulation of miR-335-5p in different cancer types was shown to be associated with aberrant DNA methylation [16,62,63]. As lung fibroblasts had differential miRNA expression after isolation and the in vitro culturing of the fibroblasts, it is conceivable that epigenetic changes are involved in the persistent change in miR-335-5p expression. Pilot data using 5-aza-2’-deoxycytidine (not shown) suggested that miR-335-5p expression in fibroblasts indeed may be regulated by DNA methylation. In our study, we focused on a specific CpG island in the miR-335 host gene enhancer region that was reported by Dohi et al. [16]. However, we did not find differences in the methylation status in lung tissue from current smokers and ex-smokers. Moreover, the methylation status of this specific region was also unchanged in lung fibroblasts from ex-smokers after CSE treatment. Thus, our findings suggest that the smoking-related downregulation of miR-335-5p in lung fibroblasts is not due to aberrant DNA methylation at this specific region. However, we cannot exclude that aberrant DNA methylation is present at other regions, which also may affect miR-335-5p expression. In addition to aberrant DNA methylation, cigarette smoke-induced histone modification in the lung has been reported [64], and thus is worthwhile to investigate.
We showed lower miR-335-5p levels in fibroblasts and lung tissue from current smokers compared to ex-smokers and in bronchial biopsies, as well as compared to never-smokers. However, in lung tissue, we did not observe a difference between current smokers and never-smokers. This could be due to lack of power, as the never-smoking group only consisted of 14 subjects.
In conclusion, we showed a lower miR-335-5p expression in lung fibroblasts and tissues from current smokers compared to ex-smokers, and in bronchial biopsies from current smokers compared to never-smokers, without a change in the regional methylation pattern of its host gene. The decreased expression of miR-335-5p in lung fibroblasts from current smokers may have an effect on the cell function via targeting Rb1, CARF, and SGK3.
4. Materials and Methods
4.1. Subjects
Small RNA sequencing was performed on human parenchymal lung fibroblasts isolated from the tissue samples of nine ex-smokers and six current smokers with normal lung function who underwent lung tumour resection surgery. Left-over, macroscopically normal lung tissue samples located far away from the tumor were used for the isolation of lung fibroblasts [65,66].
As validation cohorts, we analyzed lung tissue samples of 33 ex-smoking and 20 current smoking individuals with normal lung function by RT-qPCR. These lung tissue samples were also derived from ex-smokers and current smokers with normal lung function who underwent lung tumour resection surgery. In addition, we analyzed data from the bronchial biopsies of 42 never-smoking and of 40 currently smoking healthy individuals with normal lung function and no respiratory symptoms (Clinical Trials Identifier = NCT00848406 [67]).
This study was performed in accordance with the national ethical and professional guidelines on the use of human body material (“Code of conduct; Dutch federation of biomedical scientific societies”; https://www.federa.org/codes-conduct) and the Research Code of the University Medical Center Groningen (https://www.umcg.nl/SiteCollectionDocuments/English/Researchcode/umcg- research-code-2018-en.pdf). At the time of the experiments, the use of left-over lung tissue to isolate fibroblasts or to replicate the results did not fall within the scope of medical research involving human subjects in the Netherlands. Therefore, an ethics waiver was provided by the Medical Ethical Committee of the University Medical Center Groningen (METc UMCG). All samples and clinical information were de-identified before the start of the experimental procedures in this study.
4.2. Isolation, Cell Culture, and CSE Treatment of Primary Lung Fibroblasts
Primary lung fibroblasts were isolated and grown in complete Ham’s F12 medium supplemented with 10% (v/v) fetal calf serum (FCS), 100 U/mL penicillin/streptomycin, and 200 mM L-glutamine (all from Lonza, Breda, The Netherlands) and stored in liquid nitrogen until further use as previously described [17,68]. Fibroblast cultures were restored, cultured until passage 5, grown to around 90–100% confluence in complete Ham’s F12 culture medium, and then serum-starved (0.5% (v/v) FCS) for 24 hours before harvesting of the cells for RNA isolation.
Fibroblasts of four ex-smoking individuals were treated with 0%, 2.5%, and 5% CSE for 21 days to determine long-term smoke-exposure effects. Two 3R4F research-reference filterless cigarettes (Tobacco Research Institute, University of Kentucky, 12/2006) were bubbled into 25 ml complete Ham’s F12 medium supplemented with 10% (v/v) FCS, 100 U/mL penicillin/streptomycin, and 200 mM l-glutamine (all from Lonza) using a peristaltic pump. This was considered as 100% CSE, which was then diluted to 2.5% and 5% CSE in complete medium. The CSE treatment started at passage 5, and lasted until the cells had reached a minimum of three cell divisions. During cell culturing, half of the medium with and without CSE was replaced with fresh medium whenever there was an obvious change in color. The fibroblasts were harvested at passage 7 for RNA and DNA isolation.
4.3. RNA and DNA Isolation
Total RNA was isolated from primary lung fibroblasts and lung tissue samples using TRIzol (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. Genomic DNA was isolated using salt–chloroform extraction and isopropanol precipitation using standard procedures. The RNA and DNA concentrations were measured with a NanoDrop 1000 Spectrophotometer (Thermo Scientific, Wilmington, DE, USA). For small RNA sequencing, the RNA quantity and quality were determined using the LabChip GX (Perkin Elmer, Waltham, MA, USA).
4.4. Small RNA Sequencing
Total RNA (approximately 1 μg) was used to generate libraries with the NEXTflex Small RNA-seq kit V3 (Bioo Scientific, Uden, The Netherlands). Sequencing was performed on the NextSeq 500 sequencing system (Illumina, San Diego, CA, USA) according to the protocol of the manufacturer. TrimGAlore 0.3.7 was used to trim the adapter sequences of the raw reads. Subsequently, the reads were allocated to the known human miRNAs allowing one mismatch using the miRDeep2 V2.0.0.8 software [69] and miRBase Release 21 (http://www.mirbase.org/). The reads of miRNAs with the same mature sequence were summed up. Using the default filtering setting of the DESeq2 package in R, miRNAs not expressed in all samples were removed. This resulted in 1339 miRNAs for further analyses. The small RNA sequencing dataset generated for this manuscript is available for collaboration upon request.
4.5. RT-qPCR
To validate the differential miRNA expression, RT-qPCR was performed as described previously [17]. First, 10 ng of total RNA was reverse-transcribed using a multiplex approach with TaqMan primers (reference gene: RNU48 (Assay ID: 001006) or RNU44 (Assay ID: 001094), ssc-miR-335-5p (Assay ID: 244560_mat) and hsa-miR-335* (hsa-miR-335-3p, Assay ID:002185); Applied Biosystems, Carlsbad, CA, USA) [70]. Subsequently, qPCR was done using TaqMan microRNA assays (Applied Biosystems) and a LightCycler®480 Probes Master (Roche Diagnostics GmbH, Mannheim, Germany).
The reactions were run in triplicate on the LightCycler®480 Real-Time PCR system (Roche Diagnostics GmbH). The LightCycler®480 software release 1.5.0 (Roche Diagnostics GmbH) was used to analyze the data. The relative miRNA expression levels were calculated using the formula 2−ΔCp.
4.6. Bisulfite Treatment and Methylation-Specific qPCR
DNA from primary lung fibroblasts and lung tissue samples was treated with bisulfite using the EZ DNA Methylation-Gold™ Kit (Zymo Research, Irvine, CA, USA) according to the protocol of the manufacturer. The DNA of leukocytes in vitro methylated by SssI methyltransferase was used as a positive control, and untreated DNA was used as a negative control [71]. MSP for the miR-335 host gene was done on 10 ng of bisulfite-treated DNA using SYBR green PCR master mix (Applied Biosystems) and previously published methylated-specific primers (forward 5’-TTGTAATAGGTGGCGTTGAC-3’ and reverse 5’-ACTCGAAACTAAAACGTCGC-3’) and unmethylated-specific primers (forward 5’-TTTTTGTAATAGGTGGTGTTGAT-3’ and reverse 5’-ACTCAAAACTAAAACATCACCAA-3’) [16]. For each sample, qPCR with the methylated-specific and unmethylated-specific primers (annealing temperature 58 °C for 1.20 min) were run in triplicate on the same plate. The methylation status was determined as follows: 2^ (mean Cp value methylated-specific primers—mean Cp value unmethylated-specific primers).
4.7. Identification of miR-335-5p Targets Relevant for Lung Fibroblasts
We re-analyzed our previously published Ago2-IP data of primary lung fibroblasts from two control subjects to identify miR-335-5p target genes that are Ago2-IP-enriched, and thus targeted by miRNAs in lung fibroblasts [17]. This was done for a list of predicted targets of miR-335-5p identified using TargetScan version 7.2 [72] and for a list of experimentally proven, direct targets of miR-335-5p that was generated based on validation with luciferase reporter assays.
4.8. Statistical Analyses
To compare the subject characteristics between and within the study groups, Mann–Whitney U-test was used in IBM SPSS Statistics 20 software. Differential expression analysis of the small RNA sequencing data comparing miRNA expression in the lung fibroblasts of ex-smokers and current smokers, and in bronchial biopsies of never-smokers and current smokers, was performed using the Bioconductor-DESeq2 package (version 1.14.1) in R Project software (version 3.3.2). The data were adjusted for age, gender, and library preparation batch. A Benjamini–Hochberg FDR <0.05 was considered statistically significant.
For RT-qPCR data, significant differences for miR-335-5p levels in lung fibroblasts and lung tissues between current smokers and ex-smokers were tested using the one-tailed Mann–Whitney U-test. A paired T-test was used to test for miR-335-5p expression changes in CSE-treated lung fibroblasts. A chi-square test was performed on the percentage of predicted targets in the top 1500 enriched probes compared to the percentage of predicted targets in all expressed genes to assess the enrichment of predicted target genes in the Ago2-IP. A p-value below 0.05 was considered statistically significant.
Our previously published RNA-seq data in bronchial biopsies [56] were used to validate the proposed target genes. Differential expression analysis comparing current smokers with never-smokers was performed using the Bioconductor-DESeq2 package (version 1.14.1) in R Project software (version 3.3.2). The data were adjusted for age, gender, and batch.
Supplementary Materials
The Supplementary materials can be found at https://www.mdpi.com/1422-0067/20/20/5176/s1.
Author Contributions
Conceptualization, A.v.d.B., W.T., M.v.d.B., C-A.B., and J.K.; Formal analysis, J.O., A.F., I.M.B., C.J.V., and M.M.T.; Funding acquisition, W.T., B.G.O., and C.-A.B.; Methodology, J.O., A.v.d.B., A.F., I.M.B., W.T., C.J.V., B.G.O., K.K., M.M.T., M.v.d.B., C.-A.B., and J.K.; Resources, A.v.d.B., W.T., B.G.O., M.v.d.B., and C.-A.B.; Supervision, A.v.d.B., W.T., C.-A.B., and J.K.; Validation, J.O. and I.M.B.; Visualization, J.O.; Writing—original draft, J.O. and C.-A.B.; Writing—review and editing, J.O., A.v.d.B., A.F., I.M.B., W.T., C.J.V., B.G.O., K.K., M.M.T., M.v.d.B., C.-A.B., and J.K.
Funding
This research was funded by the Lung Foundation Netherlands (Longfonds), grant number 3.2.12.044, and by the Australian National Health and Medical Research Council (NHMRC), grant number APP1104704.
Acknowledgments
We thank the UMCG Genomics Coordination center, the UG Center for Information Technology and their sponsors BBMRI-NL and TarGet for storage and compute infrastructure. In addition, we would like to thank Wierd Kooistra for performing qMSP.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| miRNA | microRNA |
| CSE | cigarette smoke extract |
| FEV1 | forced expiratory volume in one second |
| FVC | forced vital capacity |
| FC | fold change |
| FDR | false discovery rate |
| MSP | methylation specific qPCR |
| Ago2-IP | argonaute 2-immunoprecipitation |
References
- Goldkorn, T.; Filosto, S.; Chung, S. Lung injury and lung cancer caused by cigarette smoke-induced oxidative stress: Molecular mechanisms and therapeutic opportunities involving the ceramide-generating machinery and epidermal growth factor receptor. Antioxid. Redox Signal. 2014, 21, 2149–2174. [Google Scholar] [CrossRef] [PubMed]
- Demedts, I.K.; Demoor, T.; Bracke, K.R.; Joos, G.F.; Brusselle, G.G. Role of apoptosis in the pathogenesis of COPD and pulmonary emphysema. Respir. Res. 2006, 7, 53. [Google Scholar] [CrossRef] [PubMed]
- Spira, A.; Beane, J.; Shah, V.; Liu, G.; Schembri, F.; Yang, X.; Palma, J.; Brody, J.S. Effects of cigarette smoke on the human airway epithelial cell transcriptome. Proc. Natl. Acad. Sci. USA 2004, 101, 10143–10148. [Google Scholar] [CrossRef] [PubMed]
- Vink, J.M.; Jansen, R.; Brooks, A.; Willemsen, G.; van Grootheest, G.; de Geus, E.; Smit, J.H.; Penninx, B.W.; Boomsma, D.I. Differential gene expression patterns between smokers and non-smokers: Cause or consequence? Addict. Biol. 2017, 22, 550–560. [Google Scholar] [CrossRef]
- Li, Z.; Rana, T.M. Therapeutic targeting of microRNAs: Current status and future challenges. Nat. Rev. Drug Discov. 2014, 13, 622–638. [Google Scholar] [CrossRef]
- Schembri, F.; Sridhar, S.; Perdomo, C.; Gustafson, A.M.; Zhang, X.; Ergun, A.; Lu, J.; Liu, G.; Zhang, X.; Bowers, J.; et al. MicroRNAs as modulators of smoking-induced gene expression changes in human airway epithelium. Proc. Natl. Acad. Sci. USA 2009, 106, 2319–2324. [Google Scholar] [CrossRef]
- Wang, G.; Wang, R.; Strulovici-Barel, Y.; Salit, J.; Staudt, M.R.; Ahmed, J.; Tilley, A.E.; Yee-Levin, J.; Hollmann, C.; Harvey, B.G.; et al. Persistence of smoking-induced dysregulation of miRNA expression in the small airway epithelium despite smoking cessation. PLoS ONE 2015, 10, e0120824. [Google Scholar] [CrossRef]
- Willinger, C.M.; Rong, J.; Tanriverdi, K.; Courchesne, P.L.; Huan, T.; Wasserman, G.A.; Lin, H.; Dupuis, J.; Joehanes, R.; Jones, M.R.; et al. MicroRNA Signature of Cigarette Smoking and Evidence for a Putative Causal Role of MicroRNAs in Smoking-Related Inflammation and Target Organ Damage. Circ. Cardiovasc. Genet. 2017, 10, e001678. [Google Scholar] [CrossRef]
- Van Pottelberge, G.R.; Mestdagh, P.; Bracke, K.R.; Thas, O.; van Durme, Y.M.; Joos, G.F.; Vandesompele, J.; Brusselle, G.G. MicroRNA expression in induced sputum of smokers and patients with chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2011, 183, 898–906. [Google Scholar] [CrossRef]
- Ambatipudi, S.; Cuenin, C.; Hernandez-Vargas, H.; Ghantous, A.; Le Calvez-Kelm, F.; Kaaks, R.; Barrdahl, M.; Boeing, H.; Aleksandrova, K.; Trichopoulou, A.; et al. Tobacco smoking-associated genome-wide DNA methylation changes in the EPIC study. Epigenomics 2016, 8, 599–618. [Google Scholar] [CrossRef]
- Rennard, S.I.; Togo, S.; Holz, O. Cigarette smoke inhibits alveolar repair: A mechanism for the development of emphysema. Proc. Am. Thorac. Soc. 2006, 3, 703–708. [Google Scholar] [CrossRef] [PubMed]
- Carnevali, S.; Nakamura, Y.; Mio, T.; Liu, X.; Takigawa, K.; Romberger, D.J.; Spurzem, J.R.; Rennard, S.I. Cigarette smoke extract inhibits fibroblast-mediated collagen gel contraction. Am. J. Physiol. 1998, 274 (Pt 1), L591–L598. [Google Scholar] [CrossRef]
- Miglino, N.; Roth, M.; Lardinois, D.; Sadowski, C.; Tamm, M.; Borger, P. Cigarette smoke inhibits lung fibroblast proliferation by translational mechanisms. Eur. Respir. J. 2012, 39, 705–711. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Romberger, D.J.; Tate, L.; Ertl, R.F.; Kawamoto, M.; Adachi, Y.; Mio, T.; Sisson, J.H.; Spurzem, J.R.; Rennard, S.I. Cigarette smoke inhibits lung fibroblast proliferation and chemotaxis. Am. J. Respir. Crit. Care Med. 1995, 151, 1497–1503. [Google Scholar] [CrossRef] [PubMed]
- Nyunoya, T.; Monick, M.M.; Klingelhutz, A.; Yarovinsky, T.O.; Cagley, J.R.; Hunninghake, G.W. Cigarette smoke induces cellular senescence. Am. J. Respir. Cell Mol. Biol. 2006, 35, 681–688. [Google Scholar] [CrossRef]
- Dohi, O.; Yasui, K.; Gen, Y.; Takada, H.; Endo, M.; Tsuji, K.; Konishi, C.; Yamada, N.; Mitsuyoshi, H.; Yagi, N.; et al. Epigenetic silencing of miR-335 and its host gene MEST in hepatocellular carcinoma. Int. J. Oncol. 2013, 42, 411–418. [Google Scholar] [CrossRef] [PubMed]
- Ong, J.; Timens, W.; Rajendran, V.; Algra, A.; Spira, A.; Lenburg, M.E.; Campbell, J.D.; van den Berge, M.; Postma, D.S.; van den Berg, A.; et al. Identification of transforming growth factor-beta-regulated microRNAs and the microRNA-targetomes in primary lung fibroblasts. PLoS ONE 2017, 12, e0183815. [Google Scholar] [CrossRef] [PubMed]
- Scarola, M.; Schoeftner, S.; Schneider, C.; Benetti, R. miR-335 directly targets Rb1 (pRb/p105) in a proximal connection to p53-dependent stress response. Cancer Res. 2010, 70, 6925–6933. [Google Scholar] [CrossRef]
- Shi, L.; Jiang, D.; Sun, G.; Wan, Y.; Zhang, S.; Zeng, Y.; Pan, T.; Wang, Z. miR-335 promotes cell proliferation by directly targeting Rb1 in meningiomas. J. Neuro-Oncol. 2012, 110, 155–162. [Google Scholar] [CrossRef]
- Yu, Y.; Gao, R.; Kaul, Z.; Li, L.; Kato, Y.; Zhang, Z.; Groden, J.; Kaul, S.C.; Wadhwa, R. Loss-of-function screening to identify miRNAs involved in senescence: Tumor suppressor activity of miRNA-335 and its new target CARF. Sci. Rep. 2016, 6, 30185. [Google Scholar] [CrossRef]
- Yao, L.; Li, M.; Hu, J.; Wang, W.; Gao, M. MiRNA-335-5p negatively regulates granulosa cell proliferation via SGK3 in PCOS. Reproduction 2018. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Tong, L.; Meng, H.; Zhu, W.; Guo, L.; Wei, T.; Zhang, J. MiR-335 regulates the chemo-radioresistance of small cell lung cancer cells by targeting PARP-1. Gene 2017, 600, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Heyn, H.; Engelmann, M.; Schreek, S.; Ahrens, P.; Lehmann, U.; Kreipe, H.; Schlegelberger, B.; Beger, C. MicroRNA miR-335 is crucial for the BRCA1 regulatory cascade in breast cancer development. Int. J. Cancer 2011, 129, 2797–2806. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, W.; Fu, Q. miR-335 suppresses migration and invasion by targeting ROCK1 in osteosarcoma cells. Mol. Cell. Biochem. 2013, 384, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, M.; Zhang, R.; Wang, Y.; Zang, W.; Ma, Y.; Zhao, G.; Zhang, G. Effect of miR-335 upregulation on the apoptosis and invasion of lung cancer cell A549 and H1299. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2013, 34, 3101–3109. [Google Scholar] [CrossRef]
- Xu, Y.; Zhao, F.; Wang, Z.; Song, Y.; Luo, Y.; Zhang, X.; Jiang, L.; Sun, Z.; Miao, Z.; Xu, H. MicroRNA-335 acts as a metastasis suppressor in gastric cancer by targeting Bcl-w and specificity protein 1. Oncogene 2012, 31, 1398–1407. [Google Scholar] [CrossRef] [PubMed]
- Shu, M.; Zheng, X.; Wu, S.; Lu, H.; Leng, T.; Zhu, W.; Zhou, Y.; Ou, Y.; Lin, X.; Lin, Y.; et al. Targeting oncogenic miR-335 inhibits growth and invasion of malignant astrocytoma cells. Mol. Cancer 2011, 10, 59. [Google Scholar] [CrossRef]
- Lu, Y.; Yang, H.; Yuan, L.; Liu, G.; Zhang, C.; Hong, M.; Liu, Y.; Zhou, M.; Chen, F.; Li, X. Overexpression of miR-335 confers cell proliferation and tumour growth to colorectal carcinoma cells. Mol. Cell. Biochem. 2016, 412, 235–245. [Google Scholar] [CrossRef]
- Dong, Y.; Liu, Y.; Jiang, A.; Li, R.; Yin, M.; Wang, Y. MicroRNA-335 suppresses the proliferation, migration, and invasion of breast cancer cells by targeting EphA4. Mol. Cell. Biochem. 2018, 439, 95–104. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, T.; Zhang, Z.; Lu, M.; Zhao, W.; Zeng, X.; Zhang, W. Long non-coding RNA TUG1 promotes migration and invasion by acting as a ceRNA of miR-335-5p in osteosarcoma cells. Cancer Sci. 2017, 108, 859–867. [Google Scholar] [CrossRef]
- Lynch, J.; Meehan, M.H.; Crean, J.; Copeland, J.; Stallings, R.L.; Bray, I.M. Metastasis suppressor microRNA-335 targets the formin family of actin nucleators. PLoS ONE 2013, 8, e78428. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Zhu, J.; Du, W.; Liu, S.; Zeng, Y.; Ding, Z.; Zhang, Y.; Wang, X.; Liu, Z.; Huang, J. CPNE1 is a target of miR-335-5p and plays an important role in the pathogenesis of non-small cell lung cancer. J. Exp. Clin. Cancer Res. CR 2018, 37, 131. [Google Scholar] [CrossRef] [PubMed]
- Lynch, J.; Fay, J.; Meehan, M.; Bryan, K.; Watters, K.M.; Murphy, D.M.; Stallings, R.L. MiRNA-335 suppresses neuroblastoma cell invasiveness by direct targeting of multiple genes from the non-canonical TGF-beta signalling pathway. Carcinogenesis 2012, 33, 976–985. [Google Scholar] [CrossRef] [PubMed]
- Tome, M.; Lopez-Romero, P.; Albo, C.; Sepulveda, J.C.; Fernandez-Gutierrez, B.; Dopazo, A.; Bernad, A.; Gonzalez, M.A. miR-335 orchestrates cell proliferation, migration and differentiation in human mesenchymal stem cells. Cell Death Differ. 2011, 18, 985–995. [Google Scholar] [CrossRef]
- Tavazoie, S.F.; Alarcon, C.; Oskarsson, T.; Padua, D.; Wang, Q.; Bos, P.D.; Gerald, W.L.; Massague, J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature 2008, 451, 147–152. [Google Scholar] [CrossRef]
- Zhang, S.; Kim, K.; Jin, U.H.; Pfent, C.; Cao, H.; Amendt, B.; Liu, X.; Wilson-Robles, H.; Safe, S. Aryl hydrocarbon receptor agonists induce microRNA-335 expression and inhibit lung metastasis of estrogen receptor negative breast cancer cells. Mol. Cancer Ther. 2012, 11, 108–118. [Google Scholar] [CrossRef]
- Luo, L.; Xia, L.; Zha, B.; Zuo, C.; Deng, D.; Chen, M.; Hu, L.; He, Y.; Dai, F.; Wu, J.; et al. miR-335-5p targeting ICAM-1 inhibits invasion and metastasis of thyroid cancer cells. Biomed. Pharmacother. Biomed. Pharmacother. 2018, 106, 983–990. [Google Scholar] [CrossRef]
- Sun, Z.; Zhang, Z.; Liu, Z.; Qiu, B.; Liu, K.; Dong, G. MicroRNA-335 inhibits invasion and metastasis of colorectal cancer by targeting ZEB2. Med Oncol. 2014, 31, 982. [Google Scholar] [CrossRef]
- Sandoval-Borquez, A.; Polakovicova, I.; Carrasco-Veliz, N.; Lobos-Gonzalez, L.; Riquelme, I.; Carrasco-Avino, G.; Bizama, C.; Norero, E.; Owen, G.I.; Roa, J.C.; et al. MicroRNA-335-5p is a potential suppressor of metastasis and invasion in gastric cancer. Clin. Epigenet. 2017, 9, 114. [Google Scholar] [CrossRef]
- Gao, Y.; Zeng, F.; Wu, J.Y.; Li, H.Y.; Fan, J.J.; Mai, L.; Zhang, J.; Ma, D.M.; Li, Y.; Song, F.Z. MiR-335 inhibits migration of breast cancer cells through targeting oncoprotein c-Met. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2015, 36, 2875–2883. [Google Scholar] [CrossRef]
- Alfano, D.; Gorrasi, A.; Li Santi, A.; Ricci, P.; Montuori, N.; Selleri, C.; Ragno, P. Urokinase receptor and CXCR4 are regulated by common microRNAs in leukaemia cells. J. Cell. Mol. Med. 2015, 19, 2262–2272. [Google Scholar] [CrossRef] [PubMed]
- Yue, J.; Wang, P.; Hong, Q.; Liao, Q.; Yan, L.; Xu, W.; Chen, X.; Zheng, Q.; Zhang, L.; Huang, D. MicroRNA-335-5p Plays Dual Roles in Periapical Lesions by Complex Regulation Pathways. J. Endod. 2017, 43, 1323–1328. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Guo, H.; Lu, S. MiR-335-5p restores cisplatin sensitivity in ovarian cancer cells through targeting BCL2L2. Cancer Med. 2018, 7, 4598–4609. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Chen, X.; Zhan, Y.; Jiang, W.; Liu, X.; Wang, X.; Wu, B. miR-335 inhibits the proliferation and invasion of clear cell renal cell carcinoma cells through direct suppression of BCL-W. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2015, 36, 6875–6882. [Google Scholar] [CrossRef]
- An, Y.; Cai, H.; Zhang, Y.; Liu, S.; Duan, Y.; Sun, D.; Chen, X.; He, X. circZMYM2 Competed Endogenously with miR-335-5p to Regulate JMJD2C in Pancreatic Cancer. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2018, 51, 2224–2236. [Google Scholar] [CrossRef]
- Meng, Y.; Zou, Q.; Liu, T.; Cai, X.; Huang, Y.; Pan, J. microRNA-335 inhibits proliferation, cell-cycle progression, colony formation, and invasion via targeting PAX6 in breast cancer cells. Mol. Med. Rep. 2015, 11, 379–385. [Google Scholar] [CrossRef]
- Wang, J.; Wang, X.; Liu, F.; Fu, Y. microRNA-335 inhibits colorectal cancer HCT116 cells growth and epithelial-mesenchymal transition (EMT) process by targeting Twist1. Die Pharm. 2017, 72, 475–481. [Google Scholar]
- Yang, B.; Huang, J.; Liu, H.; Guo, W.; Li, G. miR-335 directly, while miR-34a indirectly modulate survivin expression and regulate growth, apoptosis, and invasion of gastric cancer cells. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2016, 37, 1771–1779. [Google Scholar] [CrossRef]
- Gong, M.; Ma, J.; Guillemette, R.; Zhou, M.; Yang, Y.; Yang, Y.; Hock, J.M.; Yu, X. miR-335 inhibits small cell lung cancer bone metastases via IGF-IR and RANKL pathways. Mol. Cancer Res. MCR 2014, 12, 101–110. [Google Scholar] [CrossRef]
- Gao, L.; Yang, Y.; Xu, H.; Liu, R.; Li, D.; Hong, H.; Qin, M.; Wang, Y. MiR-335 functions as a tumor suppressor in pancreatic cancer by targeting OCT4. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2014, 35, 8309–8318. [Google Scholar] [CrossRef]
- Liang, H.; Zhang, C.; Guan, H.; Liu, J.; Cui, Y. LncRNA DANCR promotes cervical cancer progression by upregulating ROCK1 via sponging miR-335-5p. J. Cell. Physiol. 2019, 234, 7266–7278. [Google Scholar] [CrossRef] [PubMed]
- Tang, R.; Lei, Y.; Hu, B.; Yang, J.; Fang, S.; Wang, Q.; Li, M.; Guo, L. WW domain binding protein 5 induces multidrug resistance of small cell lung cancer under the regulation of miR-335 through the Hippo pathway. Br. J. Cancer 2016, 115, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Martin, N.T.; Nakamura, K.; Davies, R.; Nahas, S.A.; Brown, C.; Tunuguntla, R.; Gatti, R.A.; Hu, H. ATM-dependent MiR-335 targets CtIP and modulates the DNA damage response. PLoS Genet. 2013, 9, e1003505. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Bian, T.; Feng, J.; Qian, L.; Zhang, J.; Jiang, D.; Zhang, Q.; Li, X.; Liu, Y.; Shi, J. miR-335 inhibited cell proliferation of lung cancer cells by target Tra2beta. Cancer Sci. 2018, 109, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.M.; Sun, R.; Luo, D.H.; Sun, J.; Zhang, M.Y.; Wang, M.H.; Yang, Y.; Wang, H.Y.; Mai, S.J. Upregulated TRIM29 promotes proliferation and metastasis of nasopharyngeal carcinoma via PTEN/AKT/mTOR signal pathway. Oncotarget 2016, 7, 13634–13650. [Google Scholar] [CrossRef]
- Ong, J.; Woldhuis, R.R.; Boudewijn, I.M.; van den Berg, A.; Kluiver, J.; Kok, K.; Terpstra, M.M.; Guryev, V.; de Vries, M.; Vermeulen, C.J.; et al. Age-related gene and miRNA expression changes in airways of healthy individuals. Sci. Rep. 2019, 9, 3765. [Google Scholar] [CrossRef]
- Herwig, S.; Strauss, M. The retinoblastoma protein: A master regulator of cell cycle, differentiation and apoptosis. Eur. J. Biochem. 1997, 246, 581–601. [Google Scholar] [CrossRef]
- Du, X.; Qi, F.; Lu, S.; Li, Y.; Han, W. Nicotine upregulates FGFR3 and RB1 expression and promotes non-small cell lung cancer cell proliferation and epithelial-to-mesenchymal transition via downregulation of miR-99b and miR-192. Biomed. Pharmacother. Biomed. Pharmacother. 2018, 101, 656–662. [Google Scholar] [CrossRef]
- Tao, X.; West, A.E.; Chen, W.G.; Corfas, G.; Greenberg, M.E. A calcium-responsive transcription factor, CaRF, that regulates neuronal activity-dependent expression of BDNF. Neuron 2002, 33, 383–395. [Google Scholar] [CrossRef]
- Prakash, Y.S.; Martin, R.J. Brain-derived neurotrophic factor in the airways. Pharmacol. Ther. 2014, 143, 74–86. [Google Scholar] [CrossRef]
- Rojas, F.; Hernandez, M.E.; Silva, M.; Li, L.; Subramanian, S.; Wilson, M.J.; Liu, P. The Oncogenic Response to MiR-335 Is Associated with Cell Surface Expression of Membrane-Type 1 Matrix Metalloproteinase (MT1-MMP) Activity. PLoS ONE 2015, 10, e0132026. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.K.; Li, Y.S.; Zhang, C.D.; Dai, D.Q. Up-regulation of CRKL by microRNA-335 methylation is associated with poor prognosis in gastric cancer. Cancer Cell Int. 2017, 17, 28. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Serra, P.; Esteller, M. DNA methylation-associated silencing of tumor-suppressor microRNAs in cancer. Oncogene 2012, 31, 1609–1622. [Google Scholar] [CrossRef] [PubMed]
- Sundar, I.K.; Yao, H.; Rahman, I. Oxidative stress and chromatin remodeling in chronic obstructive pulmonary disease and smoking-related diseases. Antioxid. Redox Signal. 2013, 18, 1956–1971. [Google Scholar] [CrossRef]
- Noordhoek, J.A.; Postma, D.S.; Chong, L.L.; Menkema, L.; Kauffman, H.F.; Timens, W.; van Straaten, J.F.; van der Geld, Y.M. Different modulation of decorin production by lung fibroblasts from patients with mild and severe emphysema. J. Chronic Obstr. Pulm. Dis. 2005, 2, 17–25. [Google Scholar] [CrossRef]
- Noordhoek, J.A.; Postma, D.S.; Chong, L.L.; Vos, J.T.; Kauffman, H.F.; Timens, W.; van Straaten, J.F. Different proliferative capacity of lung fibroblasts obtained from control subjects and patients with emphysema. Exp. Lung Res. 2003, 29, 291–302. [Google Scholar] [CrossRef]
- Hoonhorst, S.J.; ten Hacken, N.H.; Lo Tam Loi, A.T.; Koenderman, L.; Lammers, J.W.; Telenga, E.D.; Boezen, H.M.; van den Berge, M.; Postma, D.S. Lower corticosteroid skin blanching response is associated with severe COPD. PLoS ONE 2014, 9, e91788. [Google Scholar] [CrossRef]
- Brandsma, C.A.; Timens, W.; Jonker, M.R.; Rutgers, B.; Noordhoek, J.A.; Postma, D.S. Differential effects of fluticasone on extracellular matrix production by airway and parenchymal fibroblasts in severe COPD. Am. J. Physiol. Lung Cell. Mol. Physiol. 2013, 305, L582–L589. [Google Scholar] [CrossRef]
- Friedlander, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef]
- Kluiver, J.; Slezak-Prochazka, I.; van den Berg, A. Studying microRNAs in lymphoma. Methods Mol. Biol. 2013, 971, 265–276. [Google Scholar]
- Melchers, L.J.; Clausen, M.J.; Mastik, M.F.; Slagter-Menkema, L.; van der Wal, J.E.; Wisman, G.B.; Roodenburg, J.L.; Schuuring, E. Identification of methylation markers for the prediction of nodal metastasis in oral and oropharyngeal squamous cell carcinoma. Epigenetics 2015, 10, 850–860. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).