Evidences from a Systematic Review and Meta-Analysis Unveil the Role of MiRNA Polymorphisms in the Predisposition to Female Neoplasms
Abstract
:1. Introduction
2. Results and Discussion
2.1. Study Characteristics
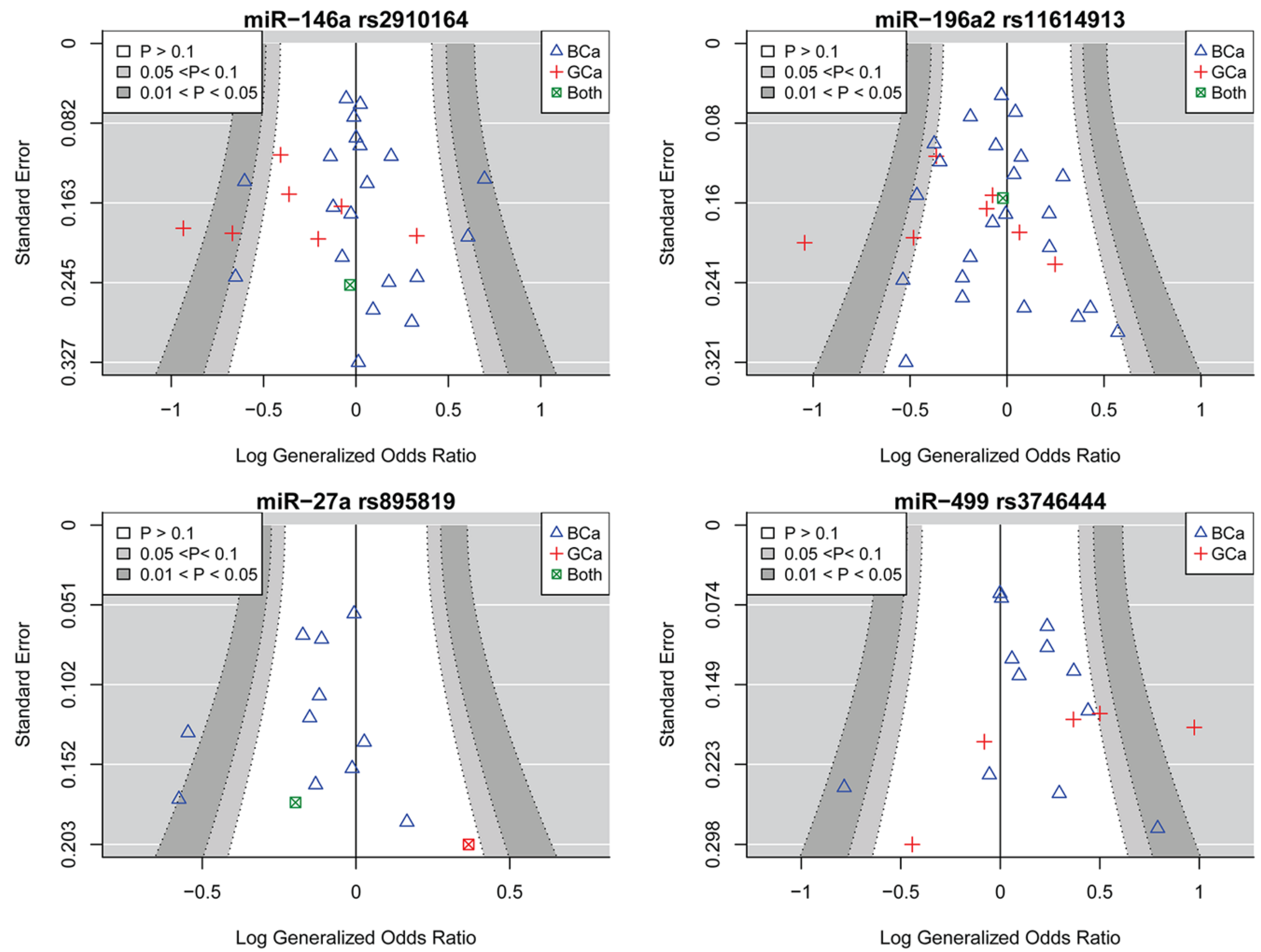
2.2. miR-146a rs2910164 and the Risk of Female Neoplasms
2.3. miR-196a2 rs11614913 and the Risk of Female Neoplasms
2.4. miR-27a rs895819 and the Risk of Female Neoplasms
2.5. miR-499 rs3746444 and the Risk of Female Neoplasms
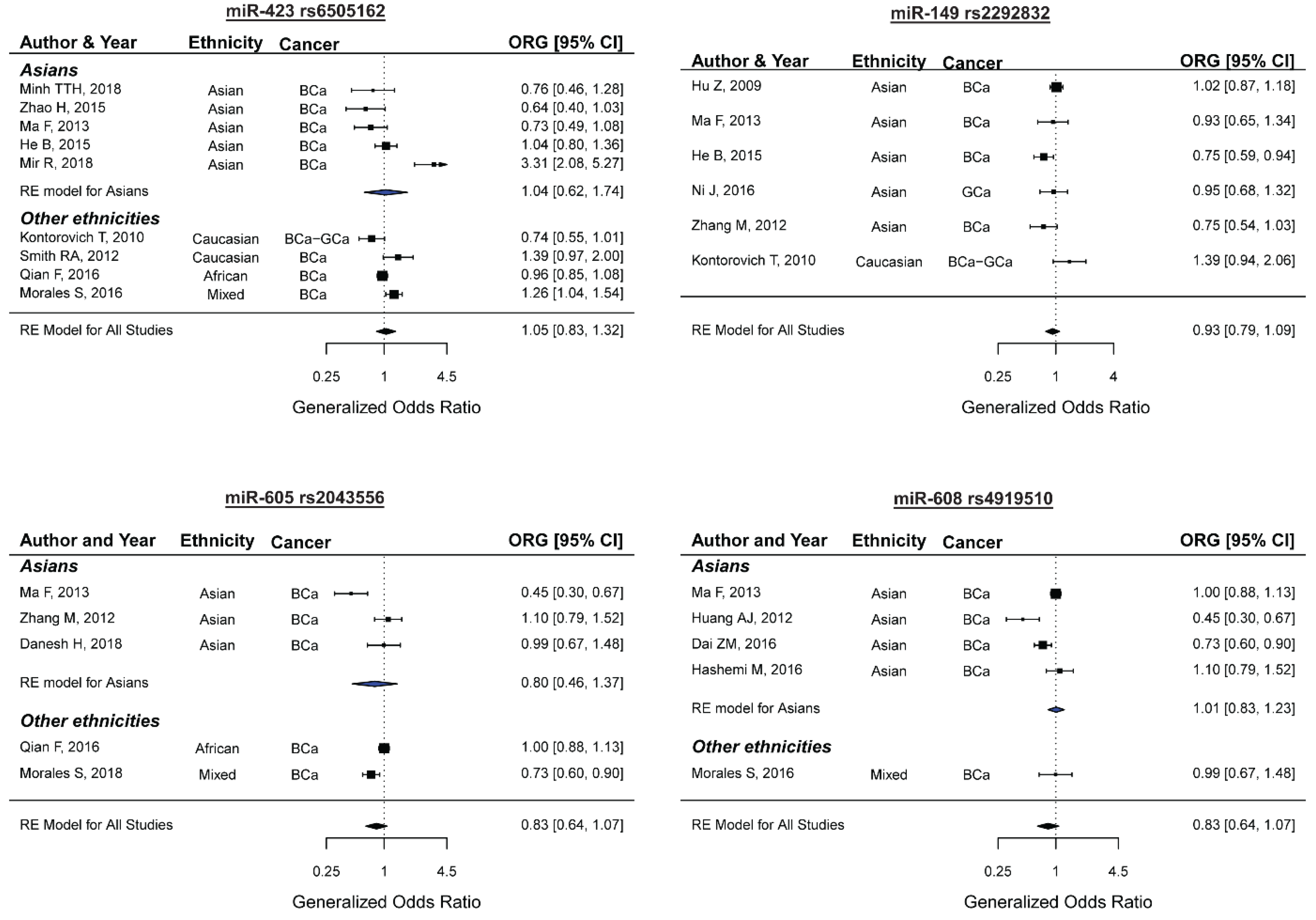
2.6. miR-423 rs6505162 and the Risk of Female Neoplasms
2.7. miR-149 rs2292832 and the Risk of Female Neoplasms
2.8. miR-605 rs2043556 and the Risk of Female Neoplasms
2.9. miR-608 rs4919510 and the Risk of Female Neoplasms
2.10. miR-100 rs1834306 and the Risk of Female Neoplasms
2.11. miR-124 rs531564 and the Risk of Female Neoplasms
2.12. miR-218 rs11134527 and the Risk of Female Neoplasms
2.13. miR-34b/c rs4938723 and the Risk of Female Neoplasms
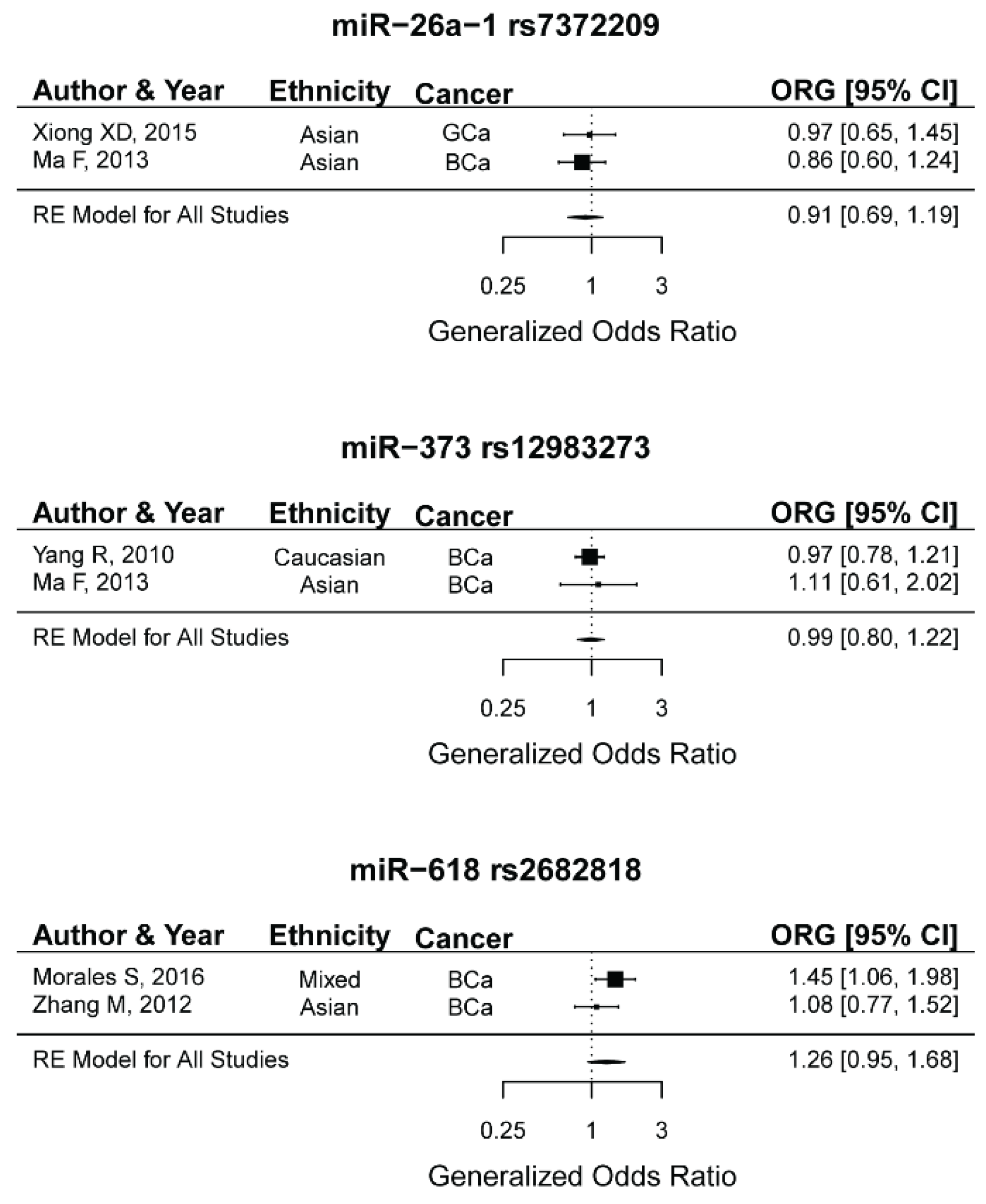
2.14. miR-26a-1 rs7372209, miR-373 rs12983273, miR-618 rs2682818 and the Risk of Female Neoplasms
3. Methods
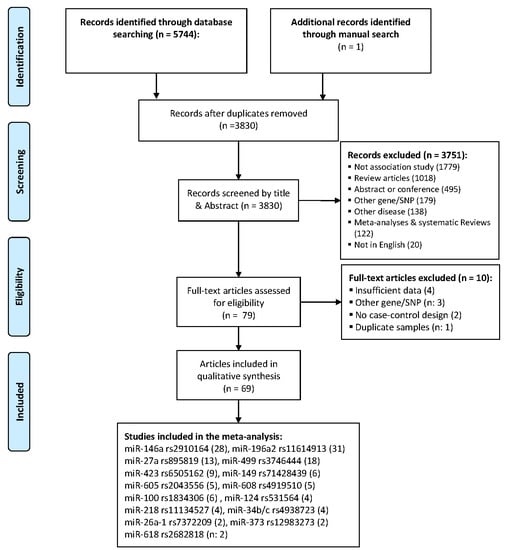
3.1. Search Strategy
3.2. Inclusion and Exclusion Criteria
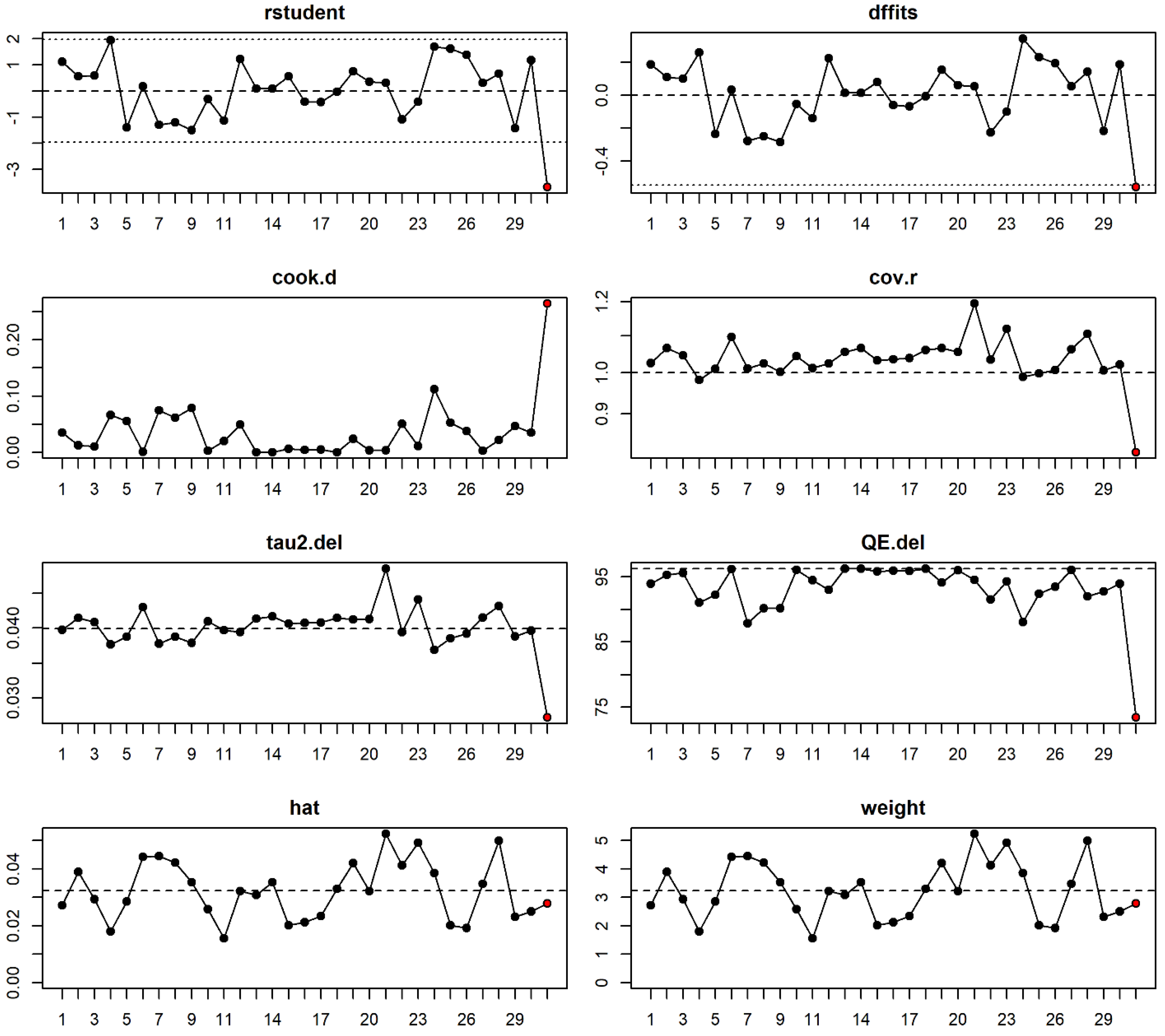
3.3. Data Extraction and Quality Assessment
3.4. Data Analysis
Author Contributions
Funding
Acknowledgement
Conflicts of Interest
Abbreviations
| BCa | Breast cancer |
| CCa | Cervical cancer |
| CI | Confidence interval |
| ECa | Endometrial Cancer |
| GCa | Gynecological cancers |
| HWE | Hardy-Weinberg equilibrium |
| OR | Odds ratio |
| ORG | Generalized odds ratio |
| OCa | Ovarian cancer |
| SNP | Single nucleotide polymorphism |
References
- Tong, C.W.S.; Wu, M.; Cho, W.C.S.; To, K.K.W. Recent Advances in the Treatment of Breast Cancer. Front. Oncol. 2018, 8, 227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fathi, E.; Farahzadi, R.; Valipour, B.; Sanaat, Z. Cytokines secreted from bone marrow derived mesenchymal stem cells promote apoptosis and change cell cycle distribution of K562 cell line as clinical agent in cell transplantation. PLoS ONE 2019, 14, e0215678. [Google Scholar] [CrossRef] [PubMed]
- Gholizadeh-Ghaleh Aziz, S.; Fathi, E.; Rahmati-Yamchi, M.; Akbarzadeh, A.; Fardyazar, Z.; Pashaiasl, M. An update clinical application of amniotic fluid-derived stem cells (AFSCs) in cancer cell therapy and tissue engineering. Artif. Cells Nanomed. Biotechnol. 2017, 45, 765–774. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- PDQ Cancer Genetics Editorial Board. Genetics Editorial Board. Genetics of Breast and Gynecologic Cancers (PDQ(R)): Health Professional Version. In PDQ Cancer Information Summaries; National Cancer Institute (US): Bethesda, MD, USA, 2002. [Google Scholar]
- Daraei, A.; Izadi, P.; Khorasani, G.; Nafissi, N.; Naghizadeh, M.M.; Younosi, N.; Meysamie, A.; Mansoori, Y.; Nariman-Saleh-Fam, Z.; Bastami, M.; et al. Methylation of progesterone receptor isoform A promoter in normal breast tissue: An epigenetic link between early age at menarche and risk of breast cancer? J. Cell Biochem. 2019. [Google Scholar] [CrossRef] [PubMed]
- Cha, S.Y.; Choi, Y.H.; Hwang, S.; Jeong, J.-Y.; An, H.J. Clinical Impact of microRNAs Associated with Cancer Stem Cells as a Prognostic Factor in Ovarian Carcinoma. J. Cancer 2017, 8, 3538–3547. [Google Scholar] [CrossRef] [PubMed]
- Choupani, J.; Nariman-Saleh-Fam, Z.; Saadatian, Z.; Ouladsahebmadarek, E.; Masotti, A.; Bastami, M. Association of mir-196a-2 rs11614913 and mir-149 rs2292832 Polymorphisms with Risk of Cancer: An Updated Meta-Analysis. Front. Genet. 2019, 10, 186. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Hayes, J.; Peruzzi, P.P.; Lawler, S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol. Med. 2014, 20, 460–469. [Google Scholar] [CrossRef]
- Malhotra, P.; Read, G.H.; Weidhaas, J.B. Breast Cancer and miR-SNPs: The Importance of miR Germ-Line Genetics. Non-Coding RNA 2019, 5, 27. [Google Scholar] [CrossRef]
- Ferracin, M.; Querzoli, P.; Calin, G.A.; Negrini, M. MicroRNAs: Toward the clinic for breast cancer patients. Semin. Oncol. 2011, 38, 764–775. [Google Scholar] [CrossRef] [PubMed]
- Mu, K.; Wu, Z.-Z.; Yu, J.-P.; Guo, W.; Wu, N.; Wei, L.-J.; Zhang, H.; Zhao, J.; Liu, J.-T. Meta-analysis of the association between three microRNA polymorphisms and breast cancer susceptibility. Oncotarget 2017, 8, 68809–68824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Y.; Yu, C.-Y.; Wang, J.-L.; Guan, J.; Chen, H.-Y.; Fang, J.-Y. MicroRNA sequence polymorphisms and the risk of different types of cancer. Sci. Rep. 2014, 4, 3648. [Google Scholar] [CrossRef] [PubMed]
- Pelletier, C.; Speed, W.C.; Paranjape, T.; Keane, K.; Blitzblau, R.; Hollestelle, A.; Safavi, K.; van den Ouweland, A.; Zelterman, D.; Slack, F.J.; et al. Rare BRCA1 haplotypes including 3’UTR SNPs associated with breast cancer risk. Cell Cycle 2011, 10, 90–99. [Google Scholar] [CrossRef]
- Bensen, J.T.; Graff, M.; Young, K.L.; Sethupathy, P.; Parker, J.; Pecot, C.V.; Currin, K.; Haddad, S.A.; Ruiz-Narváez, E.A.; Haiman, C.A.; et al. A survey of microRNA single nucleotide polymorphisms identifies novel breast cancer susceptibility loci in a case-control, population-based study of African-American women. Breast Cancer Res. 2018, 20. [Google Scholar] [CrossRef]
- Ma, L.; Hong, Y.; Lu, C.; Chen, Y.; Ma, C. The occurrence of cervical cancer in Uygur women in Xinjiang Uygur Autonomous Region is correlated to microRNA-146a and ethnic factor. Int. J. Clin. Exp. Pathol. 2015, 8, 9368–9375. [Google Scholar]
- Peterlongo, P.; Caleca, L.; Cattaneo, E.; Ravagnani, F.; Bianchi, T.; Galastri, L.; Bernard, L.; Ficarazzi, F.; Dall’olio, V.; Marme, F.; et al. The rs12975333 variant in the miR-125a and breast cancer risk in Germany, Italy, Australia and Spain. J. Med. Genet. 2011, 48, 703–704. [Google Scholar] [CrossRef]
- Al-Terehi, M.N.; Abd-Alhaleem, I.; Abidali, M.R.; Al-Qiam, Z.H.; Al-Saadi, A.H. Estimation Micr-RNA146a gene polymorphism in breast cancer tissue. J. Pure Appl. Microbiol. 2018, 12, 59–63. [Google Scholar] [CrossRef]
- Jin, T.; Wu, X.; Yang, H.; Liu, M.; He, Y.; He, X.; Shi, X.; Wang, F.; Du, S.; Ma, Y.; et al. Association of the miR-17-5p variants with susceptibility to cervical cancer in a Chinese population. Oncotarget 2016, 7, 76647–76655. [Google Scholar] [CrossRef] [Green Version]
- Chacon-Cortes, D.; Smith, R.A.; Lea, R.A.; Youl, P.H.; Griffiths, L.R. Association of microRNA 17–92 cluster host gene (MIR17HG) polymorphisms with breast cancer. Tumor Biol. 2015, 36, 5369–5376. [Google Scholar] [CrossRef]
- Chen, J.P.; Jiang, Y.; Zhou, J.; Liu, S.J.; Qin, N.; Du, J.B.; Jin, G.F.; Hu, Z.B.; Ma, H.X.; Shen, H.B.; et al. Evaluation of CpG-SNPs in miRNA promoters and risk of breast cancer. Gene 2018, 651, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, U.H.T.; Nguyen, T.N.T.; Van Tran, T.; Nguyen, H.T. High resolution melting (HRM) optimization for genotyping the SNP Rs3746444/Mir-499 in Vietnamese breast cancer patients. In Proceedings of the 6th International Conference on the Development of Biomedical Engineering in Vietnam (BME6), Ho Chi Minh City, Vietnam, 11–14 January 2010; pp. 171–176. [Google Scholar]
- Zhao, H.H.; Xu, J.M.; Zhao, D.; Geng, M.J.; Ge, H.Z.; Fu, L.; Zhu, Z.M. Somatic Mutation of the SNP rs11614913 and Its Association with Increased MIR 196A2 Expression in Breast Cancer. DNA Cell Biol. 2016, 35, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Minh, T.T.H.; Thanh, N.T.N.; Van Thiep, T.; Hue, N.T. Association between single nucleotide polymorphism Rs11614913 (C>T) on Mir-196a2 and breast cancer in Vietnamese population. In 6th International Conference on the Development of Biomedical Engineering in Vietnam (BME6); Van, T.V., Le, T.A.N., Duc, T.N., Eds.; BME 2017. Proceedings of IFMBE Proceedings; Springer: Singapore; Volume 63, pp. 381–386.
- Belaiba, F.; Medimegh, I.; Ammar, M.; Jemni, F.; Mezlini, A.; Romdhane, K.B.; Cherni, L.; Benammar Elgaaïed, A. Expression and polymorphism of micro-RNA according to body mass index and breast cancer presentation in Tunisian patients. J. Leukoc. Biol. 2019, 105, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Kalapanida, D.; Zagouri, F.; Gazouli, M.; Zografos, E.; Dimitrakakis, C.; Marinopoulos, S.; Giannos, A.; Sergentanis, T.N.; Kastritis, E.; Terpos, E.; et al. Evaluation of pre-miR-34a RS72631823 single nucleotide polymorphism in triple negative breast cancer: A case-control study. Oncotarget 2018, 9, 36906–36913. [Google Scholar] [CrossRef] [PubMed]
- Morales, S.; De Mayo, T.; Gulppi, F.A.; Gonzalez-Hormazabal, P.; Carrasco, V.; Reyes, J.M.; Gómez, F.; Waugh, E.; Jara, L. Genetic variants in pre-miR-146a, pre-miR-499, pre-miR-125a, pre-miR-605, and pri-miR-182 are associated with breast cancer susceptibility in a south American population. Genes 2018, 9, 427. [Google Scholar] [CrossRef]
- Eslami, S.Z.; Tahmaseb, M.; Ghaderi, A. The investigation of miR-196a2 rs11614913 with breast cancer susceptibility in south of IRAN. Meta Gene 2018, 17, 43–47. [Google Scholar] [CrossRef]
- Danesh, H.; Hashemi, M.; Bizhani, F.; Hashemi, S.M.; Bahari, G. Association study of miR-100, miR-124-1, miR-218-2, miR-301b, miR-605, and miR-4293 polymorphisms and the risk of breast cancer in a sample of Iranian population. Gene 2018, 647, 73–78. [Google Scholar] [CrossRef]
- Doulah, A.; Salehzadeh, A.; Mojarrad, M. Association of single nucleotide polymorphisms in miR-499 and miR-196a with susceptibility to breast cancer. Trop. J. Pharm. Res. 2018, 17, 319–323. [Google Scholar] [CrossRef]
- Mashayekhi, S.; Saeidi Saedi, H.; Salehi, Z.; Soltanipour, S.; Mirzajani, E. Effects of miR-27a, miR-196a2 and miR-146a polymorphisms on the risk of breast cancer. Br. J. Biomed. Sci. 2018, 75, 76–81. [Google Scholar] [CrossRef]
- Nejati-Azar, A.; Alivand, M.R. MiRNA 196a2(rs11614913) & 146a(rs2910164) polymorphisms & breast cancer risk for women in an Iranian population. Pers. Med. 2018, 15, 279–289. [Google Scholar] [CrossRef]
- McVeigh, T.P.; Mulligan, R.J.; McVeigh, U.M.; Owens, P.W.; Miller, N.; Bell, M.; Sebag, F.; Guerin, C.; Quill, D.S.; Weidhaas, J.B.; et al. Investigating the association of rs2910164 with cancer predisposition in an Irish cohort. Endocr. Connect. 2017, 6, 614–624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srivastava, S.; Singh, S.; Fatima, N.; Mittal, B.; Srivastava, A.N. Pre-microrna gene polymorphisms and risk of cervical squamous cell carcinoma. J. Clin. Diagn. Res. 2017, 11, GC01–GC04. [Google Scholar] [CrossRef] [PubMed]
- Parchami Barjui, S.; Reiisi, S.; Ebrahimi, S.O.; Shekari, B. Study of correlation between genetic variants in three microRNA genes (hsa-miR-146a, hsa-miR-502 binding site, hsa-miR-27a) and breast cancer risk. Curr. Res. Transl. Med. 2017, 65, 141–147. [Google Scholar] [CrossRef]
- Kabirizadeh, S.; Azadeh, M.; Mirhosseini, M.; Ghaedi, K.; Mesrian Tanha, H. The SNP rs3746444 within mir-499a is associated with breast cancer risk in Iranian population. J. Cell. Immunother. 2016, 2, 95–97. [Google Scholar] [CrossRef] [Green Version]
- Qian, F.; Feng, Y.; Zheng, Y.; Ogundiran, T.O.; Ojengbede, O.; Zheng, W.; Blot, W.; Ambrosone, C.B.; John, E.M.; Bernstein, L.; et al. Genetic variants in microRNA and microRNA biogenesis pathway genes and breast cancer risk among women of African ancestry. Hum. Genet. 2016, 135, 1145–1159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanaei, S.; Hashemi, M.; Rezaei, M.; Hashemi, S.M.; Bahari, G.; Ghavami, S. Evaluation of the pri-miR-34b/c rs4938723 polymorphism and its association with breast cancer risk. Biomed. Rep. 2016, 125–129. [Google Scholar] [CrossRef]
- Yuan, F.; Sun, R.; Chen, P.; Liang, Y.; Ni, S.; Quan, Y.; Huang, J.; Zhang, L.; Gao, L. Combined analysis of pri-miR-34b/c rs4938723 and TP53 Arg72Pro with cervical cancer risk. Tumor Biol. 2016, 37, 6267–6273. [Google Scholar] [CrossRef]
- Hashemi, M.; Sanaei, S.; Rezaei, M.; Bahari, G.; Hashemi, S.M.; Mashhadi, M.A.; Taheri, M.; Ghavami, S. MiR-608 rs4919510 C>G polymorphism decreased the risk of breast cancer in an Iranian Subpopulation. Exp. Oncol. 2016, 38, 57–59. [Google Scholar] [CrossRef]
- Upadhyaya, A.; Smith, R.A.; Chacon-Cortes, D.; Revêchon, G.; Bellis, C.; Lea, R.A.; Haupt, L.M.; Chambers, S.K.; Youl, P.H.; Griffiths, L.R. Association of the microRNA-Single Nucleotide Polymorphism rs2910164 in miR146a with sporadic breast cancer susceptibility: A case control study. Gene 2016, 576, 256–260. [Google Scholar] [CrossRef] [Green Version]
- Song, Z.S.; Wu, Y.; Zhao, H.G.; Liu, C.X.; Cai, H.Y.; Guo, B.Z.; Xie, Y.; Shi, H.R. Association between the rs11614913 variant of miRNA-196a-2 and the risk of epithelial ovarian cancer. Oncol. Lett. 2016, 11, 194–200. [Google Scholar] [CrossRef]
- Dai, Z.M.; Kang, H.F.; Zhang, W.G.; Li, H.B.; Zhang, S.Q.; Ma, X.B.; Lin, S.; Wang, M.; Feng, Y.J.; Liu, K.; et al. The associations of single nucleotide polymorphisms in miR196a2, miR-499, and miR-608 with breast cancer susceptibility: A STROBE-compliant observational study. Medicine 2016, 95, e2826. [Google Scholar] [CrossRef] [PubMed]
- Chacon-Cortes, D.; Smith, R.A.; Haupt, L.M.; Lea, R.A.; Youl, P.H.; Griffiths, L.R. Genetic association analysis of miRNA SNPs implicates MIR145 in breast cancer susceptibility. BMC Med. Genet. 2015, 16. [Google Scholar] [CrossRef] [PubMed]
- Qi, P.; Wang, L.; Zhou, B.; Yao, W.J.; Xu, S.; Zhou, Y.; Xie, Z.B. Associations of miRNA polymorphisms and expression levels with breast cancer risk in the Chinese population. Genet. Mol. Res. 2015, 14, 6289–6296. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Pan, Y.; Xu, Y.; Deng, Q.; Sun, H.; Gao, T.; Wang, S. Associations of polymorphisms in microRNAs with female breast cancer risk in Chinese population. Tumor Biol. 2015, 36, 4575–4582. [Google Scholar] [CrossRef] [PubMed]
- Hashemi, M.; Sanaei, S.; Mashhadi, M.A.; Hashemi, S.M.; Taheri, M.; Ghavami, S. Association study of Hsa-Mir-603 rs11014002 polymorphism and risk of breast cancer in a sample of Iranian population. Cell. Mol. Biol. 2015, 61, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Zhang, J. Mir-124 rs531564 polymorphism influences genetic susceptibility to cervical cancer. Int. J. Clin. Exp. Med. 2014, 7, 5847–5851. [Google Scholar]
- Bansal, C.; Sharma, K.L.; Misra, S.; Srivastava, A.N.; Mittal, B.; Singh, U.S. Common genetic variants in pre-microRNAs and risk of breast cancer in the North Indian population. Ecancer Med Sci. 2014, 8. [Google Scholar] [CrossRef]
- Xiong, X.D.; Luo, X.P.; Cheng, J.; Liu, X.; Li, E.M.; Zeng, L.Q. A genetic variant in pre-miR-27a is associated with a reduced cervical cancer risk in southern Chinese women. Gynecol. Oncol. 2014, 132, 450–454. [Google Scholar] [CrossRef]
- Omrani, M.; Hashemi, M.; Eskandari-Nasab, E.; Hasani, S.S.; Mashhadi, M.A.; Arbabi, F.; Taheri, M. Hsa-mir-499 rs3746444 gene polymorphism is associated with susceptibility to breast cancer in an Iranian population. Biomark. Med. 2014, 8, 259–267. [Google Scholar] [CrossRef]
- Zhang, N.; Huo, Q.; Wang, X.; Chen, X.; Long, L.; Jiang, L.; Ma, T.; Yang, Q. A genetic variant in pre-miR-27a is associated with a reduced breast cancer risk in younger Chinese population. Gene 2013, 529, 125–130. [Google Scholar] [CrossRef]
- Yao, S.; Graham, K.; Shen, J.; Campbell, L.E.S.; Singh, P.; Zirpoli, G.; Roberts, M.; Ciupak, G.; Davis, W.; Hwang, H.; et al. Genetic variants in microRNAs and breast cancer risk in African American and European American women. Breast Cancer Res. Treat. 2013, 141, 447–459. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Zhang, P.; Lin, D.; Yu, D.; Yuan, P.; Wang, J.; Fan, Y.; Xu, B. There Is No Association between MicroRNA Gene Polymorphisms and Risk of Triple Negative Breast Cancer in a Chinese Han Population. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.Y.; Chen, X.J.; Zhu, M.L.; Wang, M.Y.; He, J.; Yu, K.D.; Shao, Z.M.; Sun, M.H.; Zhou, X.Y.; Cheng, X.; et al. A pri-miR-218 variant and risk of cervical carcinoma in Chinese women. BMC Cancer 2013, 13. [Google Scholar] [CrossRef] [PubMed]
- Linhares, J.J.; Azevedo, M., Jr.; Siufi, A.A.; de Carvalho, C.V.; Wolgien, M.D.C.G.M.; Noronha, E.C.; Bonetti, T.C.D.S.; da Silva, I.D.C.G. Evaluation of single nucleotide polymorphisms in microRNAs (hsa-miR-196a2 rs11614913 C/T) from Brazilian women with breast cancer. BMC Med. Genet. 2012, 13. [Google Scholar] [CrossRef]
- Catucci, I.; Verderio, P.; Pizzamiglio, S.; Bernard, L.; Dall’Olio, V.; Sardella, D.; Ravagnani, F.; Galastri, L.; Barile, M.; Peissel, B.; et al. The SNP rs895819 in miR-27a is not associated with familial breast cancer risk in Italians. Breast Cancer Res. Treat. 2012, 133, 805–807. [Google Scholar] [CrossRef]
- Huang, A.J.; Yu, K.D.; Li, J.; Fan, L.; Shao, Z.M. Polymorphism rs4919510:C> in mature sequence of human microRNA-608 contributes to the risk of HER2-positive breast cancer but not other subtypes. PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
- Alshatwi, A.A.; Shafi, G.; Hasan, T.N.; Syed, N.A.; Al-Hazzani, A.A.; Alsaif, M.A.; Alsaif, A.A. Differential expression profile and genetic variants of MicroRNAs sequences in breast cancer patients. PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
- Smith, R.A.; Jedlinski, D.J.; Gabrovska, P.N.; Weinstein, S.R.; Haupt, L.; Griffiths, L.R. A genetic variant located in miR-423 is associated with reduced breast cancer risk. Cancer Genom. Proteom. 2012, 9, 115–118. [Google Scholar]
- Jedlinski, D.J.; Gabrovska, P.N.; Weinstein, S.R.; Smith, R.A.; Griffiths, L.R. Single nucleotide polymorphism in hsa-mir-196a-2 and breast cancer risk: A case control study. Twin Res. Hum. Genet. 2011, 14, 417–421. [Google Scholar] [CrossRef]
- Zhou, B.; Wang, K.; Wang, Y.; Xi, M.; Zhang, Z.; Song, Y.; Zhang, L. Common genetic polymorphisms in pre-microRNAs and risk of cervical squamous cell carcinoma. Mol. Carcinog. 2011, 50, 499–505. [Google Scholar] [CrossRef]
- Yang, R.; Dick, M.; Marme, F.; Schneeweiss, A.; Langheinz, A.; Hemminki, K.; Sutter, C.; Bugert, P.; Wappenschmidt, B.; Varon, R.; et al. Genetic variants within miR-126 and miR-335 are not associated with breast cancer risk. Breast Cancer Res. Treat. 2011, 127, 549–554. [Google Scholar] [CrossRef] [PubMed]
- Kontorovich, T.; Levy, A.; Korostishevsky, M.; Nir, U.; Friedman, E. Single nucleotide polymorphisms in miRNA binding sites and miRNA genes as breast/ovarian cancer risk modifiers in jewish high-risk women. Int. J. Cancer 2010, 127, 589–597. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Schlehe, B.; Hemminki, K.; Sutter, C.; Bugert, P.; Wappenschmidt, B.; Volkmann, J.; Varon, R.; Weber, B.H.F.; Niederacher, D.; et al. A genetic variant in the pre-miR-27a oncogene is associated with a reduced familial breast cancer risk. Breast Cancer Res. Treat. 2010, 121, 693–702. [Google Scholar] [CrossRef] [PubMed]
- Catucci, I.; Yang, R.; Verderio, P.; Pizzamiglio, S.; Heesen, L.; Hemminki, K.; Sutter, C.; Wappenschmidt, B.; Dick, M.; Arnold, N.; et al. Evaluation of SNPs in miR-146a, miR196a2 and miR-499 as low-penetrance alleles in German and Italian familial breast cancer cases. Hum. Mutat. 2010, 31, E1052–E1057. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Liang, J.; Wang, Z.; Tian, T.; Zhou, X.; Chen, J.; Miao, R.; Wang, Y.; Wang, X.; Shen, H. Common genetic variants in pre-microRNAs were associated with increased risk of breast cancer in Chinese women. Hum. Mutat. 2009, 30, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Pastrello, C.; Polesel, J.; Della Puppa, L.; Viel, A.; Maestro, R. Association between hsa-mir-146a genotype and tumor age-of-onset in BRCA1/BRCA2-negative familial breast and ovarian cancer patients. Carcinogenesis 2010, 31, 2124–2126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bodal, V.K.; Sangwan, S.; Bal, M.S.; Kaur, M.; Sharma, S.; Kaur, B. Association between Microrna 146a and Microrna 196a2 Genes Polymorphism and Breast Cancer Risk in North Indian Women. Asian Pac. J. Cancer Prev. 2017, 18, 2345–2348. [Google Scholar] [CrossRef]
- Liang, Y.; Sun, R.; Li, L.; Yuan, F.; Liang, W.; Wang, L.; Nie, X.; Chen, P.; Zhang, L.; Gao, L. A Functional Polymorphism in the Promoter of MiR-143/145 Is Associated with the Risk of Cervical Squamous Cell Carcinoma in Chinese Women: A Case-Control Study. Medine 2015, 94, e1289. [Google Scholar] [CrossRef]
- Mir, R.; Al Balawi, I.A.; Duhier, F.M.A. Involvement of microRNA-423 Gene Variability in Breast Cancer Progression in Saudi Arabia. Asian Pac. J. Cancer Prev. 2018, 19, 2581–2589. [Google Scholar] [CrossRef]
- Sun, X.C.; Zhang, A.C.; Tong, L.L.; Wang, K.; Wang, X.; Sun, Z.Q.; Zhang, H.Y. miR-146a and miR-196a2 polymorphisms in ovarian cancer risk. Genet. Mol. Res. GMR 2016, 15. [Google Scholar] [CrossRef]
- Yue, C.; Wang, M.; Ding, B.; Wang, W.; Fu, S.; Zhou, D.; Zhang, Z.; Han, S. Polymorphism of the pre-miR-146a is associated with risk of cervical cancer in a Chinese population. Gynecol. Oncol. 2011, 122, 33–37. [Google Scholar] [CrossRef] [PubMed]
- Naderi, N.; Peymani, M.; Ghaedi, K. The protective role of rs56103835 against breast cancer onset in the Iranian population. Mol. Genet. Genom. Med. 2019. [Google Scholar] [CrossRef] [PubMed]
- Meshkat, M.; Tanha, H.M.; Ghaedi, K.; Meshkat, M. Association of a potential functional mir-520f rs75598818 G > A polymorphism with breast cancer. J. Genet. 2018, 97, 1307–1313. [Google Scholar] [CrossRef] [PubMed]
- Minh, T.T.H.; Thanh, N.T.N.; Hue, N.T. Association between Selected microRNA SNPs and Breast Cancer Risk in a Vietnamese Population. Int. J. Hum. Genet. 2018, 18, 238–246. [Google Scholar] [CrossRef]
- Afsharzadeh, S.M.; Ardebili, S.M.M.; Seyedi, S.M.; Fathi, N.K.; Mojarrad, M. Association between rs11614913, rs3746444, rs2910164 and occurrence of breast cancer in Iranian population. Meta Gene 2017, 11, 20–25. [Google Scholar] [CrossRef]
- Huynh, L.H.; Bui, P.T.K.; Nguyen, T.T.N.; Nguyen, H.T. Developing a high resolution melting method for genotyping and predicting association of SNP rs353291 with breast cancer in the Vietnamese population. Biomed. Res. Ther. 2017, 4, 1812–1831. [Google Scholar] [CrossRef]
- Ni, J.H.; Huang, Y.G. Role of polymorphisms in miR-146a, miR-149, miR-196a2 and miR-499 in the development of ovarian cancer in a Chinese population. Int. J. Clin. Exp. Pathol. 2016, 9, 5706–5711. [Google Scholar]
- Liu, X.Y.; Xu, B.H.; Li, S.J.; Zhang, B.; Mao, P.M.; Qian, B.B.; Guo, L.; Ni, P.H. Association of SNPs in miR-146a, miR-196a2, and miR-499 with the risk of endometrial/ovarian cancer. Acta Biochim. Biophys. Sin. 2015, 47, 564–566. [Google Scholar] [CrossRef] [Green Version]
- Hoffman, A.E.; Zheng, T.Z.; Yi, C.H.; Leaderer, D.; Weidhaas, J.; Slack, F.; Zhang, Y.W.; Paranjape, T.; Zhu, Y. microRNA miR-196a-2 and Breast Cancer: A Genetic and Epigenetic Association Study and Functional Analysis. Cancer Res. 2009, 69, 5970–5977. [Google Scholar] [CrossRef]
- Garcia, A.I.; Cox, D.G.; Barjhoux, L.; Verny-Pierre, C.; Barnes, D.; Antoniou, A.C.; Stoppa-Lyonnet, D.; Sinilnikova, O.M.; Mazoyer, S. The rs2910164:G>C SNP in the MIR146A gene is not associated with breast cancer risk in BRCA1 and BRCA2 mutation carriers. Hum. Mutat. 2011, 32, 1004–1007. [Google Scholar] [CrossRef]
- Li, W.; Duan, R.; Kooy, F.; Sherman, S.L.; Zhou, W.; Jin, P. Germline mutation of microRNA-125a is associated with breast cancer. J. Med. Genet. 2009, 46, 358–360. [Google Scholar] [CrossRef] [PubMed]
- Morales, S.; Gulppi, F.; Gonzalez-Hormazabal, P.; Fernandez-Ramires, R.; Bravo, T.; Reyes, J.M.; Gomez, F.; Waugh, E.; Jara, L. Association of single nucleotide polymorphisms in Pre-miR-27a, Pre-miR-196a2, Pre-miR-423, miR-608 and Pre-miR-618 with breast cancer susceptibility in a South American population. BMC Genet. 2016, 17. [Google Scholar] [CrossRef] [PubMed]
- Salimi, Z.; Sadeghi, S.; Tabatabaeian, H.; Ghaedi, K.; Fazilati, M. rs11895168 C allele and the increased risk of breast cancer in Isfahan population. Breast 2016, 28, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Jin, M.; Yu, Y.; Zhang, S.; Wu, Y.; Liu, H.; Liu, H.; Chen, B.; Li, Q.; Ma, X.; et al. Associations of miRNA polymorphisms and female physiological characteristics with breast cancer risk in Chinese population. Eur. J. Cancer Care 2012, 21, 274–280. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Gao, A.; Zhang, Z.; Tian, R.; Luo, A.; Li, M.; Zhao, D.; Fu, L.; Fu, L.; Dong, J.T.; et al. Genetic analysis and preliminary function study of miR-423 in breast cancer. Tumor Biol. 2015, 36, 4763–4771. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Chen, X.; Hu, L.; Han, S.; Qiang, F.; Wu, Y.; Pan, L.; Shen, H.; Li, Y.; Hu, Z. Polymorphisms involved in the miR-218-LAMB3 pathway and susceptibility of cervical cancer, a case-control study in Chinese women. Gynecol. Oncol. 2010, 117, 287–290. [Google Scholar] [CrossRef] [PubMed]
- Nguyen-Dien, G.T.; Smith, R.A.; Haupt, L.M.; Griffiths, L.R.; Nguyen, H.T. Genetic polymorphisms in miRNAs targeting the estrogen receptor and their effect on breast cancer risk. Meta Gene 2014, 2, 226–236. [Google Scholar] [CrossRef] [Green Version]
- Bensen, J.T.; Tse, C.K.; Nyante, S.J.; Barnholtz-Sloan, J.S.; Cole, S.R.; Millikan, R.C. Association of germline microRNA SNPs in pre-miRNA flanking region and breast cancer risk and survival: The Carolina Breast Cancer Study. Cancer Causes Control CCC 2013, 24, 1099–1109. [Google Scholar] [CrossRef]
- Thakur, N.; Singhal, P.; Mehrotra, R.; Bharadwaj, M. Impacts of single nucleotide polymorphisms in three microRNAs (miR-146a, miR-196a2 and miR-499) on the susceptibility to cervical cancer among Indian women. Biosci. Rep. 2019, 39, BSR20180723. [Google Scholar] [CrossRef]
- Li, C.; Wang, X.; Yan, Z.; Zhang, Y.; Liu, S.; Yang, J.; Hong, C.; Shi, L.; Yang, H.; Yao, Y. The association between polymorphisms in microRNA genes and cervical cancer in a Chinese Han population. Oncotarget 2017, 8, 87914–87927. [Google Scholar] [CrossRef] [Green Version]
- Zintzaras, E. The power of generalized odds ratio in assessing association in genetic studies with known mode of inheritance. J. Appl. Stat. 2012, 39, 2569–2581. [Google Scholar] [CrossRef]
- Pereira, T.V.; Mingroni-Netto, R.C. A note on the use of the generalized odds ratio in meta-analysis of association studies involving bi- and tri-allelic polymorphisms. BMC Res. Notes 2011, 4, 172. [Google Scholar] [CrossRef] [PubMed]
- Zintzaras, E. The generalized odds ratio as a measure of genetic risk effect in the analysis and meta-analysis of association studies. Stat. Appl. Genet. Mol. Biol. 2010, 9, 21. [Google Scholar] [CrossRef] [PubMed]
- Bastami, M.; Choupani, J.; Saadatian, Z.; Zununi Vahed, S.; Mansoori, Y.; Daraei, A.; Samadi Kafil, H.; Masotti, A.; Nariman-Saleh-Fam, Z. miRNA Polymorphisms and Risk of Cardio-Cerebrovascular Diseases: A Systematic Review and Meta-Analysis. Int. J. Mol. Sci. 2019, 20, 293. [Google Scholar] [CrossRef]
- Ramkaran, P.; Khan, S.; Phulukdaree, A.; Moodley, D.; Chuturgoon, A.A. miR-146a polymorphism influences levels of miR-146a, IRAK-1, and TRAF-6 in young patients with coronary artery disease. Cell Biochem. Biophys. 2014, 68, 259–266. [Google Scholar] [CrossRef]
- Vinci, S.; Gelmini, S.; Pratesi, N.; Conti, S.; Malentacchi, F.; Simi, L.; Pazzagli, M.; Orlando, C. Genetic variants in miR-146a, miR-149, miR-196a2, miR-499 and their influence on relative expression in lung cancers. Clin. Chem. Lab. Med. 2011, 49, 2073–2080. [Google Scholar] [CrossRef]
- Xiong, X.D.; Cho, M.; Cai, X.P.; Cheng, J.; Jing, X.; Cen, J.M.; Liu, X.; Yang, X.L.; Suh, Y. A common variant in pre-miR-146 is associated with coronary artery disease risk and its mature miRNA expression. Mutat. Res. 2014, 761, 15–20. [Google Scholar] [CrossRef]
- Jazdzewski, K.; de la Chapelle, A. Genomic sequence matters: A SNP in microRNA-146a can turn anti-apoptotic. Cell Cycle 2009, 8, 1642–1643. [Google Scholar] [CrossRef]
- Sakaue, S.; Hirata, J.; Maeda, Y.; Kawakami, E.; Nii, T.; Kishikawa, T.; Ishigaki, K.; Terao, C.; Suzuki, K.; Akiyama, M.; et al. Integration of genetics and miRNA-target gene network identified disease biology implicated in tissue specificity. Nucleic Acids Res. 2018, 46, 11898–11909. [Google Scholar] [CrossRef]
- Gao, W.; Hua, J.; Jia, Z.; Ding, J.; Han, Z.; Dong, Y.; Lin, Q.; Yao, Y. Expression of miR-146a-5p in breast cancer and its role in proliferation of breast cancer cells. Oncol. Lett. 2018, 15, 9884–9888. [Google Scholar] [CrossRef]
- Sandhu, R.; Rein, J.; D’Arcy, M.; Herschkowitz, J.I.; Hoadley, K.A.; Troester, M.A. Overexpression of miR-146a in basal-like breast cancer cells confers enhanced tumorigenic potential in association with altered p53 status. Carcinogenesis 2014, 35, 2567–2575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Wang, C.; Guo, J.; Fan, J.; Zhang, Z. Expression of miR-146a in triple negative breast cancer and its clinical significance. Int. J. Clin. Exp. Pathol. 2016, 9, 11832–11837. [Google Scholar]
- Bastami, M.; Ghaderian, S.M.; Omrani, M.D.; Mirfakhraie, R.; Vakili, H.; Parsa, S.A.; Nariman-Saleh-Fam, Z.; Masotti, A. MiRNA-Related Polymorphisms in miR-146a and TCF21 Are Associated with Increased Susceptibility to Coronary Artery Disease in an Iranian Population. Genet. Test. Mol. Biomark. 2016, 20, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Tsai, K.-W.; Leung, C.-M.; Lo, Y.-H.; Chen, T.-W.; Chan, W.-C.; Yu, S.-Y.; Tu, Y.-T.; Lam, H.-C.; Li, S.-C.; Ger, L.-P.; et al. Arm Selection Preference of MicroRNA-193a Varies in Breast Cancer. Sci. Rep. 2016, 6, 28176. [Google Scholar] [CrossRef] [Green Version]
- Viechtbauer, W.; Cheung, M.W.-L. Outlier and influence diagnostics for meta-analysis. Res. Synth Methods 2010, 1, 112–125. [Google Scholar] [CrossRef]
- Harrer, M.; Cuijpers, P.; Furukawa, T.A.; Ebert, D.D. Doing Meta-Analysis in R: A Hands-on Guide; PROTECT Lab Erlangen: Germany, 2019. Available online: https://bookdown.org/MathiasHarrer/Doing_Meta_Analysis_in_R/ (accessed on 18 July 2019). [CrossRef]
- Karabegovic, I.; Maas, S.; Medina-Gomez, C.; Zrimsek, M.; Reppe, S.; Gautvik, K.M.; Uitterlinden, A.G.; Rivadeneira, F.; Ghanbari, M. Genetic Polymorphism of miR-196a-2 is Associated with Bone Mineral Density (BMD). Int. J. Mol. Sci. 2017, 18, 2529. [Google Scholar] [CrossRef]
- Hu, Z.; Chen, J.; Tian, T.; Zhou, X.; Gu, H.; Xu, L.; Zeng, Y.; Miao, R.; Jin, G.; Ma, H.; et al. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J. Clin. Investig. 2008, 118, 2600–2608. [Google Scholar] [CrossRef]
- Bell, M.L.; Buvoli, M.; Leinwand, L.A. Uncoupling of Expression of an Intronic MicroRNA and Its Myosin Host Gene by Exon Skipping. Mol. Cell. Biol. 2010, 30, 1937. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2018, 47, D155–D162. [Google Scholar] [CrossRef]
- Ro, S.; Park, C.; Young, D.; Sanders, K.M.; Yan, W. Tissue-dependent paired expression of miRNAs. Nucleic Acids Res. 2007, 35, 5944–5953. [Google Scholar] [CrossRef]
- Xu, Z.; Han, Y.; Liu, J.; Jiang, F.; Hu, H.; Wang, Y.; Liu, Q.; Gong, Y.; Li, X. MiR-135b-5p and MiR-499a-3p Promote Cell Proliferation and Migration in Atherosclerosis by Directly Targeting MEF2C. Sci. Rep. 2015, 5, 12276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalter, J.; Sweegers, M.G.; Verdonck-de Leeuw, I.M.; Brug, J.; Buffart, L.M. Development and use of a flexible data harmonization platform to facilitate the harmonization of individual patient data for meta-analyses. BMC Res. Notes 2019, 12, 164. [Google Scholar] [CrossRef] [PubMed]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; The, P.G. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef] [PubMed]
- Thakkinstian, A.; McKay, G.J.; McEvoy, M.; Chakravarthy, U.; Chakrabarti, S.; Silvestri, G.; Kaur, I.; Li, X.; Attia, J. Systematic review and meta-analysis of the association between complement component 3 and age-related macular degeneration: A HuGE review and meta-analysis. Am. J. Epidemiol. 2011, 173, 1365–1379. [Google Scholar] [CrossRef]
- Xue, W.; Zhu, M.; Wang, Y.; He, J.; Zheng, L. Association between PLCE1 rs2274223 A > G polymorphism and cancer risk: Proof from a meta-analysis. Sci. Rep. 2015, 5, 7986. [Google Scholar] [CrossRef]
- DerSimonian, R.; Laird, N. Meta-analysis in clinical trials. Control. Clin. Trials 1986, 7, 177–188. [Google Scholar] [CrossRef]
- Hartung, J.; Knapp, G. A refined method for the meta-analysis of controlled clinical trials with binary outcome. Stat. Med. 2001, 20, 3875–3889. [Google Scholar] [CrossRef]
- Sterne, J.A.C.; Sutton, A.J.; Ioannidis, J.P.A.; Terrin, N.; Jones, D.R.; Lau, J.; Carpenter, J.; Rücker, G.; Harbord, R.M.; Schmid, C.H.; et al. Recommendations for examining and interpreting funnel plot asymmetry in meta-analyses of randomised controlled trials. BMJ 2011, 343. [Google Scholar] [CrossRef]
- Harbord, R.M.; Egger, M.; Sterne, J.A. A modified test for small-study effects in meta-analyses of controlled trials with binary endpoints. Stat. Med. 2006, 25, 3443–3457. [Google Scholar] [CrossRef]
- Rucker, G.; Schwarzer, G.; Carpenter, J. Arcsine test for publication bias in meta-analyses with binary outcomes. Stat. Med. 2008, 27, 746–763. [Google Scholar] [CrossRef]
- Viechtbauer, W. Conducting Meta-Analyses in R with the metafor Package. J. Stat. Softw. 2010, 36, 48. [Google Scholar] [CrossRef]
- Schwarzer, G. meta: An R package for meta-analysis. R News 2007, 7, 40–45. [Google Scholar]


| miRNA Polymorphism | Author | Cancer | Ethnicity | Genotyping | Source a | Cases b | Controls b | Quality c |
|---|---|---|---|---|---|---|---|---|
| miR-100-rs1834306 | [93] | CCa | As. | TaqMan | HB | 171/299/139 | 168/289/126 | 11 |
| miR-100-rs1834306 | [30] | BCa | As. | RFLP | HB | 52/207/5 | 46/226/9 | 9 |
| miR-100-rs1834306 | [55] | BCa | As. | MA | BD | 60/93/38 | 55/87/48 | 11 |
| miR-100-rs1834306 | [51] | CCa | As. | LDR | HB | 38/38/27 | 127/203/87 | 9 |
| miR-100-rs1834306 | [54] | BCa | A.A | Illumina | PB | 473 | 412 | 14 |
| miR-100-rs1834306 | [54] | BCa | E.A | Illumina | PB | 329 | 309 | 14 |
| miR-124-rs531564 | [93] | CCa | As. | TaqMan | HB | 17/144/448 | 7/118/458 | 11 |
| miR-124-rs531564 | [30] | BCa | As. | RFLP | HB | 227/37/0 | 245/34/1 | 10 |
| miR-124-rs531564 | [55] | BCa | As. | MA | BD | 126/52/4 | 136/45/8 | 11 |
| miR-124-rs531564 | [49] | CCa | As. | PCR-LDR | HB | 134/22/2 | 184/66/10 | 8 |
| miR-146a-rs2910164 | [78] | BCa | As. | ARMS | NA | 33/61/6 | 57/84/9 | 4 |
| miR-146a-rs2910164 | [60] | BCa | As. | TaqMan | NA | 48/50/2 | 51/46/3 | 5 |
| miR-146a-rs2910164 | [50] | BCa | As. | RFLP | PB | 82/35/4 | 84/72/8 | 10 |
| miR-146a-rs2910164 | [26] | BCa | Af. | SQ | PB | 46/29/8 | 28/17/5 | 8 |
| miR-146a-rs2910164 | [70] | BCa | As. | RFLP | NA | 52/46/0 | 60/39/0 | 6 |
| miR-146a-rs2910164 | [67] | BCa | Ca. | TaqMan/SQ | BD | 860/590/109 | 1186/838/123 | 12 |
| miR-146a-rs2910164 | [83] | BCa | Ca. | TaqMan | NA | 676/388/66 | 352/220/24 | 9 |
| miR-146a-rs2910164 | [47] | BCa | As. | MA | HB | 75/242/133 | 72/225/153 | 9 |
| miR-146a-rs2910164 | [68] | BCa | As. | RFLP | PB | 165/515/329 | 180/551/362 | 14 |
| miR-146a-rs2910164 | [81] | ECa, OCa | As. | RFLP | NA | 101/62/53 | 12/55/33 | 7 |
| miR-146a-rs2910164 | [55] | BCa | As. | MA | BD | 35/94/63 | 34/93/64 | 11 |
| miR-146a-rs2910164 | [32] | BCa | As. | RFLP | HB | 130/178/45 | 190/145/18 | 10 |
| miR-146a-rs2910164 | [34] | BCa | Ca. | TaqMan | BD | 324/171/28 | 445/199/27 | 9 |
| miR-146a-rs2910164 | [77] | BCa | As. | HRM | BD | 7/54/39 | 22/49/41 | 9 |
| miR-146a-rs2910164 | [28] | BCa | SA | TaqMan | PB | 236/165/39 | 561/410/77 | 14 |
| miR-146a-rs2910164 | [33] | BCa | As. | RFLP | HB | 74/84/42 | 64/94/42 | 7 |
| miR-146a-rs2910164 | [80] | OCa | As. | RFLP | HB | 24/75/56 | 50/161/131 | 9 |
| miR-146a-rs2910164 | [52] | BCa | As. | ARMS | PB | 183/45/8 | 155/39/9 | 9 |
| miR-146a-rs2910164 | [36] | BCa | As. | RFLP | NA | 153/76/11 | 178/45/8 | 5 |
| miR-146a-rs2910164 | [69] | BCa, OCa | Ca. | SQ | NA | 60/36/5 | 90/59/6 | 5 |
| miR-146a-rs2910164 | [46] | BCa | As. | TaqMan | HB | 146/132/43 | 126/144/20 | 6 |
| miR-146a-rs2910164 | [38] | BCa | Af. | Illumina | BD | 567/802/284 | 678/972/378 | 11 |
| miR-146a-rs2910164 | [35] | CCa | As. | RFLP | HB | 81/85/18 | 84/72/8 | 8 |
| miR-146a-rs2910164 | [73] | OCa | As. | RFLP | HB | 29/62/43 | 19/103/105 | 6 |
| miR-146a-rs2910164 | [92] | CCa | As. | RFLP | NA | 80/49/21 | 73/49/28 | 6 |
| miR-146a-rs2910164 | [42] | BCa | Ca. | HRM | PB | 325/193/28 | 112/99/35 | 12 |
| miR-146a-rs2910164 | [74] | CCa | As. | RFLP | HB | 118/224/105 | 87/206/150 | 10 |
| miR-146a-rs2910164 | [63] | BCa | As. | RFLP | HB | 43/113/70 | 34/159/116 | 10 |
| miR-149-rs2292832 | [47] | BCa | As. | MA | HB | 231/183/36 | 202/188/60 | 9 |
| miR-149-rs2292832 | [68] | BCa | As. | RFLP | PB | 99/460/450 | 108/503/482 | 14 |
| miR-149-rs2292832 | [65] | BCa, OCa | Ca. | MA | HB | 40/40/87 | 39/30/53 | 6 |
| miR-149-rs2292832 | [55] | BCa | As. | MA | BD | 99/69/17 | 100/60/26 | 10 |
| miR-149-rs2292832 | [80] | OCa | As. | RFLP | HB | 26/82/47 | 55/179/108 | 9 |
| miR-149-rs2292832 | [87] | BCa | As. | RFLP | PB | 120/102/23 | 92/113/24 | 13 |
| miR-196a2-rs11614913 | [78] | BCa | As. | ARMS | NA | 34/52/14 | 38/93/19 | 4 |
| miR-196a2-rs11614913 | [60] | BCa | As. | TaqMan | NA | 35/63/2 | 46/50/4 | 4 |
| miR-196a2-rs11614913 | [50] | BCa | As. | RFLP | PB | 68/41/12 | 85/59/21 | 9 |
| miR-196a2-rs11614913 | [70] | BCa | As. | RFLP | NA | 48/47/0 | 64/35/0 | 6 |
| miR-196a2-rs11614913 | [67] | BCa | Ca. | TaqMan/SQ | BD | 766/842/244 | 1116/1246/377 | 12 |
| miR-196a2-rs11614913 | [44] | BCa | As. | MA | HB | 197/265/98 | 155/284/144 | 10 |
| miR-196a2-rs11614913 | [31] | BCa | As. | ARMS | NA | 33/51/14 | 25/62/13 | 1 |
| miR-196a2-rs11614913 | [29] | BCa | As. | RFLP | HB | 53/42/5 | 56/38/6 | 7 |
| miR-196a2-rs11614913 | [47] | BCa | As. | MA | HB | 81/233/136 | 93/223/134 | 9 |
| miR-196a2-rs11614913 | [82] | BCa | Ca. | MA | HB | 181/209/36 | 166/229/71 | 11 |
| miR-196a2-rs11614913 | [68] | BCa | As. | RFLP | PB | 239/483/287 | 218/517/358 | 14 |
| miR-196a2-rs11614913 | [62] | BCa | Ca. | RFLP | PB | 68/86/33 | 58/82/31 | 12 |
| miR-196a2-rs11614913 | [65] | BCa, OCa | Ca. | MA | HB | 106/110/53 | 78/88/39 | 7 |
| miR-196a2-rs11614913 | [57] | BCa | Ca. | TaqMan | HB | 83/148/94 | 94/144/66 | 8 |
| miR-196a2-rs11614913 | [57] | BCa | Non-Ca. | TaqMan | HB | 11/29/23 | 33/21/30 | 5 |
| miR-196a2-rs11614913 | [81] | ECa, OCa | As. | RFLP | NA | 25/133/58 | 23/49/28 | 7 |
| miR-196a2-rs11614913 | [55] | BCa | As. | MA | BD | 44/92/54 | 49/79/59 | 10 |
| miR-196a2-rs11614913 | [32] | BCa | As. | RFLP | HB | 142/169/42 | 149/158/46 | 10 |
| miR-196a2-rs11614913 | [77] | BCa | As. | HRM | BD | 37/49/14 | 29/55/28 | 9 |
| miR-196a2-rs11614913 | [85] | BCa | SA | TaqMan | PB | 192/191/57 | 342/351/114 | 15 |
| miR-196a2-rs11614913 | [33] | BCa | As. | RFLP | HB | 36/128/36 | 26/160/14 | 6 |
| miR-196a2-rs11614913 | [80] | OCa | As. | RFLP | HB | 32/82/41 | 66/176/100 | 9 |
| miR-196a2-rs11614913 | [52] | BCa | As. | ARMS | PB | 218/18/0 | 178/25/0 | 10 |
| miR-196a2-rs11614913 | [46] | BCa | As. | TaqMan | HB | 34/119/168 | 17/88/185 | 7 |
| miR-196a2-rs11614913 | [38] | BCa | Af. | Illumina | BD | 1120/503/34 | 1395/579/54 | 11 |
| miR-196a2-rs11614913 | [43] | OCa | As. | RFLP | HB | 121/247/111 | 86/203/142 | 10 |
| miR-196a2-rs11614913 | [35] | CCa | As. | RFLP | HB | 20/93/71 | 21/81/62 | 8 |
| miR-196a2-rs11614913 | [73] | OCa | As. | RFLP | HB | 29/66/29 | 34/116/77 | 6 |
| miR-196a2-rs11614913 | [92] | CCa | As. | RFLP | NA | 75/58/17 | 42/51/57 | 6 |
| miR-196a2-rs11614913 | [87] | BCa | As. | RFLP | PB | 11/89/148 | 17/93/133 | 13 |
| miR-196a2-rs11614913 | [63] | BCa | As. | RFLP | HB | 46/123/57 | 58/169/82 | 10 |
| miR-218-rs11134527 | [93] | CCa | As. | TaqMan | HB | 233/294/92 | 242/273/68 | 11 |
| miR-218-rs11134527 | [30] | BCa | As. | RFLP | HB | 206/59/0 | 269/10/0 | 10 |
| miR-218-rs11134527 | [56] | CCa | As. | TaqMan | HB | 588/752/225 | 512/638/241 | 11 |
| miR-218-rs11134527 | [89] | CCa | As. | RFLP | PB | 268/316/101 | 247/339/127 | 13 |
| miR-26a-1-rs7372209 | [55] | BCa | As. | MA | BD | 109/64/19 | 99/74/18 | 11 |
| miR-26a-1-rs7372209 | [51] | CCa | As. | LDR | HB | 57/36/10 | 221/167/29 | 9 |
| miR-27a-rs895819 | [58] | BCa | Ca. | TaqMan | BD | 547/388/90 | 803/633/157 | 10 |
| miR-27a-rs895819 | [47] | BCa | As. | MA | HB | 251/165/34 | 232/181/37 | 9 |
| miR-27a-rs895819 | [65] | BCa, OCa | Ca. | MA | HB | 141/112/14 | 91/82/15 | 8 |
| miR-27a-rs895819 | [55] | BCa | As. | MA | BD | 97/76/16 | 106/70/14 | 11 |
| miR-27a-rs895819 | [32] | BCa | As. | RFLP | HB | 167/156/30 | 127/155/71 | 10 |
| miR-27a-rs895819 | [85] | BCa | SA | TaqMan | PB | 245/166/29 | 432/298/77 | 14 |
| miR-27a-rs895819 | [36] | BCa | As. | RFLP | NA | 156/68/16 | 113/99/19 | 6 |
| miR-27a-rs895819 | [46] | BCa | As. | TaqMan | HB | 101/159/61 | 95/139/56 | 7 |
| miR-27a-rs895819 | [38] | BCa | Af. | Illumina | BD | 376/833/448 | 455/1025/548 | 11 |
| miR-27a-rs895819 | [51] | CCa | As. | LDR | HB | 48/40/15 | 223/170/24 | 9 |
| miR-27a-rs895819 | [66] | BCa | Ca. | TaqMan/SQ | HB | 576/486/127 | 605/660/151 | 11 |
| miR-27a-rs895819 | [87] | BCa | As. | RFLP | PB | 60/144/41 | 75/109/59 | 13 |
| miR-27a-rs895819 | [53] | BCa | As. | TaqMan | HB | 152/96/16 | 137/103/15 | 10 |
| miR-34b-c-rs4938723 | [91] | BCa | A.A | SQ | PB | 362/317/63 | 343/257/58 | 14 |
| miR-34b-c-rs4938723 | [91] | BCa | Ca. | SQ | PB | 496/563/144 | 430/503/155 | 14 |
| miR-34b-c-rs4938723 | [39] | BCa | As. | RFLP | PB | 125/115/23 | 100/106/15 | 8 |
| miR-34b-c-rs4938723 | [40] | CCa | As. | RFLP | HB | 117/175/36 | 242/258/68 | 10 |
| miR-373-rs12983273 | [55] | BCa | As. | MA | BD | 161/25/1 | 160/21/2 | 11 |
| miR-373-rs12983273 | [66] | BCa | Ca. | TaqMan/SQ | HB | 566/184/18 | 540/175/22 | 11 |
| miR-423-rs6505162 | [47] | BCa | As. | MA | HB | 292/142/16 | 299/129/22 | 9 |
| miR-423-rs6505162 | [65] | BCa, OCa | Ca. | MA | HB | 97/114/56 | 55/92/49 | 8 |
| miR-423-rs6505162 | [55] | BCa | As. | MA | BD | 127/57/8 | 110/69/10 | 11 |
| miR-423-rs6505162 | [77] | BCa | As. | HRM | BD | 67/34/5 | 64/49/3 | 9 |
| miR-423-rs6505162 | [72] | BCa | As. | ARMS | NA | 25/52/23 | 81/25/18 | 5 |
| miR-423-rs6505162 | [85] | BCa | SA | TaqMan | PB | 125/229/86 | 284/385/138 | 15 |
| miR-423-rs6505162 | [38] | BCa | Af. | Illumina | BD | 90/625/942 | 119/727/1182 | 11 |
| miR-423-rs6505162 | [61] | BCa | Ca. | HRM | HB | 24/95/60 | 42/80/52 | 7 |
| miR-423-rs6505162 | [88] | BCa | As. | SQ | HB | 79/30/5 | 110/69/10 | 8 |
| miR-499-rs3746444 | [78] | BCa | As. | ARMS | NA | 63/33/4 | 66/65/19 | 5 |
| miR-499-rs3746444 | [60] | BCa | As. | TaqMan | NA | 30/62/8 | 45/40/15 | 5 |
| miR-499-rs3746444 | [50] | BCa | As. | RFLP | PB | 80/30/11 | 106/43/15 | 9 |
| miR-499-rs3746444 | [67] | BCa | Ca. | TaqMan/SQ | BD | 950/545/84 | 1305/742/120 | 12 |
| miR-499-rs3746444 | [44] | BCa | As. | MA | HB | 407/135/18 | 463/109/11 | 10 |
| miR-499-rs3746444 | [31] | BCa | As. | ARMS | NA | 35/35/10 | 63/33/4 | 2 |
| miR-499-rs3746444 | [47] | BCa | As. | MA | HB | 184/177/89 | 203/188/59 | 9 |
| miR-499-rs3746444 | [68] | BCa | As. | RFLP | PB | 707/258/44 | 816/248/29 | 14 |
| miR-499-rs3746444 | [37] | BCa | As. | ASP | HB | 43 | 48 | 3 |
| miR-499-rs3746444 | [81] | ECa, OCa | As. | RFLP | NA | 181/35/0 | 77/23/0 | 7 |
| miR-499-rs3746444 | [28] | BCa | S.A | TaqMan | PB | 319/111/10 | 772/254/22 | 14 |
| miR-499-rs3746444 | [80] | OCa | As. | RFLP | HB | 84/53/18 | 213/110/19 | 9 |
| miR-499-rs3746444 | [52] | BCa | As. | ARMS | PB | 131/44/61 | 130/48/25 | 9 |
| miR-499-rs3746444 | [46] | BCa | As. | TaqMan | HB | 152/117/52 | 141/112/37 | 7 |
| miR-499-rs3746444 | [38] | BCa | Af. | Illumina | BD | 1124/463/70 | 1374/582/72 | 11 |
| miR-499-rs3746444 | [35] | CCa | As. | RFLP | HB | 26/78/80 | 54/76/34 | 8 |
| miR-499-rs3746444 | [92] | CCa | As. | RFLP | NA | 25/47/78 | 21/49/80 | 6 |
| miR-499-rs3746444 | [63] | BCa | As. | RFLP | HB | 134/84/8 | 223/71/15 | 9 |
| miR-605-rs2043556 | [30] | BCa | As. | RFLP | HB | 38/211/15 | 42/221/18 | 9 |
| miR-605-rs2043556 | [55] | BCa | As. | MA | BD | 42/51/4 | 68/81/60 | 10 |
| miR-605-rs2043556 | [28] | BCa | S.A | TaqMan | PB | 208/182/50 | 376/571/101 | 13 |
| miR-605-rs2043556 | [38] | BCa | Af. | Illumina | BD | 975/574/108 | 1186/726/116 | 11 |
| miR-605-rs2043556 | [87] | BCa | As. | RFLP | PB | 131/90/27 | 125/102/11 | 13 |
| miR-608-rs4919510 | [44] | BCa | As. | MA | HB | 157/296/107 | 183/287/113 | 10 |
| miR-608-rs4919510 | [41] | BCa | As. | RFLP | PB | 140/20/0 | 149/43/0 | 8 |
| miR-608-rs4919510 | [59] | BCa | As. | SNPstream | PB | 128/381/254 | 277/684/456 | 14 |
| miR-608-rs4919510 | [55] | BCa | As. | MA | BD | 37/98/57 | 47/82/61 | 11 |
| miR-608-rs4919510 | [85] | BCa | S.A | TaqMan | PB | 226/174/40 | 431/310/66 | 15 |
| miR-618-rs2682818 | [85] | BCa | S.A | TaqMan | PB | 359/78/3 | 699/102/6 | 15 |
| miR-618-rs2682818 | [87] | BCa | As. | RFLP | PB | 132/99/13 | 130/91/11 | 13 |
| Genetic Models | N a | Samples | ORb (95% CI) | Pc | PHetd | I2 | τ | Pbiase |
|---|---|---|---|---|---|---|---|---|
| miR-146a rs2910164 | ||||||||
| Homozygote(CC vs. GG) | 28 | 11071/12312 | 0.92 (0.7–1.19) | 0.5 | <0.01 | 76.8 (66.8–83.8) | 0.47 | 0.66 |
| Heterozygote(GC vs. GG) | 28 | 11071/12312 | 0.94 (0.79–1.12) | 0.49 | <0.01 | 72.1 (59.4–80.9) | 0.27 | 0.54 |
| Dominant(CC+GC vs. GG) | 28 | 11071/12312 | 0.93 (0.77–1.12) | 0.43 | <0.01 | 77.6 (68.1–84.3) | 0.3 | 0.47 |
| Recessive(CC vs. GC+GG) | 28 | 11071/12312 | 0.96 (0.81–1.14) | 0.63 | <0.01 | 63.2 (44.8–75.2) | 0.28 | 0.66 |
| Allelic(C vs. G) | 28 | 11071/12312 | 0.95 (0.84–1.08) | 0.45 | <0.01 | 79.5 (71–85.5) | 0.22 | 0.73 |
| ORG f | 28 | 11071/12312 | 0.94 (0.84–1.05) | 0.29 | <0.01 | 78.34 | 0.25 | - |
| miR-196a2 rs11614913 | ||||||||
| Homozygote(TT vs. CC) | 31 | 11034/12955 | 0.82 (0.68–0.99) | 0.04 | <0.01 | 65.2 (49.1–76.2) | 0.34 | 0.90 |
| Heterozygote(CT vs. CC) | 31 | 11034/12955 | 0.97 (0.86–1.09) | 0.55 | <0.01 | 48.3 (21.5–66) | 0.17 | 0.90 |
| Dominant(CT+TT vs. CC) | 31 | 11034/12955 | 0.92 (0.8–1.06) | 0.23 | <0.01 | 62.6 (45–74.6) | 0.22 | 0.87 |
| Recessive(TT vs. CC+CT) | 31 | 11034/12955 | 0.85 (0.73–0.98) | 0.03 | <0.01 | 60.8 (42.0 –73.5) | 0.25 | 0.91 |
| Allelic(T vs. C) | 31 | 11034/12955 | 0.94 (0.87–1.01) | 0.11 | <0.01 | 59.2 (39.4–72.5) | 0.14 | 0.74 |
| ORG | 31 | 11034/12955 | 0.91 (0.83–0.99) | 0.03 | <0.01 | 68.83 | 0.19 | - |
| miR-27a rs895819 | ||||||||
| Homozygote(GG vs. AA) | 13 | 6743/8461 | 0.85 (0.65–1.11) | 0.2 | <0.05 | 64.2 (35.3–80.2) | 0.28 | 0.83 |
| Heterozygote(AG vs. AA) | 13 | 6743/8461 | 0.91 (0.79–1.05) | 0.18 | <0.05 | 54.2 (14.3–75.5) | 0.15 | 0.54 |
| Dominant(AG+GG vs. AA) | 13 | 6743/8461 | 0.89 (0.77–1.03) | 0.11 | <0.05 | 57.9 (22–77.2) | 0.15 | 0.67 |
| Recessive(GG vs. AA+AG) | 13 | 6743/8461 | 0.87 (0.68–1.11) | 0.24 | <0.05 | 64.5 (35.8–80.4) | 0.26 | 0.64 |
| Allelic(G vs. A) | 13 | 6743/8461 | 0.9 (0.8–1.02) | 0.09 | <0.05 | 65.7 (38.3–80.9) | 0.13 | 0.73 |
| ORG | 13 | 6743/8461 | 0.89 (0.80–0.98) | 0.02 | <0.01 | 62.2 | 0.14 | - |
| miR-499 rs3746444 | ||||||||
| Homozygote(CC vs. TT) | 17 | 7584/9441 | 1.40 (1.01–1.96) | 0.046 | <0.01 | 69.1 (49.1–81.2) | 0.43 | 0.62 |
| Heterozygote(TC vs. TT) | 17 | 7584/9441 | 1.13 (0.95–1.35) | 0.16 | <0.01 | 63.1 (37.8–78.1) | 0.2 | 0.37 |
| Dominant(TC+CC vs. TT) | 17 | 7584/9441 | 1.2 (0.98–1.46) | 0.07 | <0.01 | 72 (54.4–82.8) | 0.23 | 0.24 |
| Recessive(CC vs. TT+TC) | 17 | 7584/9441 | 1.33 (0.99–1.77) | 0.05 | <0.01 | 65.3 (41.9–79.2) | 0.37 | 0.74 |
| Allelic(C vs. T) | 18 | 7627/9489 | 1.23 (1.03–1.46) | 0.02 | <0.01 | 77.9 (65.6–85.9) | 0.23 | 0.23 |
| ORG | 17 | 7584/9441 | 1.20 (1.05–1.38) | <0.01 | <0.01 | 76.42 | 0.23 | - |
| miR-423 rs6505162 | ||||||||
| Homozygote(AA vs. CC) | 9 | 3505/4273 | 1.18 (0.74–1.88) | 0.43 | <0.01 | 67.6 (34.8–83.9) | 0.41 | - |
| Heterozygote(AC vs. CC) | 9 | 3505/4273 | 1.15 (0.67–2) | 0.57 | <0.01 | 84.9 (73.1–91.5) | 0.48 | - |
| Dominant(AC+AA vs. CC) | 9 | 3505/4273 | 1.14 (0.68–1.91) | 0.58 | <0.01 | 85.5 (74.4–91.8) | 0.47 | - |
| Recessive(AA vs. CC+AC) | 9 | 3505/4273 | 0.98 (0.86–1.12) | 0.78 | 0.43 | 1 (0–65.2) | 0.02 | - |
| Allelic(A vs. C) | 9 | 3505/4273 | 1.05 (0.77–1.44) | 0.72 | <0.01 | 82.4 (68–90.4) | 0.26 | - |
| ORG | 9 | 3505/4273 | 1.05 (0.83–1.32) | 0.69 | <0.01 | 82.15 | 0.30 | - |
| miR-149 rs2292832 | ||||||||
| Homozygote(CC vs. TT) | 6 | 2211/2422 | 0.86 (0.57–1.29) | 0.39 | 0.05 | 55.9 (0–82.3) | 0.29 | - |
| Heterozygote(CT vs. TT) | 6 | 2211/2422 | 0.92 (0.75–1.13) | 0.35 | 0.43 | 0 (0–74.2) | 0 | - |
| Dominant(CT+CC vs. TT) | 6 | 2211/2422 | 0.92 (0.71–1.18) | 0.42 | 0.17 | 35.2 (0–74.1) | 0.14 | - |
| Recessive(CC vs. TT+CT) | 6 | 2211/2422 | 0.9 (0.64–1.26) | 0.45 | 0.06 | 52.7 (0–81.1) | 0.21 | - |
| Allelic(C vs. T) | 6 | 2211/2422 | 0.94 (0.76–1.16) | 0.48 | 0.03 | 61 (4.5–84) | 0.15 | - |
| ORG | 6 | 2211/2422 | 0.92 (0.78–1.09) | 0.36 | 0.06 | 52.34 | 0.14 | - |
| miR-605 rs2043556 | ||||||||
| Homozygote(GG vs. AA) | 5 | 2706/3804 | 0.85 (0.24–2.97) | 0.74 | <0.01 | 82.3 (59.4–92.3) | 0.57 | - |
| Heterozygote(GA vs. AA) | 5 | 2706/3804 | 0.85 (0.62–1.16) | 0.22 | <0.01 | 72.5 (30.9–89) | 0.24 | - |
| Dominant(GA+GG vs. AA) | 5 | 2706/3804 | 0.84 (0.61–1.15) | 0.19 | <0.01 | 72.1 (29.9–88.9) | 0.23 | - |
| Recessive(GG vs. AA+GA) | 5 | 2706/3804 | 0.91 (0.25–3.35) | 0.85 | <0.01 | 84.3 (64.8–93) | 0.58 | - |
| Allelic(G vs. A) | 5 | 2706/3804 | 0.88 (0.59–1.3) | 0.41 | <0.01 | 81.3 (56.5–92) | 0.21 | - |
| ORG | 5 | 2706/3804 | 0.83 (0.64–1.07) | 0.15 | <0.01 | 79.97 | 0.25 | - |
| miR-608 rs4919510 | ||||||||
| Homozygote(GG vs. CC) | 5 | 2115/3189 | 1.16 (0.97–1.39) | 0.08 | 0.98 | 0 (0–0) | 0 | - |
| Heterozygote(GC vs. CC) | 5 | 2115/3189 | 1.09 (0.72–1.65) | 0.59 | 0.05 | 58.8 (0–84.6) | 0.19 | - |
| Dominant(GG+GC vs. CC) | 5 | 2115/3189 | 1.09 (0.75–1.58) | 0.57 | 0.06 | 55.2 (0–83.5) | 0.17 | - |
| Recessive(GG vs. GC+CC) | 5 | 2115/3189 | 1.02 (0.89–1.17) | 0.71 | 0.94 | 0 (0–0) | 0 | - |
| Allelic(G vs. C) | 5 | 2115/3189 | 1.04 (0.86–1.26) | 0.57 | 0.18 | 35.7 (0–75.9) | 0.08 | - |
| ORG | 5 | 2115/3189 | 1.04 (0.90–1.19) | 0.57 | 0.14 | 40.73 | 0.09 | - |
| miR-100 rs1834306 | ||||||||
| Homozygote(AA vs. GG) | 4 | 1167/1471 | 0.96 (0.65–1.42) | 0.77 | 0.42 | 0 (0–83.9) | 0 | - |
| Heterozygote(AG vs. GG) | 4 | 1167/1471 | 0.9 (0.65–1.24) | 0.37 | 0.37 | 5.1 (0–85.5) | 0.05 | - |
| Dominant(AA+AG vs. GG) | 4 | 1167/1471 | 0.92 (0.72–1.17) | 0.35 | 0.55 | 0 (0–78) | 0 | - |
| Recessive(AA vs. AG+GG) | 6 | 1969/2192 | 0.94 (0.71–1.24) | 0.59 | 0.15 | 38.6 (0–75.6) | 0.16 | - |
| Allelic(A vs. G) | 4 | 1167/1471 | 0.97 (0.84–1.12) | 0.55 | 0.62 | 0 (0–73.9) | 0 | - |
| ORG | 4 | 1167/1471 | 0.98 (0.84–1.15) | 0.84 | 0.41 | 0 | 0 | - |
| miR-124 rs531564 | ||||||||
| Homozygote(GG vs. CC) | 4 | 1213/1312 | 0.41 (0.27–0.61) | 0.01 | 0.93 | 0 (0–1.2) | 0 | - |
| Heterozygote(GC vs. CC) | 4 | 1213/1312 | 0.8 (0.34–1.88) | 0.47 | 0.01 | 71.9 (20.4–90.1) | 0.46 | - |
| Dominant(GG+GC vs. CC) | 4 | 1213/1312 | 0.74 (0.3–1.8) | 0.35 | <0.01 | 74.8 (30.1–90.9) | 0.48 | - |
| Recessive(GG vs. GC+CC) | 4 | 1213/1312 | 0.72 (0.53–0.99) | 0.04 | 0.63 | 0 (0–73.5) | 0 | - |
| Allelic(G vs. C) | 4 | 1213/1312 | 0.79 (0.43–1.44) | 0.3 | 0.03 | 67.9 (6.5–88.9) | 0.28 | - |
| ORG | 4 | 1213/1312 | 0.80 (0.55–1.17) | 0.25 | 0.01 | 70.02 | 0.31 | - |
| miR-218 rs11134527 G>A | ||||||||
| Homozygote(AA vs. GG) | 4 | 3134/2966 | 1.08 (0.47–2.5) | 0.73 | 0.02 | 75.8 (20.3–92.6) | 0.26 | - |
| Heterozygote(AG vs. GG) | 4 | 3134/2966 | 1.58 (0.35–7.02) | 0.4 | <0.01 | 90.9 (79.8–95.9) | 0.53 | - |
| Dominant(AA+AG vs. GG) | 4 | 3134/2966 | 1.57 (0.35–7.03) | 0.41 | <0.01 | 91.5 (81.3–96.1) | 0.52 | - |
| Recessive(AA vs. AG+GG) | 4 | 3134/2966 | 1.03 (0.68–1.56) | 0.82 | 0.09 | 58.2 (0–88.1) | 0.12 | - |
| Allelic(A vs. G) | 4 | 3134/2966 | 1.31 (0.4–4.24) | 0.52 | <0.01 | 91.8 (82.3–96.2) | 0.29 | - |
| ORG | 4 | 3134/2966 | 1.38 (0.95–2.01) | 0.08 | <0.01 | 92.09 | 0.34 | - |
| miR-34b/c rs4938723 | ||||||||
| Homozygote(CC vs. TT) | 4 | 2536/2535 | 0.93 (0.7–1.23) | 0.45 | 0.47 | 0 (0–81.7) | 0 | - |
| Heterozygote(CT vs. TT) | 4 | 2536/2535 | 1.09 (0.8–1.48) | 0.44 | 0.1 | 52.3 (0–84.2) | 0.13 | - |
| Dominant(CT+CC vs. TT) | 4 | 2536/2535 | 1.06 (0.81–1.4) | 0.52 | 0.11 | 50.5 (0–83.6) | 0.12 | - |
| Recessive(CC vs. TT+CT) | 4 | 2536/2535 | 0.89 (0.71–1.12) | 0.21 | 0.59 | 0 (0–75.9) | 0 | - |
| Allelic(C vs. T) | 4 | 2536/2535 | 1.01 (0.86–1.19) | 0.83 | 0.23 | 30.7 (0–74.9) | 0.06 | - |
| ORG | 4 | 2536/2535 | 1.02 (0.89–1.18) | 0.67 | 0.16 | 41.53 | 0.08 | - |
| miR-26a-1 rs7372209 | ||||||||
| Homozygote(TT vs. CC) | 2 | 295/608 | 1.11 (0.14–9.1) | 0.63 | 0.53 | 0 | 0 | - |
| Heterozygote(CT vs. CC) | 2 | 295/608 | 0.81 (0.55–1.2) | 0.09 | 0.85 | 0 | 0 | - |
| Dominant(CT+TT vs. CC) | 2 | 295/608 | 0.86 (0.44–1.67) | 0.21 | 0.73 | 0 | 0 | - |
| Recessive(TT vs. CC+CT) | 2 | 295/608 | 1.21 (0.17–8.58) | 0.43 | 0.55 | 0 | 0 | - |
| Allelic(T vs. C) | 2 | 295/608 | 0.95 (0.44–2.05) | 0.53 | 0.61 | 0 | 0 | - |
| ORG | 2 | 295/608 | 0.90 (0.69–1.18) | 0.48 | 0.66 | 0 | 0 | - |
| miR-373 rs12983273 | ||||||||
| Homozygote(TT vs. CC) | 2 | 955/920 | 0.76 (0.18–3.11) | 0.24 | 0.72 | 0 | 0 | - |
| Heterozygote(CT vs. CC) | 2 | 955/920 | 1.02 (0.51–2.07) | 0.74 | 0.63 | 0 | 0 | - |
| Dominant(CT+TT vs. CC) | 2 | 955/920 | 0.99 (0.56–1.78) | 0.94 | 0.67 | 0 | 0 | - |
| Recessive(TT vs. CC+CT) | 2 | 955/920 | 0.76 (0.17–3.29) | 0.25 | 0.71 | 0 | 0 | - |
| Allelic(T vs. C) | 2 | 955/920 | 0.97 (0.64–1.47) | 0.51 | 0.74 | 0 | 0 | - |
| ORG | 2 | 955/920 | 0.98 (0.80–1.21) | 0.9 | 0.67 | 0 | 0 | - |
| miR-618 rs2682818 | ||||||||
| Homozygote(AA vs. CC) | 2 | 684/1039 | 1.11 (0.41–3.03) | 0.41 | 0.83 | 0 | 0 | - |
| Heterozygote(CA vs. CC) | 2 | 684/1039 | 1.28(0.16–10.28) | 0.37 | 0.19 | 41.7 | 0.15 | - |
| Dominant(CA+AA vs. CC) | 2 | 684/1039 | 1.27 (0.19–8.52) | 0.35 | 0.22 | 33.7 | 0.12 | - |
| Recessive(AA vs. CC+CA) | 2 | 684/1039 | 1.07 (0.33–3.45) | 0.59 | 0.8 | 0 | 0 | - |
| Allelic(A vs. C) | 2 | 684/1039 | 1.22 (0.23–6.39) | 0.37 | 0.22 | 33.8 | 0.11 | - |
| ORG | 2 | 684/1039 | 1.26 (0.94–1.68) | 0.11 | 0.21 | 35.50 | 0.12 | |
| Models | N a | Samples b | OR b (95% CI) c | Pd | PHe | I2 | τ |
|---|---|---|---|---|---|---|---|
| Ethnicity: Asians | |||||||
| Homozygote | 20 | 5032/5371 | 0.89 (0.63–1.27) | 0.5 | <0.01 | 77.6 (65.8–85.3) | 0.57 |
| Heterozygote | 20 | 5032/5371 | 0.93 (0.7–1.23) | 0.58 | <0.01 | 78.7 (67.7–85.9) | 0.43 |
| Dominant | 20 | 5032/5371 | 0.91 (0.69–1.21) | 0.51 | <0.01 | 81.9 (73.1–87.9) | 0.46 |
| Recessive | 20 | 5032/5371 | 0.94 (0.77–1.15) | 0.52 | <0.01 | 58.2 (31.4–74.5) | 0.28 |
| Allelic | 20 | 5032/5371 | 0.95 (0.8–1.13) | 0.57 | <0.01 | 81.7 (72.8–87.8) | 0.29 |
| ORGf | 20 | 5032/5371 | 0.94 (0.79–1.11) | 0.48 | <0.01 | 81.51 | 0.33 |
| Ethnicity: Caucasians | |||||||
| Homozygote | 5 | 3859/3815 | 0.96 (0.39–2.35) | 0.9 | <0.01 | 85.4 (67.9–93.4) | 0.61 |
| Heterozygote | 5 | 3859/3815 | 0.94 (0.75–1.19) | 0.5 | 0.11 | 46.8 (0–80.5) | 0.11 |
| Dominant | 5 | 3859/3815 | 0.93 (0.66–1.31) | 0.58 | <0.01 | 74.1 (35.8–89.6) | 0.2 |
| Recessive | 5 | 3859/3815 | 0.99 (0.44–2.22) | 0.98 | <0.01 | 83.1 (61.4–92.6) | 0.55 |
| Allelic | 5 | 3859/3815 | 0.94 (0.65–1.35) | 0.66 | <0.01 | 84.9 (66.3–93.2) | 0.23 |
| ORG | 5 | 3859/3815 | 0.92 (0.74–1.16) | 0.52 | <0.01 | 80.78 | 0.22 |
| Ethnicity: Africans | |||||||
| Homozygote | 2 | 1740/2078 | 0.9 (0.77–1.05) | 0.07 | 0.9 | 0 | 0 |
| Heterozygote | 2 | 1740/2078 | 0.99 (0.89–1.11) | 0.55 | 0.91 | 0 | 0 |
| Dominant | 2 | 1740/2078 | 0.97 (0.84–1.11) | 0.2 | 0.87 | 0 | 0 |
| Recessive | 2 | 1740/2078 | 0.9 (0.81–1.01) | 0.05 | 0.92 | 0 | 0 |
| Allelic | 2 | 1740/2078 | 0.96 (0.86–1.06) | 0.12 | 0.86 | 0 | 0 |
| ORG | 2 | 1740/2078 | 0.95 (0.85–1.05) | 0.35 | 0.84 | 0 | 0 |
| Breast Cancer | |||||||
| Homozygote | 20 | 9458/10422 | 1.1 (0.85–1.43) | 0.45 | <0.01 | 68.7 (50.3–80.2) | 0.36 |
| Heterozygote | 20 | 9458/10422 | 1.03 (0.89–1.2) | 0.66 | <0.01 | 62.6 (39.4–76.9) | 0.2 |
| Dominant | 20 | 9458/10422 | 1.04 (0.88–1.22) | 0.62 | <0.01 | 70.3 (53.1–81.1) | 0.22 |
| Recessive | 20 | 9458/10422 | 1.06 (0.87–1.3) | 0.54 | <0.01 | 60.8 (36.2–75.9) | 0.26 |
| Allelic | 20 | 9458/10422 | 1.03 (0.91–1.17) | 0.62 | <0.01 | 74.1 (59.8–83.3) | 0.18 |
| ORG | 20 | 9458/10422 | 1.03 (0.92–1.15) | 0.58 | <0.01 | 72.68 | 0.19 |
| Breast Cancer: in Caucasians | |||||||
| Homozygote | 4 | 3758/3660 | 0.92 (0.26–3.23) | 0.85 | <0.01 | 89 (74.7–95.3) | 0.64 |
| Heterozygote | 4 | 3758/3660 | 0.94 (0.68–1.3) | 0.58 | 0.06 | 59.9 (0–86.6) | 0.13 |
| Dominant | 4 | 3758/3660 | 0.92 (0.57–1.5) | 0.64 | <0.01 | 80.6 (48.9–92.6) | 0.21 |
| Recessive | 4 | 3758/3660 | 0.96 (0.31–2.94) | 0.91 | <0.01 | 87.2 (69.5–94.7) | 0.58 |
| Allelic | 4 | 3758/3660 | 0.93 (0.56–1.56) | 0.69 | <0.01 | 88.7 (73.5–95.1) | 0.24 |
| ORG | 4 | 3758/3660 | 0.92 (0.71–1.19) | 0.53 | <0.01 | 85.58 | 0.23 |
| Breast Cancer: in Asians | |||||||
| Homozygote | 13 | 3520/3636 | 1.22 (0.87–1.72) | 0.22 | <0.01 | 59.2 (24.9–77.9) | 0.39 |
| Heterozygote | 13 | 3520/3636 | 1.12 (0.86–1.45) | 0.37 | <0.01 | 69.6 (46.2–82.8) | 0.33 |
| Dominant | 13 | 3520/3636 | 1.13 (0.87–1.46) | 0.33 | <0.01 | 72.7 (52.4–84.3) | 0.33 |
| Recessive | 13 | 3520/3636 | 1.11 (0.87–1.41) | 0.36 | 0.04 | 45.5 (0–71.4) | 0.23 |
| Allelic | 13 | 3520/3636 | 1.09 (0.91–1.29) | 0.32 | <0.01 | 72.1 (51.2–84) | 0.22 |
| ORG | 13 | 3520/3636 | 1.09 (0.91–1.31) | 0.30 | <0.01 | 73.42 | 0.26 |
| Gynecological Cancers | |||||||
| Homozygote | 7 | 1512/1735 | 0.55 (0.27–1.11) | 0.08 | <0.01 | 76.7 (51.2–88.9) | 0.55 |
| Heterozygote | 7 | 1512/1735 | 0.62 (0.32–1.21) | 0.13 | <0.01 | 82.5 (65.3–91.2) | 0.55 |
| Dominant | 7 | 1512/1735 | 0.59 (0.31–1.13) | 0.09 | <0.01 | 84 (68.7–91.8) | 0.55 |
| Recessive | 7 | 1512/1735 | 0.73 (0.54–0.99) | 0.04 | 0.11 | 42.3 (0–75.7) | 0.19 |
| Allelic | 7 | 1512/1735 | 0.73 (0.51–1.05) | 0.08 | <0.01 | 79.8 (58.7–90.1) | 0.3 |
| ORG | 7 | 1286/1426 | 0.71 (0.54–0.93) | 0.01 | <0.01 | 78.16 | 0.31 |
| High quality studies (score ≥ 9) | |||||||
| Homozygote | 16 | 9145/10396 | 0.96 (0.69–1.33) | 0.78 | <0.01 | 79.6 (67.7–87.2) | 0.42 |
| Heterozygote | 16 | 9145/10396 | 0.97 (0.82–1.15) | 0.68 | <0.01 | 64.4 (39.4–79.1) | 0.19 |
| Dominant | 16 | 9145/10396 | 0.95 (0.77–1.16) | 0.57 | <0.01 | 76.1 (61.3–85.2) | 0.24 |
| Recessive | 16 | 9145/10396 | 0.95 (0.76–1.2) | 0.66 | <0.01 | 71.7 (53.1–82.9) | 0.28 |
| Allelic | 16 | 9145/10396 | 0.95 (0.82–1.1) | 0.48 | <0.01 | 81.2 (70.4–88) | 0.19 |
| ORG | 16 | 9145/10396 | 0.93 (0.82–1.06) | 0.32 | <0.01 | 79.73 | 0.21 |
| Low quality studies (score < 9) | |||||||
| Homozygote | 12 | 1927/1916 | 0.85 (0.51–1.41) | 0.5 | <0.01 | 73.5 (52.9–85.1) | 0.72 |
| Heterozygote | 12 | 1927/1916 | 0.87 (0.57–1.33) | 0.49 | <0.01 | 79.8 (65.5–88.2) | 0.51 |
| Dominant | 12 | 1927/1916 | 0.88 (0.58–1.33) | 0.5 | <0.01 | 81.1 (67.9–88.8) | 0.51 |
| Recessive | 12 | 1927/1916 | 0.99 (0.72–1.38) | 0.97 | 0.02 | 51.2 (2.9–75.5) | 0.36 |
| Allelic | 12 | 1927/1916 | 0.97 (0.75–1.25) | 0.79 | <0.01 | 78.9 (63.7–87.7) | 0.36 |
| ORG | 12 | 1927/1916 | 0.95 (0.74–1.23) | 0.74 | <0.01 | 78.23 | 0.39 |
| Studies compatible with HWE | |||||||
| ORG | 22 | 9926/11189 | 0.90 (0.79–1.02) | 0.10 | <0.01 | 80.91 | 0.25 |
| Models | N a | Samples b | OR b (95% CI) c | Pd | PHete | I2 | τ |
|---|---|---|---|---|---|---|---|
| Ethnicity: Asians | |||||||
| Homozygote | 23 | 5815/6151 | 0.77 (0.6–0.97) | 0.03 | <0.01 | 62.9 (41.9–76.3) | 0.38 |
| Heterozygote | 23 | 5815/6151 | 0.94 (0.8–1.09) | 0.39 | <0.01 | 47.6 (14.7–67.8) | 0.22 |
| Dominant | 23 | 5815/6151 | 0.88 (0.73–1.05) | 0.14 | <0.01 | 61.9 (40.2–75.8) | 0.28 |
| Recessive | 23 | 5815/6151 | 0.82 (0.67–0.98) | 0.03 | <0.01 | 62 (40.3–75.8) | 0.28 |
| Allelic | 23 | 5815/6151 | 0.91 (0.83–1) | 0.06 | <0.01 | 55.1 (28.2–71.9) | 0.15 |
| ORGf | 23 | 5815/6151 | 0.87 (0.77–0.98) | 0.02 | <0.01 | 68.30 | 0.23 |
| Ethnicity: Caucasians | |||||||
| Homozygote | 5 | 3059/3885 | 0.92 (0.53–1.59) | 0.69 | <0.01 | 73.9 (35.3–89.5) | 0.34 |
| Heterozygote | 5 | 3059/3885 | 0.97 (0.87–1.08) | 0.44 | 0.7 | 0 (0–62.1) | 0 |
| Dominant | 5 | 3059/3885 | 0.95 (0.76–1.19) | 0.58 | 0.17 | 37.4 (0–76.7) | 0.11 |
| Recessive | 5 | 3059/3885 | 0.95 (0.59–1.51) | 0.75 | <0.01 | 70.9 (26–88.5) | 0.28 |
| Allelic | 5 | 3059/3885 | 0.97 (0.76–1.23) | 0.73 | 0.01 | 70.3 (24.4–88.4) | 0.15 |
| ORG | 5 | 3059/3885 | 0.96 (0.80–1.16) | 0.67 | 0.01 | 68.51 | 0.16 |
| Breast Cancer | |||||||
| Homozygote | 24 | 9420/11232 | 0.88 (0.73–1.04) | 0.13 | <0.01 | 53.3 (25.7–70.6) | 0.26 |
| Heterozygote | 24 | 9420/11232 | 0.97 (0.85–1.11) | 0.68 | <0.01 | 48.9 (18–68.2) | 0.16 |
| Dominant | 24 | 9420/11232 | 0.95 (0.83–1.09) | 0.43 | <0.01 | 57.2 (32.6–72.8) | 0.18 |
| Recessive | 24 | 9420/11232 | 0.9 (0.78–1.04) | 0.15 | <0.01 | 50.6 (20.9–69.1) | 0.21 |
| Allelic | 24 | 9420/11232 | 0.95 (0.87–1.04) | 0.28 | <0.01 | 61.8 (40.5–75.4) | 0.14 |
| ORG | 24 | 9646/11541 | 0.94 (0.86–1.04) | 0.26 | <0.01 | 61.66 | 0.16 |
| Breast Cancer: among Asians | |||||||
| Homozygote | 16 | 4281/4428 | 0.82 (0.65–1.02) | 0.07 | 0.04 | 41 (0–67.4) | 0.23 |
| Heterozygote | 16 | 4281/4428 | 0.93 (0.77–1.12) | 0.42 | 0.02 | 48.7 (8.5–71.2) | 0.2 |
| Dominant | 16 | 4281/4428 | 0.9 (0.74–1.09) | 0.24 | <0.01 | 55.6 (22.1–74.7) | 0.23 |
| Recessive | 16 | 4281/4428 | 0.89 (0.72–1.09) | 0.23 | 0.01 | 50.6 (12.3–72.2) | 0.21 |
| Allelic | 16 | 4281/4428 | 0.92 (0.81–1.04) | 0.18 | <0.01 | 59.5 (29.8–76.6) | 0.15 |
| ORG | 16 | 4507/4737 | 0.91 (0.79–1.04) | 0.19 | <0.01 | 60.65 | 0.19 |
| Breast Cancer: among Caucasians | |||||||
| Homozygote | 5 | 2979/3885 | 0.93 (0.53–1.63) | 0.74 | <0.01 | 74.3 (36.3–89.6) | 0.35 |
| Heterozygote | 5 | 2979/3885 | 0.97 (0.87–1.09) | 0.53 | 0.7 | 0 (0–62) | 0 |
| Dominant | 5 | 2979/3885 | 0.96 (0.76–1.22) | 0.68 | 0.16 | 38.9 (0–77.4) | 0.11 |
| Recessive | 5 | 2979/3885 | 0.95 (0.59–1.52) | 0.77 | <0.01 | 71 (26.4–88.6) | 0.29 |
| Allelic | 5 | 2979/3885 | 0.98 (0.76–1.25) | 0.81 | <0.01 | 70.9 (26.1–88.5) | 0.15 |
| ORG | 5 | 2979/3885 | 0.97 (0.80–1.17) | 0.76 | 0.1 | 69.08 | 0.17 |
| Gynecological Cancers | |||||||
| Homozygote | 8 | 1614/1928 | 0.69 (0.38–1.26) | 0.19 | <0.01 | 78.3 (57.4–89) | 0.62 |
| Heterozygote | 8 | 1614/1928 | 0.93 (0.66–1.29) | 0.6 | 0.06 | 47.9 (0–76.8) | 0.3 |
| Dominant | 8 | 1614/1928 | 0.83 (0.55–1.26) | 0.33 | <0.01 | 70 (37.6–85.6) | 0.42 |
| Recessive | 8 | 1614/1928 | 0.71 (0.47–1.09) | 0.1 | <0.01 | 71.9 (42.3–86.3) | 0.4 |
| Allelic | 8 | 1614/1928 | 0.89 (0.75–1.05) | 0.13 | 0.11 | 40.3 (0–73.6) | 0.15 |
| ORG | 8 | 1614/1928 | 0.78 (0.61–0.99) | 0.04 | <0.01 | 74.97 | 0.30 |
| Gynecological Cancers: among Asians | |||||||
| Homozygote | 7 | 1534/1723 | 0.68 (0.33–1.37) | 0.23 | <0.01 | 81.2 (62.1–90.7) | 0.6 |
| Heterozygote | 7 | 1534/1723 | 0.96 (0.65–1.41) | 0.8 | 0.047 | 52.8 (0–79.9) | 0.27 |
| Dominant | 7 | 1534/1723 | 0.84 (0.51–1.39) | 0.44 | <0.01 | 74.2 (45–87.9) | 0.41 |
| Recessive | 7 | 1534/1723 | 0.69 (0.43–1.12) | 0.11 | <0.01 | 75.1 (47.1–88.2) | 0.38 |
| Allelic | 7 | 1534/1723 | 0.89 (0.74–1.09) | 0.21 | 0.06 | 48.8 (0–78.3) | 0.14 |
| ORG | 7 | 1534/1723 | 0.77 (0.59–1.02) | 0.06 | <0.01 | 78.46 | 0.32 |
| Gynecological Cancers: OCa | |||||||
| Homozygote | 5 | 913/1305 | 0.77 (0.35–1.69) | 0.41 | 0.02 | 64.7 (7.3–86.5) | 0.42 |
| Heterozygote | 5 | 913/1305 | 0.94 (0.48–1.85) | 0.82 | 0.05 | 58.5 (0–84.6) | 0.32 |
| Dominant | 5 | 913/1305 | 0.87 (0.44–1.74) | 0.62 | 0.03 | 63.5 (3.6–86.2) | 0.34 |
| Recessive | 5 | 913/1305 | 0.73 (0.54–1) | 0.05 | 0.32 | 14.5 (0–82.2) | 0.1 |
| Allelic | 5 | 913/1305 | 0.85 (0.63–1.17) | 0.23 | 0.06 | 56.7 (0–84) | 0.17 |
| ORG | 5 | 913/1305 | 0.82 (0.64–1.05) | 0.12 | 0.05 | 57.01 | 0.20 |
| High quality studies (score ≥ 9) | |||||||
| Homozygote | 17 | 8689/10682 | 0.79 (0.67–0.93) | <0.01 | 0.01 | 48 (8.7–70.4) | 0.2 |
| Heterozygote | 17 | 8689/10682 | 0.96 (0.88–1.04) | 0.26 | 0.35 | 8.8 (0–45.2) | 0.05 |
| Dominant | 17 | 8689/10682 | 0.9 (0.81–0.99) | 0.049 | 0.04 | 41.8 (0–67.2) | 0.12 |
| Recessive | 17 | 8689/10682 | 0.83 (0.73–0.93) | <0.01 | 0.08 | 33.9 (0–63.2) | 0.12 |
| Allelic | 17 | 8689/10682 | 0.9 (0.83–0.98) | 0.01 | <0.01 | 56.5 (25.3–74.7) | 0.11 |
| ORG | 17 | 8689/10682 | 0.88 (0.81–0.96) | <0.01 | <0.01 | 57.12 | 0.12 |
| Low quality studies (score < 9) | |||||||
| Homozygote | 14 | 2345/2273 | 0.89 (0.58–1.37) | 0.57 | <0.01 | 76 (59.7–85.7) | 0.67 |
| Heterozygote | 14 | 2345/2273 | 1.04 (0.75–1.44) | 0.78 | <0.01 | 67.6 (43.4–81.5) | 0.41 |
| Dominant | 14 | 2345/2273 | 0.99 (0.7–1.39) | 0.94 | <0.01 | 75.2 (58.3–85.3) | 0.47 |
| Recessive | 14 | 2345/2273 | 0.9 (0.63–1.28) | 0.53 | <0.01 | 74.9 (57.6–85.1) | 0.5 |
| Allelic | 14 | 2345/2273 | 1.03 (0.88–1.2) | 0.71 | <0.01 | 59.9 (27.9–77.7) | 0.21 |
| ORG | 14 | 2345/2273 | 0.96 (0.77–1.21) | 0.78 | <0.01 | 77.76 | 0.37 |
| Studies compatible with HWE | |||||||
| ORG | 22 | 9917/11720 | 0.89 (0.82–0.98) | 0.02 | <0.01 | 64.15 | 0.15 |
| Models | N a | Samples b | OR b (95% CI) c | Pd | PHete | I2 | τ |
|---|---|---|---|---|---|---|---|
| Ethnicity: Asians | |||||||
| Homozygote | 8 | 2165/2429 | 0.9 (0.53–1.5) | 0.63 | <0.01 | 76.1 (52.2–88) | 0.53 |
| Heterozygote | 8 | 2165/2429 | 0.93 (0.69–1.24) | 0.57 | <0.01 | 67.2 (30.7–84.4) | 0.27 |
| Dominant | 8 | 2165/2429 | 0.91 (0.68–1.22) | 0.47 | <0.01 | 71.6 (41.4–86.2) | 0.28 |
| Recessive | 8 | 2165/2429 | 0.89 (0.56–1.44) | 0.6 | <0.01 | 74.7 (49–87.5) | 0.47 |
| Allelic | 8 | 2165/2429 | 0.91 (0.72–1.16) | 0.38 | <0.01 | 76.9 (54–88.4) | 0.24 |
| ORGf | 8 | 2165/2429 | 0.89 (0.72–1.09) | 0.26 | <0.01 | 73.73 | 0.25 |
| Ethnicity: Caucasians | |||||||
| Homozygote | 3 | 2481/3197 | 0.85 (0.65–1.1) | 0.11 | 0.66 | 0 (0–75.4) | 0 |
| Heterozygote | 3 | 2481/3197 | 0.84 (0.67–1.05) | 0.08 | 0.43 | 0 (0–87.7) | 0 |
| Dominant | 3 | 2481/3197 | 0.84 (0.71–0.99) | 0.04 | 0.61 | 0 (0–79.1) | 0 |
| Recessive | 3 | 2481/3197 | 0.92 (0.66–1.29) | 0.41 | 0.48 | 0 (0–85.7) | 0 |
| Allelic | 3 | 2481/3197 | 0.89 (0.83–0.95) | 0.02 | 0.86 | 0 (0–28.5) | 0 |
| ORG | 3 | 2481/3197 | 0.86 (0.78–0.95) | <0.01 | 0.79 | 0 | 0 |
| Breast Cancer | |||||||
| Homozygote | 12 | 6556/8044 | 0.8 (0.65–0.99) | 0.04 | 0.02 | 50.1 (3.2–74.3) | 0.2 |
| Heterozygote | 12 | 6556/8044 | 0.9 (0.77–1.05) | 0.17 | <0.01 | 56.8 (17.8–77.3) | 0.15 |
| Dominant | 12 | 6556/8044 | 0.87 (0.76–1.01) | 0.07 | <0.01 | 56.2 (16.4–77) | 0.14 |
| Recessive | 12 | 6556/8044 | 0.83 (0.68–1) | 0.05 | 0.02 | 51.9 (7.2–75.1) | 0.19 |
| Allelic | 12 | 6556/8044 | 0.88 (0.79–0.98) | 0.03 | <0.01 | 59.1 (22.8–78.4) | 0.11 |
| ORG | 12 | 6556/8044 | 0.86 (0.78–0.95) | <0.01 | <0.01 | 57.55 | 0.12 |
| Breast Cancer: in Asians | |||||||
| Homozygote | 7 | 2062/2012 | 0.76 (0.5–1.17) | 0.17 | 0.01 | 63.6 (17.7–83.9) | 0.38 |
| Heterozygote | 7 | 2062/2012 | 0.91 (0.65–1.28) | 0.53 | <0.01 | 70.9 (36.6–86.7) | 0.28 |
| Dominant | 7 | 2062/2012 | 0.87 (0.63–1.19) | 0.32 | <0.01 | 71.4 (37.9–86.8) | 0.27 |
| Recessive | 7 | 2062/2012 | 0.76 (0.53–1.11) | 0.13 | 0.03 | 58.2 (3.4–81.9) | 0.32 |
| Allelic | 7 | 2062/2012 | 0.86 (0.69–1.07) | 0.14 | <0.01 | 71 (36.7–86.7) | 0.2 |
| ORG | 7 | 2062/2012 | 0.83 (0.68–1.02) | 0.08 | <0.01 | 69.57 | 0.22 |
| Breast Cancer: in Caucasians | |||||||
| Homozygote | 3 | 2401/3197 | 0.85 (0.72–1.02) | 0.06 | 0.83 | 0 (0–42.4) | 0 |
| Heterozygote | 3 | 2401/3197 | 0.84 (0.67–1.05) | 0.08 | 0.43 | 0 (0–87.6) | 0 |
| Dominant | 3 | 2401/3197 | 0.84 (0.71–0.99) | 0.045 | 0.61 | 0 (0–79.2) | 0 |
| Recessive | 3 | 2401/3197 | 0.93 (0.72–1.21) | 0.36 | 0.64 | 0 (0–76.4) | 0 |
| Allelic | 3 | 2401/3197 | 0.89 (0.84–0.94) | 0.01 | 0.92 | 0 (0–0) | 0 |
| ORG | 3 | 2401/3197 | 0.86 (0.78–0.95) | <0.01 | 0.82 | 0 | 0 |
| High quality studies (score ≥ 9) | |||||||
| Homozygote | 10 | 5915/7752 | 0.87 (0.61–1.23) | 0.38 | <0.01 | 71.2 (45–84.9) | 0.31 |
| Heterozygote | 10 | 5915/7752 | 0.93 (0.81–1.07) | 0.26 | 0.06 | 45.2 (0–73.7) | 0.12 |
| Dominant | 10 | 5915/7752 | 0.91 (0.79–1.06) | 0.2 | 0.03 | 52.6 (2.9–76.9) | 0.13 |
| Recessive | 10 | 5915/7752 | 0.88 (0.63–1.22) | 0.39 | <0.01 | 72.4 (47.9–85.4) | 0.29 |
| Allelic | 10 | 5915/7752 | 0.92 (0.8–1.06) | 0.21 | <0.01 | 67 (35.8–83.1) | 0.12 |
| ORG | 10 | 5915/7752 | 0.90 (0.81–1.00) | 0.07 | <0.01 | 60.97 | 0.12 |
| Low quality studies (score < 9) | |||||||
| Homozygote | 3 | 828/709 | 0.81 (0.36–1.78) | 0.36 | 0.34 | 8.5 (0–90.5) | 0.1 |
| Heterozygote | 3 | 828/709 | 0.78 (0.29–2.11) | 0.4 | 0.01 | 76.3 (22.5–92.8) | 0.35 |
| Dominant | 3 | 828/709 | 0.77 (0.31–1.93) | 0.35 | 0.02 | 75.3 (18.3–92.5) | 0.32 |
| Recessive | 3 | 828/709 | 0.87 (0.53–1.44) | 0.36 | 0.59 | 0 (0–80.1) | 0 |
| Allelic | 3 | 828/709 | 0.82 (0.44–1.52) | 0.3 | 0.04 | 69.9 (0–91.2) | 0.21 |
| ORG | 3 | 828/709 | 0.78 (0.55–1.12) | 0.18 | 0.02 | 72.78 | 0.26 |
| Studies compatible with HWE | |||||||
| ORG | 12 | 6303/7654 | 0.88 (0.78–0.99) | 0.35 | <0.01 | 65.17 | 0.15 |
| Models | N a | Samples b | OR (95% CI) c | Pd | PHete | I2 | τ |
|---|---|---|---|---|---|---|---|
| Ethnicity: Asians | |||||||
| Homozygote | 14 | 3908/4198 | 1.5 (0.98–2.3) | 0.06 | <0.01 | 68.5 (45.2–81.9) | 0.50 |
| Heterozygote | 14 | 3908/4198 | 1.18 (0.93–1.5) | 0.16 | <0.01 | 64.8 (37.8–80.1) | 0.27 |
| Dominant | 14 | 3908/4198 | 1.26 (0.97–1.64) | 0.07 | <0.01 | 70.8 (49.7–83.1) | 0.29 |
| Recessive | 14 | 3908/4198 | 1.4 (0.97–2.02) | 0.07 | <0.01 | 65.5 (39.2–80.4) | 0.43 |
| Allelic | 15 | 3951/4246 | 1.3 (1.05–1.61) | 0.02 | <0.01 | 74.2 (57.1–84.5) | 0.26 |
| ORGf | 14 | 3908/4198 | 1.26 (1.05–1.51) | 0.01 | <0.01 | 74.91 | 0.29 |
| Breast Cancer | |||||||
| Homozygote | 12 | 6653/8376 | 1.32 (0.93–1.87) | 0.11 | <0.01 | 61.6 (28.1–79.5) | 0.34 |
| Heterozygote | 12 | 6653/8376 | 1.08 (0.91–1.28) | 0.35 | 0.01 | 54.1 (11.9–76.1) | 0.15 |
| Dominant | 12 | 6653/8376 | 1.14 (0.94–1.38) | 0.17 | <0.01 | 65.1 (35.6–81.1) | 0.18 |
| Recessive | 12 | 6653/8376 | 1.28 (0.91–1.78) | 0.14 | <0.01 | 61.4 (27.6–79.4) | 0.33 |
| Allelic | 13 | 6696/8424 | 1.17 (0.97–1.41) | 0.09 | <0.01 | 74.5 (55.9–85.2) | 0.19 |
| ORG | 12 | 6653/8376 | 1.14 (1.00–1.30) | 0.04 | <0.01 | 69.38 | 0.18 |
| Breast Cancer: in Asians | |||||||
| Homozygote | 9 | 2977/3133 | 1.43 (0.85–2.41) | 0.15 | 0.01 | 62.2 (22.1–81.7) | 0.41 |
| Heterozygote | 9 | 2977/3133 | 1.13 (0.85–1.51) | 0.36 | 0.01 | 60.9 (18.9–81.1) | 0.23 |
| Dominant | 9 | 2977/3133 | 1.21 (0.9–1.64) | 0.18 | <0.01 | 66.2 (31.5–83.3) | 0.24 |
| Recessive | 9 | 2977/3133 | 1.36 (0.83–2.24) | 0.19 | 0.01 | 63.2 (24.3–82.1) | 0.4 |
| Allelic | 10 | 3020/3181 | 1.24 (0.96–1.62) | 0.09 | <0.01 | 72.9 (48.7–85.6) | 0.24 |
| ORG | 9 | 2977/3133 | 1.21 (0.99–1.47) | 0.05 | <0.01 | 69.35 | 0.23 |
| Gynecological cancer | |||||||
| Homozygote | 5 | 931/1065 | 1.65 (0.56–4.88) | 0.27 | <0.01 | 79.1 (50.3–91.2) | 0.8 |
| Heterozygote | 5 | 931/1065 | 1.26 (0.66–2.4) | 0.37 | <0.01 | 72.4 (30.7–89) | 0.42 |
| Dominant | 5 | 931/1065 | 1.35 (0.64–2.83) | 0.32 | <0.01 | 78.8 (49.4–91.1) | 0.47 |
| Recessive | 5 | 931/1065 | 1.46 (0.64–3.35) | 0.27 | <0.01 | 74.8 (37.8–89.8) | 0.59 |
| Allelic | 5 | 931/1065 | 1.42 (0.8–2.54) | 0.17 | <0.01 | 74.7 (37.5–89.8) | 0.35 |
| ORG | 5 | 931/1065 | 1.34 (0.88–2.06) | 0.17 | <0.01 | 82.77 | 0.44 |
| High quality studies (score ≥ 9) | |||||||
| Homozygote | 10 | 6433/8387 | 1.43 (1.1–1.85) | 0.01 | 0.04 | 49.7 (0–75.6) | 0.25 |
| Heterozygote | 10 | 6433/8387 | 1.12 (0.97–1.3) | 0.11 | 0.03 | 50.2 (0–75.8) | 0.13 |
| Dominant | 10 | 6433/8387 | 1.19 (1.03–1.36) | 0.02 | 0.02 | 55.4 (9.2–78.1) | 0.13 |
| Recessive | 10 | 6433/8387 | 1.39 (1.07–1.82) | 0.02 | 0.03 | 51.8 (1–76.5) | 0.26 |
| Allelic | 10 | 6433/8387 | 1.21 (1.06–1.39) | 0.01 | <0.01 | 68.2 (38.5–83.6) | 0.14 |
| ORG | 10 | 6433/8387 | 1.20 (1.07–1.35) | <0.01 | <0.01 | 61.00 | 0.13 |
| Low quality studies (score < 9) | |||||||
| Homozygote | 7 | 1151/1054 | 1.25 (0.44–3.52) | 0.62 | <0.01 | 82.2 (64.5–91.1) | 0.89 |
| Heterozygote | 7 | 1151/1054 | 1.13 (0.65–1.98) | 0.6 | <0.01 | 76.2 (50.1–88.7) | 0.5 |
| Dominant | 7 | 1151/1054 | 1.18 (0.62–2.23) | 0.55 | <0.01 | 83.7 (68.1–91.7) | 0.59 |
| Recessive | 7 | 1151/1054 | 1.13 (0.49–2.59) | 0.74 | <0.01 | 78.1 (54.6–89.4) | 0.66 |
| Allelic | 8 | 1194/1102 | 1.22 (0.75–1.98) | 0.36 | <0.01 | 85.1 (72.5–91.9) | 0.51 |
| ORG | 7 | 1151/1054 | 1.13 (0.73–1.76) | 0.56 | <0.01 | 86.53 | 0.54 |
| Studies compatible with HWE | |||||||
| ORG | 13 | 6851/8615 | 1.19 (1.01–1.39) | 0.03 | <0.01 | 79.42 | 0.24 |
| Genetic Models | N a | Samples b | OR (95% CI) c | Pd | PHe | I2 | τ |
|---|---|---|---|---|---|---|---|
| miR-423 rs6505162: HWE compatible studies | |||||||
| Homozygote(AA vs. CC) | 8 | 3405/4149 | 1.04 (0.73–1.47) | 0.8 | 0.06 | 47.9 (0–76.8) | 0.26 |
| Heterozygote(AC vs. CC) | 8 | 3405/4149 | 0.97 (0.7–1.36) | 0.86 | 0 | 69.5 (36.3–85.3) | 0.29 |
| Dominant(AC+AA vs. CC) | 8 | 3405/4149 | 0.97 (0.7–1.34) | 0.82 | 0 | 70.9 (39.8–85.9) | 0.29 |
| Recessive(AA vs. CC+AC) | 8 | 3405/4149 | 0.97 (0.86–1.08) | 0.5 | 0.63 | 0 (0–56.4) | 0 |
| Allelic(A vs. C) | 8 | 3405/4149 | 0.96 (0.8–1.16) | 0.66 | 0.01 | 61.7 (17.1–82.3) | 0.15 |
| ORGf | 8 | 3405/4149 | 0.94 (0.80–1.12) | 0.54 | <0.01 | 64.58 | 0.18 |
| miR-423 rs6505162: BCa | |||||||
| Homozygote(AA vs. CC) | 9 | 3426/4273 | 1.17 (0.72–1.89) | 0.48 | <0.01 | 68.8 (37.5–84.4) | 0.43 |
| Heterozygote(AC vs. CC) | 9 | 3426/4273 | 1.17 (0.68–2.01) | 0.52 | <0.01 | 84.2 (71.6–91.2) | 0.47 |
| Dominant(AC+AA vs. CC) | 9 | 3426/4273 | 1.15 (0.69–1.92) | 0.55 | <0.01 | 85 (73.4–91.6) | 0.46 |
| Recessive(AA vs. CC+AC) | 9 | 3426/4273 | 0.99 (0.82–1.2) | 0.94 | 0.29 | 17 (0–59) | 0.1 |
| Allelic(A vs. C) | 9 | 3426/4273 | 1.05 (0.76–1.44) | 0.74 | <0.01 | 82.5 (68.2–90.4) | 0.27 |
| ORG | 9 | 3426/4273 | 1.04 (0.82–1.31) | 0.72 | <0.01 | 82.30 | 0.30 |
| miR-423 rs6505162 in Asians | |||||||
| Homozygote(AA vs. CC) | 5 | 962/1068 | 1.2 (0.43–3.29) | 0.65 | <0.01 | 72.2 (30.2–89) | 0.76 |
| Heterozygote(AC vs. CC) | 5 | 962/1068 | 1.14 (0.34–3.84) | 0.78 | <0.01 | 90.3 (80.2–95.2) | 0.71 |
| Dominant(AC+AA vs. CC) | 5 | 962/1068 | 1.12 (0.37–3.41) | 0.79 | <0.01 | 90.4 (80.5–95.3) | 0.68 |
| Recessive(AA vs. CC+AC) | 5 | 962/1068 | 1.04 (0.58–1.87) | 0.85 | 0.33 | 14 (0–82.1) | 0.18 |
| Allelic(A vs. C) | 5 | 962/1068 | 1.06 (0.51–2.18) | 0.85 | <0.01 | 88.1 (74.8–94.4) | 0.49 |
| ORG | 5 | 962/1068 | 1.04 (0.62–1.74) | 0.87 | <0.01 | 87.60 | 0.54 |
| miR-149 rs2292832: HWE compatible studies | |||||||
| Homozygote(CC vs. TT) | 4 | 1859/2114 | 0.79 (0.48–1.3) | 0.23 | 0.11 | 49.6 (0–83.3) | 0.24 |
| Heterozygote(CT vs. TT) | 4 | 1859/2114 | 0.87 (0.68–1.11) | 0.17 | 0.5 | 0 (0–80.5) | 0 |
| Dominant(CT+CC vs. TT) | 4 | 1859/2114 | 0.84 (0.64–1.1) | 0.14 | 0.37 | 4.4 (0–85.4) | 0.04 |
| Recessive(CC vs. TT+CT) | 4 | 1859/2114 | 0.87 (0.57–1.32) | 0.35 | 0.1 | 51.2 (0–83.9) | 0.19 |
| Allelic(C vs. T) | 4 | 1859/2114 | 0.89 (0.7–1.12) | 0.2 | 0.08 | 55.9 (0–85.4) | 0.12 |
| ORG | 4 | 1859/2114 | 0.87 (0.72–1.04) | 0.13 | 0.09 | 52.24 | 0.12 |
| miR-100 rs1834306: HWE compatible studies | |||||||
| Homozygote(AA vs. GG) | 3 | 903/1190 | 0.99 (0.62–1.6) | 0.95 | 0.47 | 0 (0–86.2) | 0 |
| Heterozygote(AG vs. GG) | 3 | 903/1190 | 0.9 (0.49–1.65) | 0.54 | 0.24 | 30.4 (0–92.8) | 0.14 |
| Dominant(AA+AG vs. GG) | 3 | 903/1190 | 0.95 (0.64–1.39) | 0.6 | 0.45 | 0 (0–87.1) | 0 |
| Recessive(AA vs. AG+GG) | 5 | 1705/1911 | 0.95 (0.69–1.32) | 0.71 | 0.12 | 45.5 (0–80) | 0.16 |
| Allelic(A vs. G) | 3 | 903/1190 | 0.99 (0.78–1.25) | 0.86 | 0.5 | 0 (0–85.1) | 0 |
| ORG | 3 | 903/1190 | 0.98 (0.84–1.14) | 0.85 | 0.52 | 0 | 0 |
| miR-605 rs2043556 in Asians | |||||||
| Homozygote(GG vs. AA) | 3 | 609/728 | 0.64 (0.01–31.22) | 0.67 | <0.01 | 90.8 (75.9–96.5) | 1.39 |
| Heterozygote(GA vs. AA) | 3 | 609/728 | 0.94 (0.68–1.3) | 0.5 | 0.72 | 0 (0–68) | 0 |
| Dominant(GA+GG vs. AA) | 3 | 609/728 | 0.89 (0.47–1.68) | 0.51 | 0.27 | 23.4 (0–92) | 0.12 |
| Recessive(GG vs. AA+GA) | 3 | 609/728 | 0.64 (0.01–33.78) | 0.68 | <0.01 | 92 (79.7–96.8) | 1.4 |
| Allelic(G vs. A) | 3 | 609/728 | 0.83 (0.26–2.69) | 0.57 | <0.01 | 87.6 (65.1–95.6) | 0.39 |
| ORG | 3 | 609/728 | 0.79 (0.46–1.36) | 0.40 | <0.01 | 84.24 | 0.43 |
| miR-608 rs4919510 in Asians | |||||||
| Homozygote(GG vs. CC) | 4 | 1675/2382 | 1.16 (0.96–1.41) | 0.11 | 0.98 | 0 (0–0) | 0 |
| Heterozygote(GC vs. CC) | 4 | 1675/2382 | 1.08 (0.55–2.13) | 0.75 | 0.02 | 68.4 (8.4–89.1) | 0.27 |
| Dominant(GG+GC vs. CC) | 4 | 1675/2382 | 1.06 (0.57–1.99) | 0.78 | 0.03 | 66.1 (0.5–88.4) | 0.24 |
| Recessive(GG vs. GC+CC) | 4 | 1675/2382 | 1.01 (0.87–1.17) | 0.85 | 0.91 | 0 (0–12.7) | 0 |
| Allelic(G vs. C) | 4 | 1675/2382 | 1.02 (0.73–1.42) | 0.86 | 0.1 | 51.4 (0–83.9) | 0.11 |
| ORG | 4 | 1675/2382 | 1.00 (0.83–1.22) | 0.93 | 0.08 | 55.24 | 0.14 |
| miR-218 rs11134527: CC | |||||||
| Homozygote(AA vs. GG) | 3 | 2869/2687 | 1.08 (0.47–2.5) | 0.73 | 0.02 | 75.8 (20.3–92.6) | 0.26 |
| Heterozygote(AG vs. GG) | 3 | 2869/2687 | 1.09 (0.6–1.96) | 0.6 | 0.09 | 59.1 (0–88.3) | 0.17 |
| Dominant(AA+AG vs. GG) | 3 | 2869/2687 | 1.08 (0.54–2.16) | 0.68 | 0.03 | 71.2 (2.2–91.5) | 0.22 |
| Recessive(AA vs. AG+GG) | 3 | 2869/2687 | 1.03 (0.68–1.56) | 0.82 | 0.09 | 58.2 (0–88.1) | 0.12 |
| Allelic(A vs. G) | 3 | 2869/2687 | 1.03 (0.7–1.52) | 0.74 | 0.02 | 74.7 (15.8–92.4) | 0.12 |
| ORG | 3 | 2869/2687 | 1.03 (0.86–1.24) | 0.69 | 0.02 | 72.87 | 0.13 |
| miR-34b/c rs4938723: BCa | |||||||
| Homozygote(CC vs. TT) | 3 | 2208/1967 | 0.9 (0.58–1.4) | 0.4 | 0.39 | 0 (0–89.1) | 0 |
| Heterozygote(CT vs. TT) | 3 | 2208/1967 | 1.02 (0.73–1.42) | 0.82 | 0.29 | 19.5 (0–91.6) | 0.06 |
| Dominant(CT+CC vs. TT) | 3 | 2208/1967 | 1 (0.74–1.36) | 0.99 | 0.29 | 19.9 (0–91.7) | 0.06 |
| Recessive(CC vs. TT+CT) | 3 | 2208/1967 | 0.89 (0.59–1.35) | 0.35 | 0.39 | 0 (0–89) | 0 |
| Allelic(C vs. T) | 3 | 2208/1967 | 0.98 (0.78–1.22) | 0.7 | 0.3 | 16.1 (0–91.3) | 0.04 |
| ORG | 3 | 2208/1967 | 1.01 (0.89–1.15) | 0.77 | 0.27 | 22.79 | 0.05 |
| miR-124 rs531564: high quality studies | |||||||
| Homozygote(GG vs. CC) | 3 | 1055/1052 | 0.44 (0.28–0.68) | 0.01 | 0.92 | 0 (0–0) | 0 |
| Heterozygote(GC vs. CC) | 3 | 1055/1052 | 1.04 (0.37–2.86) | 0.88 | 0.2 | 36.9 (0–80.0) | 0.05 |
| Dominant(GG+GC vs. CC) | 3 | 1055/1052 | 0.93 (0.27–3.13) | 0.82 | 0.12 | 52.5 (0–86.4) | 0.09 |
| Recessive(GG vs. GC+CC) | 3 | 1055/1052 | 0.74 (0.57–0.96) | 0.02 | 0.73 | 0 (0–65.7) | 0 |
| Allelic(G vs. C) | 3 | 1055/1052 | 0.88 (0.52–1.51) | 0.44 | 0.20 | 36.0 (0–79.5) | 0.02 |
| ORG | 3 | 1055/1052 | 1.07 (0.80–1.44) | 0.63 | 0.17 | 43.47 | 0.17 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bastami, M.; Choupani, J.; Saadatian, Z.; Zununi Vahed, S.; Ouladsahebmadarek, E.; Mansoori, Y.; Daraei, A.; Samadi Kafil, H.; Yousefi, B.; Mahdipour, M.; et al. Evidences from a Systematic Review and Meta-Analysis Unveil the Role of MiRNA Polymorphisms in the Predisposition to Female Neoplasms. Int. J. Mol. Sci. 2019, 20, 5088. https://doi.org/10.3390/ijms20205088
Bastami M, Choupani J, Saadatian Z, Zununi Vahed S, Ouladsahebmadarek E, Mansoori Y, Daraei A, Samadi Kafil H, Yousefi B, Mahdipour M, et al. Evidences from a Systematic Review and Meta-Analysis Unveil the Role of MiRNA Polymorphisms in the Predisposition to Female Neoplasms. International Journal of Molecular Sciences. 2019; 20(20):5088. https://doi.org/10.3390/ijms20205088
Chicago/Turabian StyleBastami, Milad, Jalal Choupani, Zahra Saadatian, Sepideh Zununi Vahed, Elaheh Ouladsahebmadarek, Yasser Mansoori, Abdolreza Daraei, Hossein Samadi Kafil, Bahman Yousefi, Mahdi Mahdipour, and et al. 2019. "Evidences from a Systematic Review and Meta-Analysis Unveil the Role of MiRNA Polymorphisms in the Predisposition to Female Neoplasms" International Journal of Molecular Sciences 20, no. 20: 5088. https://doi.org/10.3390/ijms20205088
APA StyleBastami, M., Choupani, J., Saadatian, Z., Zununi Vahed, S., Ouladsahebmadarek, E., Mansoori, Y., Daraei, A., Samadi Kafil, H., Yousefi, B., Mahdipour, M., Masotti, A., & Nariman-Saleh-Fam, Z. (2019). Evidences from a Systematic Review and Meta-Analysis Unveil the Role of MiRNA Polymorphisms in the Predisposition to Female Neoplasms. International Journal of Molecular Sciences, 20(20), 5088. https://doi.org/10.3390/ijms20205088