Comparative Study of Extracellular Proteolytic, Cellulolytic, and Hemicellulolytic Enzyme Activities and Biotransformation of Palm Kernel Cake Biomass by Lactic Acid Bacteria Isolated from Malaysian Foods
Abstract
:1. Introduction
2. Results
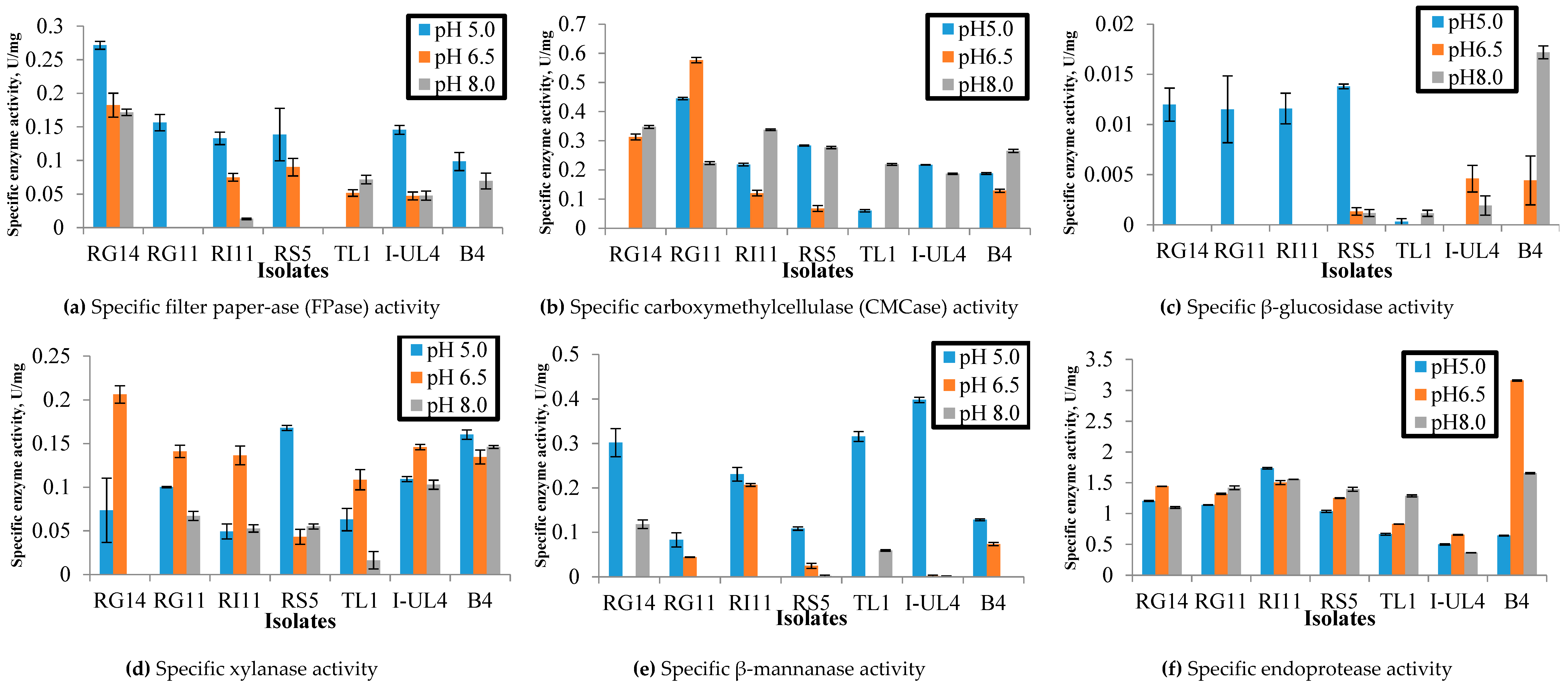
2.1. Extracellular Hydrolytic Enzyme Activities of Lactobacillus plantarum Strains
2.1.1. Effect of pH on the Extracellular Hydrolytic Enzyme Activities of Lactobacillus plantarum Strains
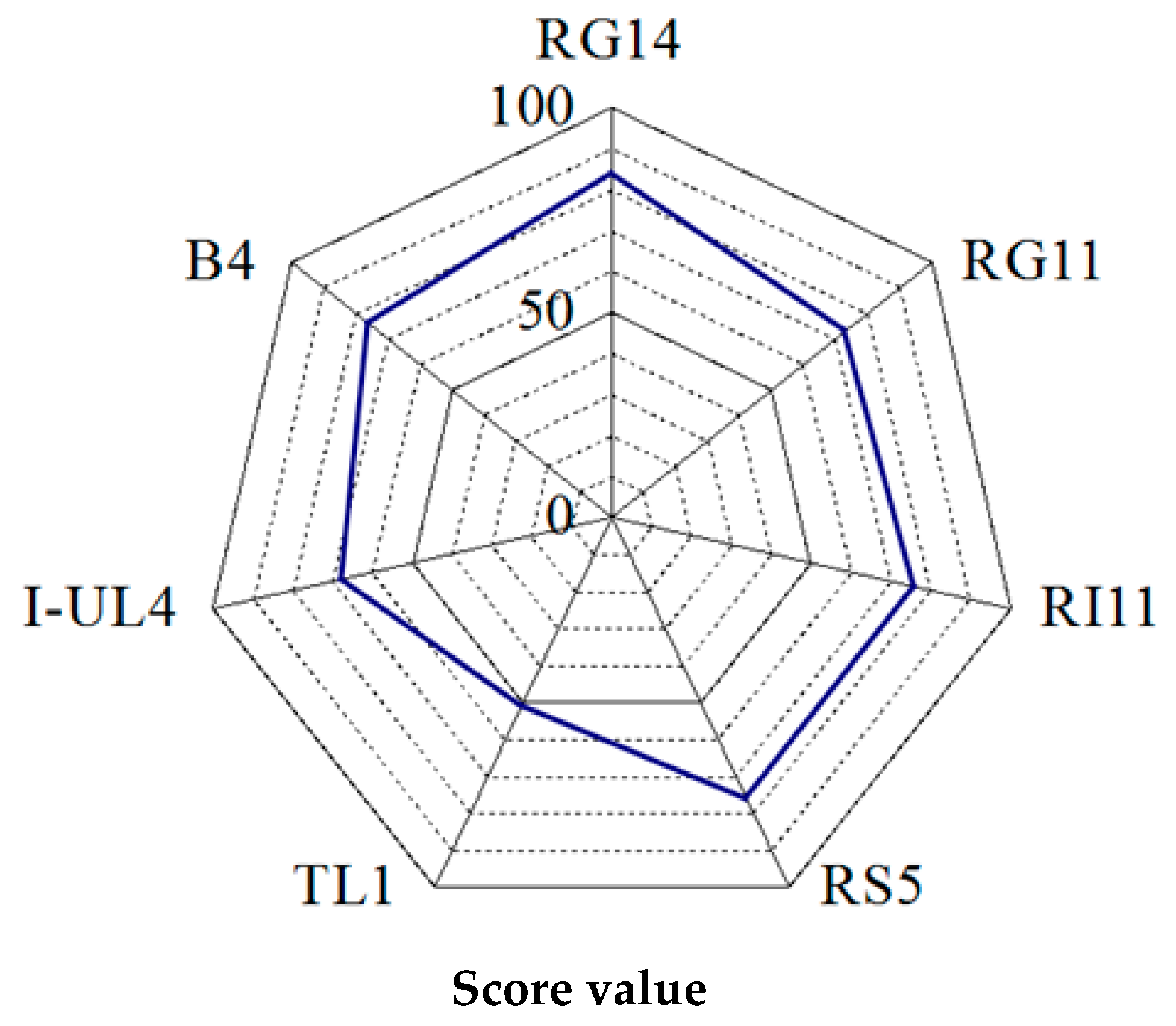
2.1.2. Rating of the Overall Extracellular Hydrolytic Enzyme Activities of Lactobacillus plantarum Strains
2.2. Solid State Fermentation of Palm Kernel Cake
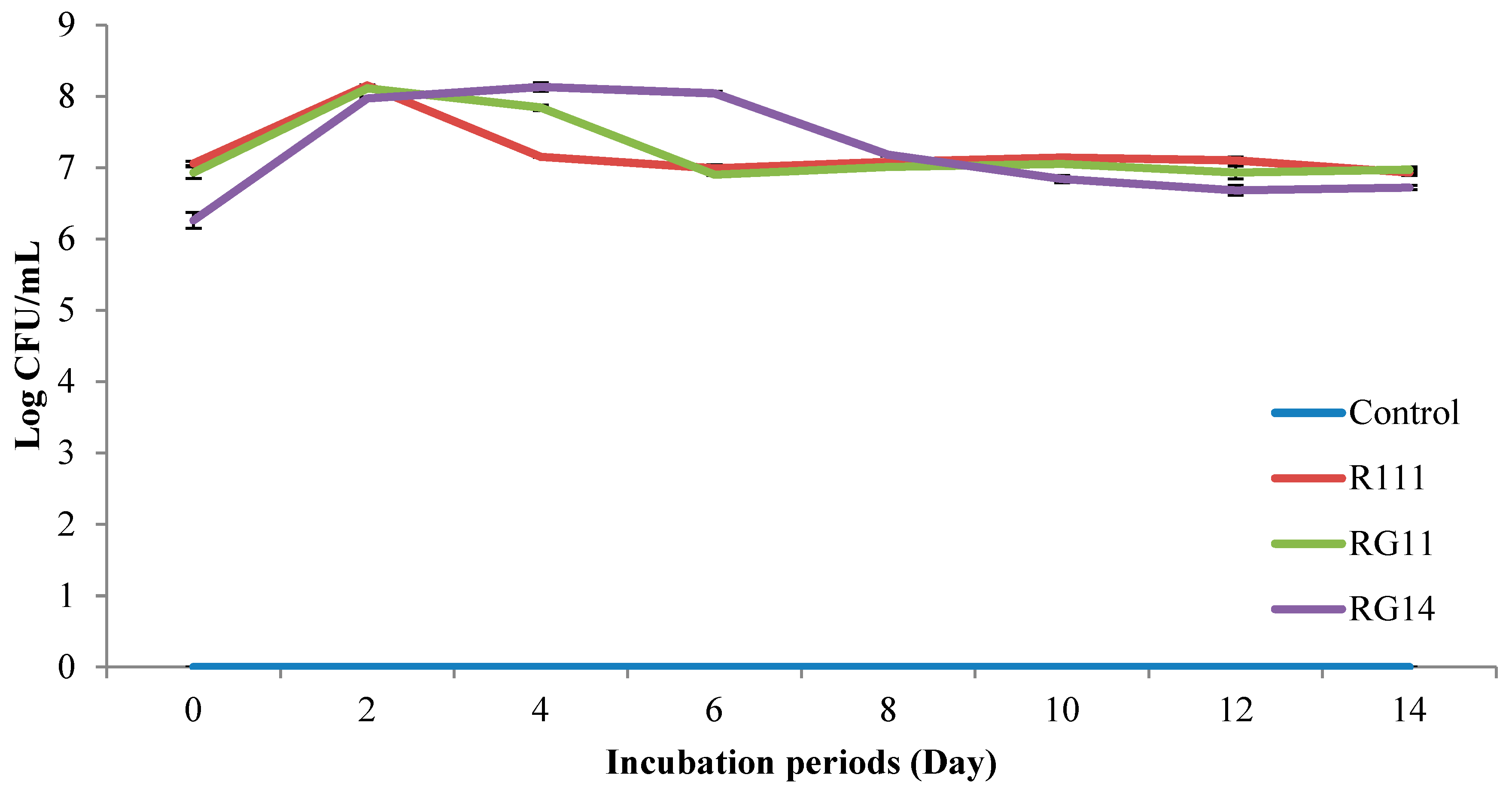
2.2.1. Viable Cell Count of Fermented Palm Kernel Cake Extract
2.2.2. Reducing Sugar Concentration of Fermented Palm Kernel Cake Extract
2.2.3. Solubilised Protein Concentration of Fermented Palm Kernel Cake Extract
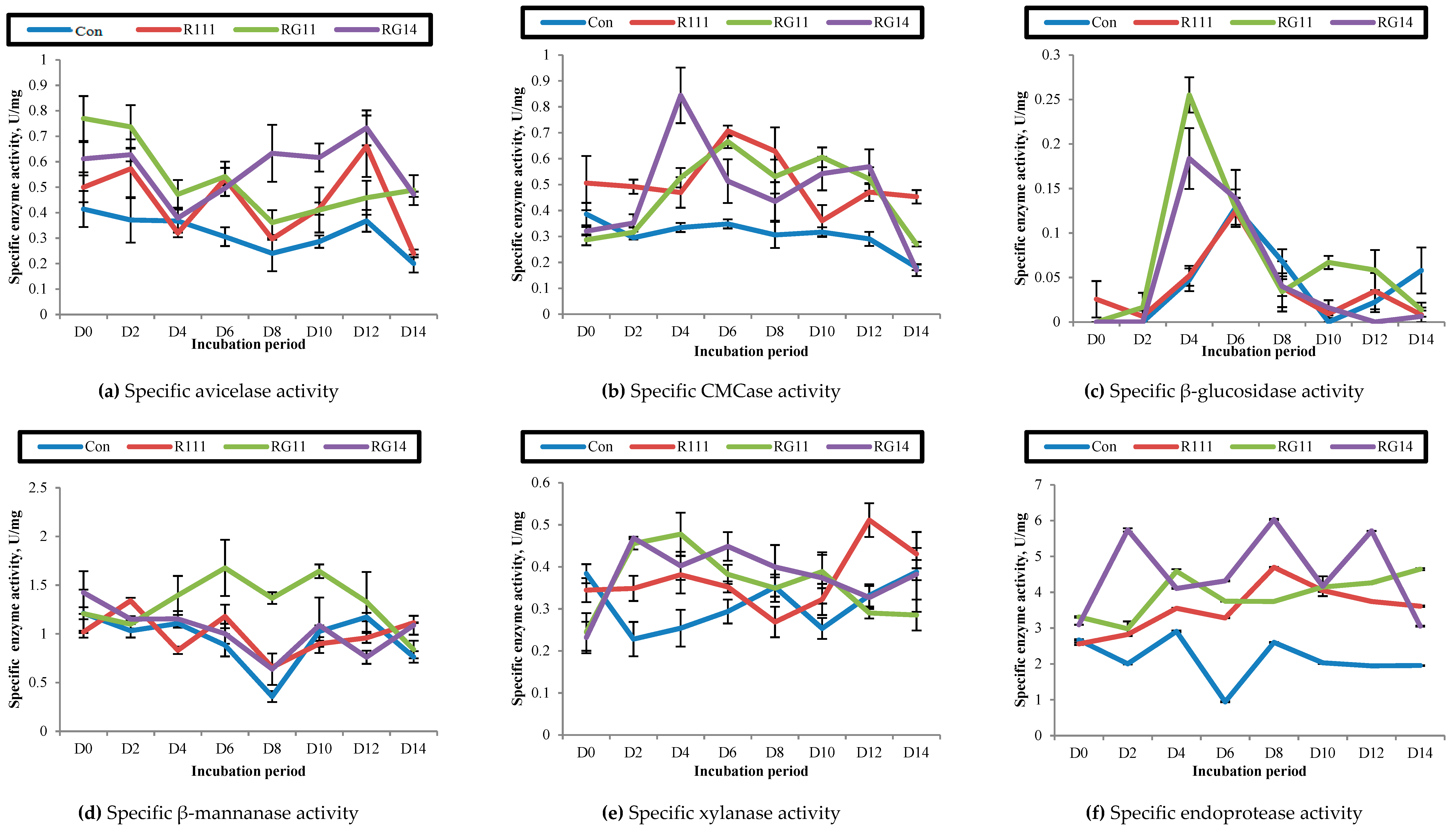
2.2.4. Hydrolytic Enzyme Activities of Fermented Palm Kernel Cake Extract
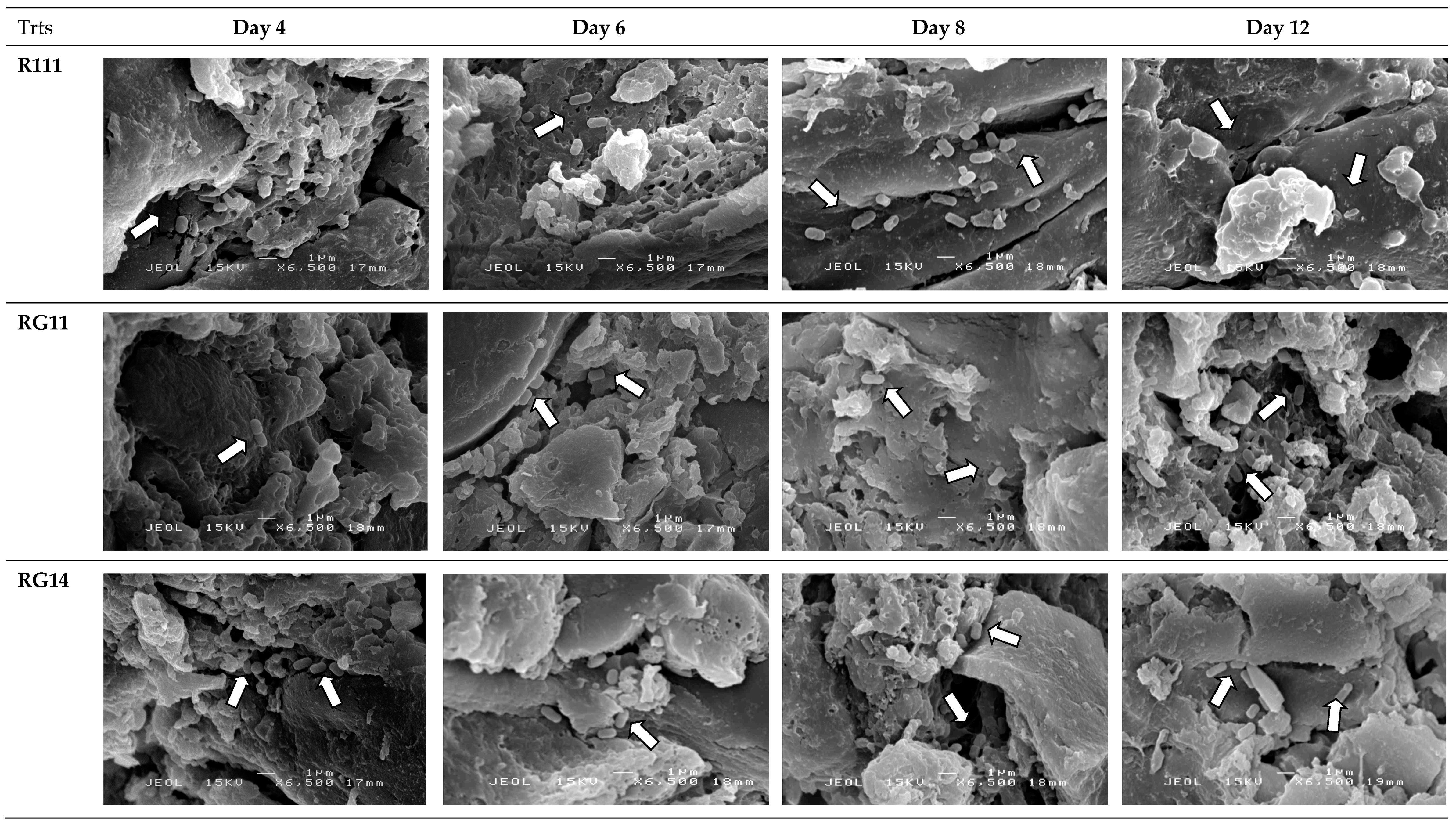
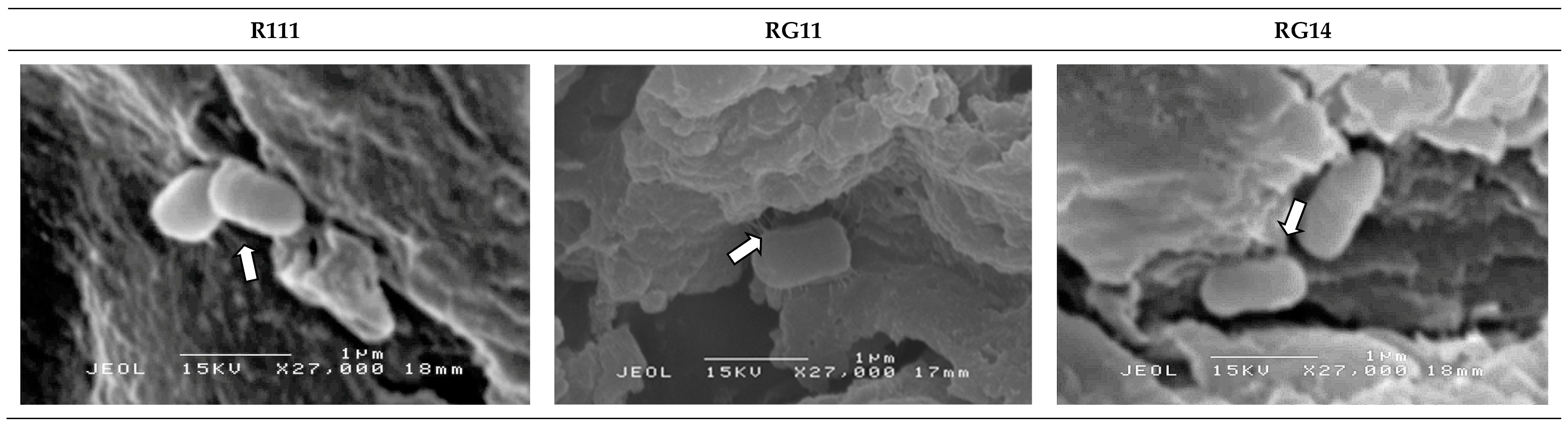
2.2.5. Attachment and Growth of Lactobacillus plantarum Strains on Palm Kernel Cake
3. Discussion
3.1. Extracellular Hydrolytic Enzyme Activities of Lactobacillus plantarum Strains
3.1.1. Effect of pH on Specific Extracellular Hydrolytic Enzyme Activities of Lactobacillus plantarum Strains
3.1.2. Rating of Extracellular Hydrolytic Enzyme Activities of Lactobacillus plantarum Strains
3.2. Solid State Fermentation of Palm Kernel Cake
3.2.1. LAB Viable Count of Fermented Palm Kernel Cake Extract
3.2.2. Solubilised Protein Concentration of Fermented Palm Kernel Cake Extract
3.2.3. Hydrolytic Enzyme Activities of Fermented Palm Kernel Cake Extract
3.2.4. Attachment and Growth of Lactobacillus plantarum Strains on Palm Kernel Cake
4. Materials and Methods
4.1. Microorganisms and Maintenance
4.2. Preparation of Extracellular Hydrolytic Enzymes
4.3. Effect of pH on Extracellular Hydrolytic Enzyme Activities
4.4. Cellulase and Hemicellulase Activities
4.5. Endoproteolytic Activity
4.6. Protein Concentration Determination
4.7. Rating of Extracellular Hydrolytic Enzyme Activities of Lactobacillus plantarum Strains
4.8. Solid State Fermentation of Palm Kernel Cake
4.9. Lactic Acid Bacteria Viable Count of Fermented Palm Kernel Cake Extract
4.10. Scanning Electron Microscopy Analyses of Fermented Palm Kernel Cake
4.11. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| PKC | Palm kernel cake |
| SSF | Solid state fermentation |
| LAB | Lactic acid bacteria |
| GRAS | Generally recognized as safe |
| QPS | Quantified presumption of safety |
| MRS | deMan, Rogosa and Sharpe |
| CFU | Colony forming unit |
| CFS | Cell free supernatant |
| CMCase | Carboxymethylcellulase |
| FPase | Filter paper-ase |
| EB | Enzyme blank |
| SB | Substrate blank |
| RM | Reaction mixture |
| DNS | 3,5-dinitrosalicyclic acid |
| NaOH | Sodium hydroxide |
| dH2O | Deionised water |
| Abs | Absorbance |
| RA | Reference absorbance |
| SEM | Standard error of mean |
| OsO4 | Osmium tetraoxide |
| ANOVA | Analysis of variance |
| L. plantarum | Lactobacillus plantarum |
References
- Garcia-Galindo, I.; Gomez-Garcia, R.; Palacios-Ponce, S.; Ventura, J.; Boone, D.; Ruiz, H.A.; Sepulveda, L.; Sabu, A.; Aguilar-Gonzalez, C.N. Chapter 31—New features and properties of microbial cellulases required for bioconversion of agro-industrial wastes. In Enzymes in Food Biotechnology: Production, Applications and Future Prospects; Mohammed, K., Ed.; Elsevier Inc.: New York, NY, USA, 2019; pp. 535–550. ISBN 978-0-128-13280-7. [Google Scholar]
- Ezjiofor, T.I.N.; Enebaku, U.E.; Ogueke, C. waste to wealth-value recovery from agro-food processing wastes using biotechnology: A review. Br. Biotechnol. J. 2014, 4, 418–481. [Google Scholar] [CrossRef]
- Fischer, C.R.; Klein-Marcuschamer, D.; Stephanopoulos, G. Selection and optimization of microbial hosts for biofuel production. Metab. Eng. 2008, 10, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Anwar, Z.; Gulfaz, M.; Irshad, M. Agro-industrial lignocellulosic biomass a key to unlock the future bio-energy: A brief review. J. Radiat. Res. Appl. Sci. 2014, 7, 163–173. [Google Scholar] [CrossRef]
- Koutinas, A.A.; Vlysidis, A.; Pleissner, D.; Kopsahelis, N.; Garcia, I.L.; Kookos, I.K.; Papanikolaou, S.; Kwan, T.H.; Lin, C.S.K. Valorization of industrial waste and by-product streams via fermentation for the production of chemicals and biopolymers. Chem. Soc. Rev. 2014, 43, 2587–2627. [Google Scholar] [CrossRef] [PubMed]
- FitzPatrick, M.; Champagne, P.; Cunningham, M.F.; Whitney, R.A. A biorefinery processing perspective: Treatment of lignocellulosic materials for the production of value-added products. Bioresour. Technol. 2010, 101, 8915–8922. [Google Scholar] [CrossRef] [PubMed]
- Deswal, D.; Khasa, Y.P.; Kuhad, R.C. Optimization of cellulase production by a brown rot fungus Fomitopsis sp. RCK2010 under solid state fermentation. Bioresour. Technol. 2011, 102, 6065–6072. [Google Scholar] [CrossRef] [PubMed]
- Mahro, B.; Timm, M. Potential of biowaste from the food industry as a biomass resource. Eng. Life Sci. 2007, 7, 457–468. [Google Scholar] [CrossRef]
- Ravinder, T.; Swamy, M.V.; Seenayya, G.; Reddy, G. Clostridium Lentocellum SG6—A potential organism for fermentation of cellulose to acetic acid. Bioresour. Technol. 2001, 80, 171–177. [Google Scholar] [CrossRef]
- Lau, M.W.; Bals, B.D.; Chundawat, S.P.S.; Jin, M.; Gunawan, C.; Balan, V.; Jones, A.D.; Dale, B.E. An integrated paradigm for cellulosic biorefineries: Utilization of lignocellulosic biomass as self-sufficient feedstock for fuel, food precursors and saccharolytic enzyme production. Energy Environ. Sci. 2012, 5, 7100–7110. [Google Scholar] [CrossRef]
- Isikgor, F.H.; Becer, C.R. Lignocellulosic biomass: A sustainable platform for the production of bio-based chemicals and polymers. Polym. Chem. 2015, 6, 4497–4559. [Google Scholar] [CrossRef]
- Srivastava, N.; Srivastava, M.; Mishra, P.K.; Gupta, V.K.; Molina, G.; Rodriguez-Couto, S.; Manikanta, A.; Ramteke, P.W. Applications of fungal cellulases in biofuel production: Advances and limitations. Renew. Sustain. Energy Rev. 2018, 82, 2379–2386. [Google Scholar] [CrossRef]
- Wilson, D.B. Cellulases and biofuels. Curr. Opin. Biotechnol. 2009, 20, 295–299. [Google Scholar] [CrossRef] [PubMed]
- Artzi, L.; Bayer, E.A.; Morais, S. Cellulosomes: Bacteria nanomachines for dismantling plant polysaccharides. Nat. Rev. Microbiol. 2017, 15, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Fontes, C.M.; Gilbert, H.J. Cellulosomes: Highly efficient nanomachines designed to deconstruct plant cell wall complex carbohydrates. Annu. Rev. Biochem. 2010, 79, 655–681. [Google Scholar] [CrossRef] [PubMed]
- Berghem, L.E.R.; Pettersson, L.G. The mechanism of enzymatic cellulosic degradation: Purification of a cellulolytic enzyme from Trichoderma viride active on highly ordered cellulose. Eur. J. Biochem. 1973, 37, 21–30. [Google Scholar] [CrossRef]
- Rahmana, N.; Shah, U.K.M.; Foo, H.L.; Rahman, N.A.A.; Ariff, A.B. Production and characterisation of cellulase from solid state fermentation of rice straw by Trichoderma harizianum SNRS3. Pertanika J. Trop. Agric. Sci. 2016, 39, 507–531. [Google Scholar]
- Rahnama, N.; Foo, H.L.; Abdul Rahman, N.A.; Ariff, A.; Shah, U.K.M. Saccharification of rice straw by cellulase from local Trichoderma harizianum SNRS3 for biobutanol production. BMC Biotechnol. 2014, 14, 1–12. [Google Scholar] [CrossRef]
- Gao, J.; Weng, H.; Zhu, D.; Yuan, M.; Guan, F.; Xi, Y. Production and characterization of cellulolytic enzymes from the thermoacidophilic fungal Aspergillus terreus M11 under solid-state cultivation of corn stover. Bioresour. Technol. 2008, 99, 7623–7629. [Google Scholar] [CrossRef]
- Renganathan, V.; Usha, S.N.; Flordeliz, L. Cellobiose-oxidizing enzymes from the lignocellulose-degrading basidiomycete Phanerochaete chrysosprium: Interaction with microcrystalline cellulose. Appl. Microbiol. Biotechnol. 1990, 32, 609–613. [Google Scholar] [CrossRef]
- Uzcategui, E.; Raices, M.; Montesino, R.; Johansson, G.; Pettersson, G.; Eriksson, K.E. Pilot-scale production and purification of the cellulolytic enzyme system from the white-rot fungus Phanerochaete chrysosporium. Biotechnol. Appl. Biochem. 1991. Available online: http://agris.fao.org/agris-search/search.do?recordID=US9156534 (accessed on 20 June 2019).
- Robson, L.M.; Chambliss, G.H. Characterization of the cellulolytic activity of a Bacillus isolate. Appl. Environ. Microbiol. 1984, 47, 1039–1046. [Google Scholar] [PubMed]
- Shinmyo, A.; Garcia-Martinez, D.V.; Demain, A.L. Studies on the extracellular cellulolytic enzyme complex produced by Clostridium thermocellum. J. Appl. Biochem. 1979, 1, 202–209. [Google Scholar]
- Kumar, M.; Revathi, K.; Khanna, S. Biodegradation of cellulosic and lignocellulosic waste by Pseudoxanthomonas sp. R-28. Carbohydr. Polym. 2015, 134, 761–766. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Sheng, P.; Zhang, H. Isolation and identification of cellulolytic bacteria from the gut of Holotrichia parallela larvae (Coleoptera: Scarabaeidae). Int. J. Mol. Sci. 2012, 13, 2563–2577. [Google Scholar] [CrossRef] [PubMed]
- Mudasir, A.D.; Kiran, D.P.; Jyoti, P.J.; Radhakrishna, S.P. Isolation of cellulolytic bacteria from the gastrointestinal tract of Achatina fulica (Gastropoda: Pulmonata) and their evaluation for cellulose biodegradation. Int. Biodeterior. Biodegrad. 2015, 98, 73–80. [Google Scholar] [CrossRef]
- Waghmare, P.R.; Krhirsagar, S.D.; Saratale, R.G.; Govindwar, P.; Saratale, G.D. Production and characterization of cellulolytic enzymes by isolated Klebsiella sp. PRW-1 using agricultural waste biomass. Emir. J. Food. Agric. 2014, 26, 44–59. [Google Scholar] [CrossRef]
- Rickard, P.A.D.; Laughlin, T.A. Detection and assay of xylanolytic enzymes in a Cellulomonas isolate. Biotechnol. Lett. 1980, 2, 363–368. [Google Scholar] [CrossRef]
- Langsford, M.L.; Gilkes, N.R.; Wakarchuk, W.W.; Kilburn, D.G.; Miller, R.C., Jr.; Warren, A.J. The cellulase system of Cellulomonas fimi. J. Gen. Microbiol. 1984, 130, 1367–1376. [Google Scholar] [CrossRef]
- Lo, Y.C.; Lu, W.C.; Chen, C.Y.; Chen, W.M.; Chang, J.S. Characterization and high-level production of xylanase from an indigenous cellulolytic bacterium Acinetobacter junii F6-02 from southern Taiwan soil. Biochem. Eng. J. 2010, 53, 77–84. [Google Scholar] [CrossRef]
- Hazlewood, G.P.; Laurie, J.I.; Ferreira, L.M.A.; Gilbert, H.J. Pseudomonas fluorescens subsp. cellulosa: An alternative model for bacterial cellulase. J. Appl. Bacteriol. 1992, 72, 244–251. [Google Scholar] [CrossRef]
- Yadav, J.S.S.; Bezawada, J.; Ajila, C.M.; Yan, S.; Tyagi, R.D.; Surampalli, R.Y. Mixed culture of Kluyveromyces marxianus and Candida krusei for single-cell protein production and organic load removal from whey. Bioresour. Technol. 2014, 164, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Loubna, E.F.; Mohamed, Z.; Abdelghani, E.A.; Mohamed, H. Assessment of biotransformation of organic matter during co-composting of sewage sludge-lignocellulosic waste by chemical, FTIR analyses, and phytotoxicity tests. Int. Biodeterior. Biodegrad. 2014, 87, 128–137. [Google Scholar] [CrossRef]
- Man, L.H.; Behera, S.K.; Park, H.S. Optimization of operation parameters for ethanol production from Korean food waste leachate. Int. J. Environ. Sci. Technol. 2010, 7, 157–164. [Google Scholar] [CrossRef]
- Philippoussis, A.; Diamantopoulou, P. Agro-food industry wastes and agricultural residues conversion into high value products by mushroom cultivation. In Proceedings of the 7th International Conference on Mushroom Biology and Mushroom Products, Arcachon, France, 4–7 October 2011; Available online: https://www.cabdirect.org/cabdirect/abstract/20123168993 (accessed on 26 June 2019).
- Van Beilen, J.B.; Li, Z. Enzyme technology: An overview. Curr. Opin. Biotechnol. 2002, 13, 338–344. [Google Scholar] [CrossRef]
- Satinder, K.B.; Gurpreet, S.D.; Carlos, R.S. Biotransformation of Waste Biomass into High Value Biochemicals; Springer Science & Business Media: New York, NY, USA, 2013. [Google Scholar]
- Bisaria, V.S.; Ghose, T.K. Biodegradation of cellulosic materials: Substrates, microorganisms, enzymes and products. Enzyme Microb. Technol. 1981, 3, 90–104. [Google Scholar] [CrossRef]
- De Silva, F.L.; de Oliveira Compos, A.; dos Santos, D.A.; de Oliveria, S.D., Jr.; de Arauja Padilha, C.E.; de Sousa, F.C., Jr.; de Macedo, G.R.; dos Santos, E.S. Pretreatments of Carnauba (Copernicia prunifera) straw residue for production of cellulolytic enzymes by Trichorderma reesei CCT-2768 by solid state fermentation. Renew. Energy 2018, 116, 299–308. [Google Scholar] [CrossRef]
- Saxena, R.; Sigh, R. Amylase production by solid-state fermentation of agro industrial wastes using Bacillus sp. Braz. J. Microbiol. 2011, 42, 1334–1342. [Google Scholar] [CrossRef]
- Joshi, V.K.; Sandu, D.K. Preparation and evaluation of an animal feed byproduct produced by solid-state fermentation of apple pomace. Bioresour. Technol. 1996, 56, 251–255. [Google Scholar] [CrossRef]
- Villas-Boas, S.G.; Esposito, E.; Mitchell, D.A. Microbial conversion of lignocellulosic residues for production of animal feeds. Anim. Feed Sci. Technol. 2002, 98, 1–12. [Google Scholar] [CrossRef]
- Bartkiene, E.; Kruogleviciute, V.; Juodeikiene, G.; Vidmantiene, D.; Maknickiene, Z. Solid state fermentation with lactic acid bacteria to improve the nutritional quality of lupin and soya bean. J. Sci. Food Agric. 2015, 95, 1336–1342. [Google Scholar] [CrossRef]
- Godoy, M.G.; Amorim, G.M.; Barreto, M.S.; Freire, D.M.G. Chapter 12- Agricultural residues as animal feed: Protein enrichment and detoxification using solid-state fermentation. In Current Developments in Biotechnology and Bioengineering: Current Advances in Solid-State Fermentation; Pandey, A., Larroche, C., Soccol, C., Eds.; Elseiver Inc.: New York, YN, USA, 2018; pp. 235–256. ISBN 978-0-444-63990-5. [Google Scholar]
- Alshelmani, M.I.; Loh, T.C.; Foo, H.L.; Sazili, A.Q.; Lau, W.H. Effect of solid state fermentation on nutrient content and ileal amino acids digestibility of palm kernel cake in broiler chickens. Indian J. Anim. Sci. 2017, 87, 1135–1140. [Google Scholar]
- Suhartatik, N.; Cahyanto, M.N.; Rahardjo, S.; Miyashita, M.; Rahayu, E.S. Isolation and identification of lactic acid bacteria producing β-glucosidase from Indonesian fermented foods. Int. Food Res. J. 2014, 21, 973–978. [Google Scholar]
- Gueguen, Y.; Chemardin, P.; Labrot, P.; Arnaud, A.; Galzy, P. Purification and characterization of an intracellular β-glucosidase from a new strain of Leuconostoc mesenteroides isolated from cassava. J. Appl. Microbiol. 1997, 82, 469–476. [Google Scholar] [CrossRef]
- Nadaroglu, H.; Adiguzel, A.; Adiguzel, G. Purification and characterisation of β-mannanase from Lactobacillus plantarum (M24) and its application in some fruit juices. Int. J. Food Sci. Technol. 2015, 50, 1158–1165. [Google Scholar] [CrossRef]
- Oda, Y.; Komaki, T.; Tonomura, K. Production of β-mannanase and β-mannosidase by Enterococcus casseliflavus FL2121 from decayed Konjac. Food Microbiol. 1993, 10, 353–358. [Google Scholar] [CrossRef]
- Loh, T.C.; Lee, T.M.; Foo, H.L.; Law, F.L.; Rajion, M.A. Growth performance and fecal microflora of rats offered metabolites from lactic acid bacteria. J. Appl. Anim. Res. 2008, 34, 61–64. [Google Scholar] [CrossRef]
- Loh, T.C.; Choe, D.W.; Foo, H.L.; Awis, Q.S.; Hair-Bejo, M. Effects of feeding different postbiotic metabolite combinations produced by Lactobacillus plantarum strains on egg quality and production performance, faecal parameters and plasma cholesterol in laying hens. BMC Vet. Res. 2014, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kareem, K.Y.; Loh, T.C.; Foo, H.L.; Asmara, S.A.; Akit, H.; Abdulla, N.R.; Ooi, M.F. Carcass, meat and bone quality of broiler chickens fed with postbiotic and prebiotic combinations. Int. J. Probiotic Prebiotic 2015, 10, 23–30. [Google Scholar]
- Loh, T.C.; Chong, S.W.; Foo, H.L.; Law, F.L. Effects on growth performance, faecal microflora and plasma cholesterol after supplementation of spray-dried metabolite to postweaning rats. Czech J. Anim. Sci. 2009, 54, 10–16. [Google Scholar] [CrossRef] [Green Version]
- Kareem, K.Y.; Loh, T.C.; Foo, H.L.; Henny, A.; Samsudin, A.A. Effects of dietary postbiotic and inulin on growth performance, IGF1 and GHR mRNA expression, faecal microbiota and volatile fatty acids in broilers. BMC Vet. Res. 2016, 12, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Alshelmani, M.I.; Loh, T.C.; Foo, H.L.; Lau, W.H.; Sazili, A.Q. Characterisation of cellulolytic bacterial cultures grown in different substrates. Sci. World. J. 2013, 689235, 1–6. [Google Scholar] [CrossRef]
- Ahmed, A.; Zulkifli, I.; Farjam, A.S.; Abdullah, N.; Liang, J.B.; Awad, E.A. Effect of solid state fermentation on nutrient content and ileal amino acids digestibility of canola meal in broiler chickens. Ital. J. Anim. Sci. 2014, 13, 410–414. [Google Scholar] [CrossRef]
- Alshelmani, M.I.; Loh, T.C.; Foo, H.L.; Sazili, A.Q.; Lau, W.H. Effect of feeding different levels of palm kernel cake fermented by Paenibacillus polymyxa ATCC 842 on nutrient digestibility, intestinal morphology, and gut microflora in broiler chickens. Anim. Feed Sci. Technol. 2016, 216, 216–224. [Google Scholar] [CrossRef]
- Alshelmani, M.I.; Loh, T.C.; Foo, H.L.; Sazili, A.Q.; Lau, W.H. Effect of feeding different levels of palm kernel cake fermented by Paenibacillus polymyxa ATCC 842 on broiler growth performance, blood biochemistry, carcass characteristics, and meat quality. Anim. Prod. Sci. 2017, 57, 839–848. [Google Scholar] [CrossRef]
- Zamani, H.U.; Loh, T.C.; Foo, H.L.; Samsudin, A.A.; Alshelmani, M.I. Effects of feeding palm kernel cake with crude enzyme supplementation on growth performance and meat quality of broiler chicken. Int. J. Microbiol. Biotechnol. 2017, 2, 22–28. [Google Scholar]
- Kareem, K.Y.; Foo, H.L.; Loh, T.C.; Ooi, M.F.; Samsudin, A.A. Inhibitory activity of postbiotic produced by strains of Lactobacillus plantarum using reconstituted media supplemented with inulin. Gut Pathog. 2014, 6, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Graminha, E.B.N.; Gonclaves, A.Z.L.; Pirota, R.D.P.B.; Balsalobre, M.A.A.; da Silva, R.; Gomes, E. Enzyme production by solid state fermentation: Application to animal production. Anim. Feed Sci. Technol. 2008, 144, 1–22. [Google Scholar] [CrossRef]
- Shrivastava, B.; Jain, K.K.; Kalra, A.; Kuhad, R.C. Bioprocessing of wheat straw into nutritionally rich and digested cattle feed. Sci. Rep. 2014, 4, 1–8. [Google Scholar] [CrossRef]
- Roger, D.D.; Jean-Justin, E.N.; Francois-Xavier, E. Cassava solid-state fermentation with a starter culture of Lactobacillus plantarum and Rhyzopus oryzae for cellulase production. Afr. J. Microbiol. 2011, 5, 4866–4872. [Google Scholar] [CrossRef]
- Hong, L.S.; Ibrahim, D.; Omar, I.C. Lignocellulolytic materials as a raw material for the production of fermented sugar via solid state fermentation. Asian J. Sci. Res. 2011, 4, 53–61. [Google Scholar] [CrossRef]
- Reddy, G.P.K.; Narasimha, G.; Kumar, K.D.; Ramanjaneyulu, G.; Ramya, A.; Kumari, B.S.S.; Reddy, B.R. Cellulase production by Aspergillus niger on different natural lignocellulosic substrates. Int. J. Curr. Microbiol. Appl. Sci. 2015, 4, 835–845. [Google Scholar]
- Lee, Y.J.; Kim, B.K.; Lee, B.H.; Jo, K.I.; Lee, N.K.; Chung, C.H.; Lee, Y.C.; Lee, J.W. Purification and characterisation of cellulase produced by Bacillus amyoliquefacien DL-3 utilising rice hull. Bioresour. Technol. 2008, 99, 378–386. [Google Scholar] [CrossRef] [PubMed]
- Waeonukul, R.; Kyu, K.L.; Sakka, K.; Ratanakhankchai, K. Isolation and characterization of a multienzyme complex (cellulosome) of the Paenibacillus curdlanolyticus B-6 grown on Avicel under aerobic condition. J. Biosci. Bioeng. 2009, 107, 610–614. [Google Scholar] [CrossRef] [PubMed]
- Nidetzky, B.; Steiner, W.; Hayn, M.; Claeyssens, M. Cellulose hydrolysis by the cellulases from Trichoderma reesei: A new model for synergistic interaction. Biochem. J. 1994, 298, 705–710. [Google Scholar] [CrossRef]
- Eriksson, K.E. Enzyme mechanisms involved in cellulose hydrolysis by the rot fungus Sporotrichum pulverulentum. Biotechnol. Bioeng. 1978, 20, 317–332. [Google Scholar] [CrossRef]
- Lin, L.; Kan, X.; Yan, H.; Wang, D. Characterization of extracellular cellulose-degrading enzymes from Bacillus thringiensis strains. Electron. J. Biotechnol. 2012, 15, 1–7. [Google Scholar] [CrossRef]
- Hutkins, R.W.; Nannen, N.L. pH homeostasis in lactic acid bacteria. J. Dairy Sci. 1993, 76, 2354–2365. [Google Scholar] [CrossRef]
- Michlmayr, H.; Kneifel, W. β-glucosidase activities of lactic acid bacteria: Mechanisms, impact on fermented food and human health. FEMS Microbiol. Lett. 2014, 352, 1–10. [Google Scholar] [CrossRef]
- Matthews, A.; Grimaldi, A.; Walker, M.; Bartowsky, E.; Grbin, P.; Jiranek, V. Lactic acid bacteria as potential source of enzymes for use in vinification. Appl. Environ. Microbiol. 2004, 70, 5715–5731. [Google Scholar] [CrossRef]
- Araki, T.; Inoue, N.; Morishita, T. Purification and characterization of β-1,3-xylanase from a marine bacterium, Alcaligene sp. XY-234. J. Gen. Appl. Microbiol. 1998, 44, 269–274. [Google Scholar] [CrossRef]
- Gilbert, H.J.; Hazlewood, G.P. Bacterial cellulases and xylanases. J. Gen. Microbiol. 1993, 139, 187–194. [Google Scholar] [CrossRef] [Green Version]
- Beg, Q.K.; Kapoor, M.; Mahajan, L.; Hoondal, G.S. Microbial xylanases and their industrial applications: A review. Appl. Microbiol. Biotechnol. 2001, 56, 326–338. [Google Scholar] [CrossRef] [PubMed]
- Scheirlinck, T.; De Meutter, J.; Arnaut, G.; Joos, H.; Claeyssens, M.; Michiels, F. Cloning and expression of cellulase and xylanase genes in Lactobacillus plantarum. Appl. Microbiol. Biotechnol. 1990, 33, 534–541. [Google Scholar] [CrossRef]
- Mawadza, C.; Hatti-Kaul, R.; Zvauyam, R.; Mattiasson, B. Purification and characterisation of cellulases produced by two Bacillus strains. J. Biotechnol. 2000, 83, 177–187. [Google Scholar] [CrossRef]
- Yasemin, C.A.F.; Ebrahim, V.; Burhan, A. Study on cold-activee and acidophilic cellulase (CMCase) from a novel psychrotropic isolat Bacillus sp. K-11. Int. J. Curr. Microbiol. Appl. Sci. 2014, 3, 16–25. [Google Scholar]
- Khasin, A.; Alchanati, I.; Shoham, Y. Purification and characterisation of a thermostable xylanase from Bacillus stearothermophilus T-6. Appl. Environ. Microbiol. 1993, 59, 1725–1730. [Google Scholar] [PubMed]
- Nakamura, S.; Wakabayashi, K.; Nakai, R.; Aono, R.; Horikoshi, K. Purification and some properties of an alkaline xylanase from alkaliphilic Bacillus sp. strain 41M-1. Appl. Environ. Microbiol. 1993, 59, 2311–2316. [Google Scholar] [PubMed]
- Talbot, G.; Sygusch, J. Purification and characterisation of thermostable β-mannanase and α-galactosidase from Bacillus stearothermophilus. Appl. Environ. Microbiol. 1990, 56, 3505–3510. [Google Scholar]
- Law, J.; Haandrikman, A. Proteolytic enzymes of lactic acid bacteria. Int. Dairy J. 1997, 7, 1–11. [Google Scholar] [CrossRef]
- Christensen, J.E.; Dudley, E.G.; Pederson, J.A.; Steele, J.L. Peptidases and amino acid catabolism in lactic acid bacteria. Antonie Van Leeuwenhoek 1999, 76, 217–246. [Google Scholar] [CrossRef] [PubMed]
- Toe, C.J.; Foo, H.L.; Loh, T.C.; Mohamad, R.; Abdul Rahim, R.; Idrus, Z. Extracellular proteolytic activity and amino acid production by lactic acid bacteria isolated from Malaysian foods. Int. J. Mol. Sci. 2019, 20, 1777. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.H.; Foo, H.L.; Loh, T.C.; Mohamad1, R.; Abdullah, N. Comparative studies of versatile extracellular proteolytic activities of lactic acid bacteria and their potential for extracellular amino acid productions as feed supplements. J. Anim. Sci. Biotechnol. 2019, 10, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muck, R.E.; Nadeau, E.M.G.; McAllister, T.A.; Contreias-Govea, F.E.; Santos, M.C.; Kung, L., Jr. Silage review: Recent advances and future use of silage addictives. J. Dairy Sci. 2018, 101, 3980–4000. [Google Scholar] [CrossRef]
- Lara, E.C.; Bragiato, U.C.; Rabelo, H.S.; Messana, J.D.; Reis, R.A. Inoculation of corn silage with Lactobacillus plantarum and Bacillus subtilis associated with amylolytic enzyme supply at feeding. 1. feed intake, apparent digestibility, and microbial protein synthesis in wethers. Anim. Feed Sci. Technol. 2018, 243, 22–34. [Google Scholar] [CrossRef]
- Fan, S.P.; Jiang, L.Q.; Chia, C.H.; Fang, Z.; Zakaria, S.; Chee, K.L. High yield production of sugar from deproteinated palm kernel cake under microwave irradiation via diluted sulfuric acid hydrolysis. Bioresour. Technol. 2014, 153, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Espejo, J.; Alcala, M.; Esteban, M.A.; Gomez, R.; Slik, S. Influence of sodium chloride concentration on the growth and survival of microorganisms isolated from Cabrales cheese. Microbiol. Aliment. Nutr. J. 1994, 12, 251–254. [Google Scholar]
- Catte, M.; Gancel, F.; Dzierszinski, F.; Tailliez, R. Effect of water activity, NaCl and smoke concentrations on the growth of Lactobacillus plantarum ATCC 12315. Int. J. Food Microbiol. 1999, 52, 105–108. [Google Scholar] [CrossRef]
- Zakaria, Z.; Hall, G.M.; Shama, G. Lactic acid fermentation of scampi waste in a rotating horizontal bioreactor for chitin recovery. Process Biochem. 1998, 33, 1–6. [Google Scholar] [CrossRef]
- Alshelmani, M.I.; Loh, T.C.; Foo, H.L.; Lau, W.H.; Sazili, A.Q. Biodegradation of palm kernel cake by cellulolytic and hemicellulolytic bacterial culture through solid state fermentation. Sci. World J. 2014, 759852, 1–8. [Google Scholar] [CrossRef]
- Shukor, H.; Abdeshahian, P.; Al-Shorgani, N.K.N.; Hamid, A.A.; Rahman, N.A.; Kalil, M.S. Enhanced mannan-derived fermentable sugars of palm kernel cake by mannanase-catalyzed hydrolysis for production of biobutanol. Bioresour. Technol. 2016, 218, 257–264. [Google Scholar] [CrossRef]
- Kalidas, N.R.; Saminathan, M.; Ismail, I.S.; Abas, F.; Maity, P.; Islam, S.S.; Manshoor, N.; Shaari, K. Structural characterization and evaluation of prebiotic activity of oil palm kernel cake mannoligosaccharides. Food Chem. 2017, 234, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Shastak, Y.; Ader, P.; Feuerstein, D.; Ruehle, R.; Matuschek, M. β-mannan and mannanase in poultry nutrition. World Poult. Sci. 2015, 71, 161–174. [Google Scholar] [CrossRef]
- Chen, J.; Tellez, G.; Richards, J.D.; Escobar, J. Identification of potential biomarkers for gut barrier failure in broiler chickens. Front. Vet. Sci. 2015, 2, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.H.; Chou, L.M.; Chien, Y.W.; Chang, J.S.; Lin, C.I. Prebiotic effects of xylooligosaccharides on the improvement of microbiota balance in human subjects. Gastroenterol. Res. Pract. 2016, 5789232, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Samanta, A.K.; Jayapal, N.; Jayaram, C.; Roy, S.; Kolte, A.P.; Senani, S.; Sridhar, M. Xylooligosaccharides as prebiotics from agricultural by-products: Production and applications. Bioact. Carbohydr. Diet. Fibre 2015, 5, 62–71. [Google Scholar] [CrossRef]
- Jain, I.; Kumar, V.; Satyanarayana, T. Xylooligosaccharides: An economical prebiotic from agroresidues and their health benefits. Indian J. Exp. Biol. 2015, 53, 131–142. [Google Scholar] [PubMed]
- De Maesschalck, C.D.; Eeckhaut, V.; Maertens, L.; de Lange, L.; Marchal, L.; Nezer, C.; de Baere, S.; Croubels, S.; Daube, G.; Dewulf, J.; et al. Effects of xylo-oligosaccharides on broiler chicken performance and microbiota. Appl. Environ. Microbiol. 2015, 81, 5880–5888. [Google Scholar] [CrossRef] [PubMed]
- Ezieshi, E.V.; Olomu, J.M. Nutritional evaluation of palm kernel meal types: 1. proximate composition and metabolizable energy values. Afr. J. Biotechnol. 2007, 6, 2484–2486. [Google Scholar] [CrossRef]
- Ramachandran, S.; Singh, S.K.; Larroche, C.; Soccol, C.R.; Pandey, A. Oil cakes and their biotechnological applications—A review. Bioresour. Technol. 2007, 98, 2000–2009. [Google Scholar] [CrossRef] [PubMed]
- Abdollahi, R.; Hosking, B.; Ravindran, V. Nutrient analysis, metabolisable energy and ileal amino acid digestibility of palm kernel meal for broilers. Anim. Feed Sci. Technol. 2015, 206, 119–125. [Google Scholar] [CrossRef]
- Leh, M.B.; Charles, M. The effect of whey protein hydrolyzate average molecular weight on the lactic acid fermentation. J. Ind. Microbiol. 1989, 4, 77–80. [Google Scholar] [CrossRef]
- Fauld, C.B.; Collins, S.; Robertson, J.A.; Treimo, J.; Eijsink, V.G.H.; Hinz, S.W.A.; Schols, H.A.; Buchert, J.; Waldron, K.W. Protease-induced solubilisation of carbohydrate from brewers’ spent grain. J. Cereal Sci. 2009, 50, 332–336. [Google Scholar] [CrossRef]
- Saw, H.Y.; Januan, J.; Kumaresan, S.; Chu, C.M. Characterization of the physical properties of palm kernel cake. Int. J. Food Prop. 2012, 15, 536–548. [Google Scholar] [CrossRef]
- Kawarai, T.; Fukukawa, S.; Ogihara, H.; Yamasaki, M. Mixed-species biofilm formation by lactic acid bacteria and rice wine yeasts. Appl. Envion. Microbiol. 2007, 73, 4673–4676. [Google Scholar] [CrossRef] [PubMed]
- Kubota, H.; Senda, S.; Nomura, N.; Tokuda, H.; Uchimaya, H. Biofilm formation by lactic acid bacteria and resistance to environmental stress. J. Biosci. Bioeng. 2008, 106, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.M.; Xie, L.S.; Zhang, W.Y.; Zhou, W.Y.; Su, J.Y.; Liu, J.C. The association of biofilm formation with antibiotic resistance in lactic acid bacteria from fermented food. J. Food Saf. 2013, 33, 114–120. [Google Scholar] [CrossRef]
- Somers, E.B.; Johnson, M.E.; Wong, A.C. Biofilm formation and contamination of cheese by nonstarter lactic acid bacteria in the dairy environment. J. Dairy Sci. 2001, 84, 1926–1936. [Google Scholar] [CrossRef]
- Jalilsood, T.; Baradaran, A.; Song, A.A.L.; Foo, H.L.; Mustafa, S.; Saad, W.Z.; Yusoff, K.; Rahim, R.A. Inhibition of pathogenic and spoilage bacteria by a novel biofilming-forming Lactobacillus isolate: A potential host for the expression of heterologous proteins. Microb. Cell Fact. 2015, 14, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Phitsuwan, P.; Morag, E.; Tachaapaikoon, C.; Pason, P.; Kosugi, A.; Ratanakhanokchai, K. Behavior and supportive evidence of a large xylanase-containing multienzyme complex of Tepidimicrobium xylanilyticum BT14. Bioresour. Technol. 2012, 7, 5934–5949. [Google Scholar] [CrossRef]
- Kenyon, W.J.; Esch, S.W.; Buller, C.S. The curdlan-type exopolysaccharide produced by Cellulomonas flavigena KU forms part of an extracellular glycocalyx involved in cellulose degradation. Antonie Van Leeuwenhoek 2005, 87, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.A. Purification and Characterisation of Bacteriocin Produced by Lactococcus Lactis Subsp. Lactis RW18 From Steamed Fish (Rastrelliger sp.). Master’s Thesis, Universiti Putra Malaysia, Selangor, Malaysia, 2002. [Google Scholar]
- Lim, Y.S. Isolation of Bacteriogenic Lactic acid Bacteria and Purification of Selected Bacteriocins from Traditional Fermented Foods. Master’s Thesis, Universiti Putra Malaysia, Selangor, Malaysia, 2003. [Google Scholar]
- Thung, T.Y. Isolation and Purification of Proteolytic Enzyme Produced by Lactic Acid Bacteria From Budu and Bambangan. Master’s Thesis, Universiti Putra Malaysia, Selangor, Malaysia, 2012. [Google Scholar]
- Foo, H.L.; Loh, T.C.; Lai, P.W.; Lim, Y.S.; Kufli, C.N.; Gulam, R. Effects of adding Lactobacillus plantarum I-UL4 metabolites in drinking water of rats. Pak. J. Nutr. 2003, 2, 283–288. [Google Scholar] [CrossRef]
- Moghadam, M.S.; Foo, H.L.; Leow, T.C.; Rahim, R.A.; Loh, T.C. Novel bacteriocinogenic Lactobacillus plantarum strains and their differentiation by sequence analysis of 16S rDNA, 16S-5S intergenic spacer region and randomly amplied polymorphic DNA analysis. Food Technol. Biotechnol. 2010, 48, 476–483. [Google Scholar]
- Ghose, T.K. Measurement of cellulase activities. Pure Appl. Chem. 1987, 59, 257–268. [Google Scholar] [CrossRef]
- Yin, L.J.; Lin, H.H.; Xiao, Z.R. Purification and characterization of a cellulase from Bacillus subtilis YJ1. J. Mar. Sci. Technol. 2010, 18, 466–471. [Google Scholar]
- Gowdhaman, D.; Manaswini, V.S.; Jayanthi, V.; Dhanasri, M.; Jeyalakshimi, G.; Gunasekar, V.; Sugumaran, K.R.; Ponnusami, V. Xylanase production from Bacillus aerophilus KGJ2 and its application in xylooligosaccharides preparation. Int. J. Biol. Macromol. 2014, 64, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.L. Use of dinitrosalicyclic acid agent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Miller, G.L. Protein determination of Large Numbers of Samples. Anal. Chem. 1959, 31, 964. [Google Scholar] [CrossRef]
| Days | Reducing Sugar Concentration (mg/mL) | |||
|---|---|---|---|---|
| Control | R111 | RG11 | RG14 | |
| Day 0 | 0.49cA | 0.68aA | 0.59aA | 0.61aA |
| Day 2 | 0.61bcA | 0.44bB | 0.19dC | 0.23bcC |
| Day 4 | 0.62bcA | 0.24cdB | 0.21cdB | 0.21cB |
| Day 6 | 0.67abcA | 0.21cdB | 0.29bcB | 0.24bcB |
| Day 8 | 0.70abcA | 0.16dC | 0.28bcB | 0.21cBC |
| Day 10 | 0.73abcA | 0.24cdBC | 0.25bcdB | 0.20cC |
| Day 12 | 0.79abA | 0.24cdB | 0.30bB | 0.23bcB |
| Day 14 | 0.93aA | 0.27cB | 0.30bB | 0.31bB |
| SEM ± | 0.05 | 0.06 | 0.04 | 0.05 |
| Days | Solubilized Protein Concentration (mg/mL) | |||
|---|---|---|---|---|
| Control | R111 | RG11 | RG14 | |
| Day 0 | 0.95bA | 0.99aA | 0.95aA | 0.94aA |
| Day 2 | 0.98bA | 1.00aA | 0.86aA | 0.92aA |
| Day 4 | 1.03abA | 0.93aA | 0.93aA | 0.91aA |
| Day 6 | 1.09abA | 1.00aA | 1.05aA | 0.99aA |
| Day 8 | 1.18abA | 1.00aA | 1.05aA | 1.03aA |
| Day 10 | 1.20abA | 1.00aA | 1.05aA | 1.00aA |
| Day 12 | 1.17abA | 1.02aA | 1.05aA | 0.98aA |
| Day 14 | 1.27aA | 1.07aA | 1.10aA | 1.11aA |
| SEM ± | 0.04 | 0.01 | 0.03 | 0.02 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, F.H.; Wan, S.Y.; Foo, H.L.; Loh, T.C.; Mohamad, R.; Abdul Rahim, R.; Idrus, Z. Comparative Study of Extracellular Proteolytic, Cellulolytic, and Hemicellulolytic Enzyme Activities and Biotransformation of Palm Kernel Cake Biomass by Lactic Acid Bacteria Isolated from Malaysian Foods. Int. J. Mol. Sci. 2019, 20, 4979. https://doi.org/10.3390/ijms20204979
Lee FH, Wan SY, Foo HL, Loh TC, Mohamad R, Abdul Rahim R, Idrus Z. Comparative Study of Extracellular Proteolytic, Cellulolytic, and Hemicellulolytic Enzyme Activities and Biotransformation of Palm Kernel Cake Biomass by Lactic Acid Bacteria Isolated from Malaysian Foods. International Journal of Molecular Sciences. 2019; 20(20):4979. https://doi.org/10.3390/ijms20204979
Chicago/Turabian StyleLee, Fu Haw, Suet Ying Wan, Hooi Ling Foo, Teck Chwen Loh, Rosfarizan Mohamad, Raha Abdul Rahim, and Zulkifli Idrus. 2019. "Comparative Study of Extracellular Proteolytic, Cellulolytic, and Hemicellulolytic Enzyme Activities and Biotransformation of Palm Kernel Cake Biomass by Lactic Acid Bacteria Isolated from Malaysian Foods" International Journal of Molecular Sciences 20, no. 20: 4979. https://doi.org/10.3390/ijms20204979
APA StyleLee, F. H., Wan, S. Y., Foo, H. L., Loh, T. C., Mohamad, R., Abdul Rahim, R., & Idrus, Z. (2019). Comparative Study of Extracellular Proteolytic, Cellulolytic, and Hemicellulolytic Enzyme Activities and Biotransformation of Palm Kernel Cake Biomass by Lactic Acid Bacteria Isolated from Malaysian Foods. International Journal of Molecular Sciences, 20(20), 4979. https://doi.org/10.3390/ijms20204979





