Efficient Selection of Antibodies Reactive to Homologous Epitopes on Human and Mouse Hepatocyte Growth Factors by Next-Generation Sequencing-Based Analysis of the B Cell Repertoire
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Chicken Immunization
4.2. Preparation of cDNA
4.3. NGS Analysis
4.4. Preprocessing
4.5. Sequence Distance Matrix Calculation
4.6. Component-Based Analysis
4.7. Phage Display of the Combinatorial scFv Library and Bio-Panning
4.8. High Throughput Retrieval of scFv-Clones
4.9. Phage ELISA
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| HGF | hepatocyte growth factor |
| NGS | next-generation sequencing |
| PBMC | peripheral blood mononuclear cell |
| LD | Levenshtein distance |
| CDR | complementary determining region |
References
- Schmidt, C.; Bladt, F.; Goedecke, S.; Brinkmann, V.; Zschiesche, W.; Sharpe, M.; Gherardi, E.; Birchmeier, C. Scatter factor/hepatocyte growth factor is essential for liver development. Nature 1995, 373, 699–702. [Google Scholar] [CrossRef] [PubMed]
- Tokunou, M.; Niki, T.; Eguchi, K.; Iba, S.; Tsuda, H.; Yamada, T.; Matsuno, Y.; Kondo, H.; Imamura, H.; Hirohashi, S. c-MET expression in myofibroblasts: Role in autocrine activation and prognostic significance in adenocarcinoma. Am. J. Pathol. 2001, 158, 1451–1463. [Google Scholar] [CrossRef]
- Mo, H.N.; Liu, P. Targeting MET in cancer therapy. Chronic. Dis. Transl. Med. 2017, 3, 148–153. [Google Scholar] [CrossRef] [PubMed]
- Lokker, N.A.; Mark, M.R.; Luis, E.A.; Bennett, G.L.; Robbins, K.A.; Baker, J.B.; Gowdowski, P.J. Structure-function analysis of hepatocyte growth factor: Identification of variants that lack mitogenic activity yet retain high affinity receptor binding. EMBO J. 1992, 11, 2503–2510. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Kim, H. Progress of antibody-based inhibitors of the HGF-cMET axis in cancer therapy. Exp. Mol. Med. 2017, 49, e307. [Google Scholar] [CrossRef] [PubMed]
- Birchmeier, C.; Birchmeier, W.; Gherardi, E.; Vande Woudge, G.F. Met, metastasis, motility and more. Nat. Rev. Mol. Cell Biol. 2003, 4, 915. [Google Scholar] [CrossRef] [PubMed]
- Burgess, T.L.; Sun, J.; Meyer, S.; Tsuruda, T.S.; Sun, J.; Elliott, G.; Chen, Q.; Haniu, M.; Barron, W.F.; Juan, T.; et al. Biochemical characterization of AMG 102: A neutralizing, fully human monoclonal antibody to human and nonhuman primate hepatocyte growth factor. Mol. Cancer. Ther. 2010, 9, 400. [Google Scholar] [CrossRef] [PubMed]
- Patnaik, A.; Weiss, G.J.; Papadopoulos, K.P.; Hofmeister, C.C.; Tibes, R.; Tolcher, A.; Isaacs, R.; Jac, J.; Han, M.; Payumos, F.C.; et al. Phase I ficlatuzumab monotherapy or with erlotinib for refractory advanced solid tumors and multiple myeloma. Br. J. Cancer. 2014, 111, 272. [Google Scholar] [CrossRef]
- Okamoto, W.; Okamoto, I.; Tanaka, K.; Hatashita, E.; Yamada, Y.; Kuwata, K.; Yamaguchi, H.; Arao, T.; Nishio, K.; Fukuoka, M.; et al. TAK-701, a humanized monoclonal antibody to hepatocyte growth factor, reverses gefitinib resistance induced by tumor-derived HGF in non-small cell lung cancer with an EGFR mutation. Mol. Cancer. Ther. 2010, 9, 2785. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Hong, S.H.; Kim, J.Y.; Kim, I.C.; Park, Y.W.; Lee, S.J.; Song, S.W.; Kim, J.J.; Park, G.; Kim, Y.H.; et al. Preclinical development of a humanized neutralizing antibody targeting HGF. Exp. Mol. Med. 2017, 49, e309. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.F.; Xie, Q.; Zhang, Y.W.; Su, Y.; Zhao, P.; Cao, B.; Furge, K.; Sun, J.; Rex, K.; Osgood, T.; et al. Therapeutic potential of hepatocyte growth factor/scatter factor neutralizing antibodies: Inhibition of tumor growth in both autocrine and paracrine hepatocyte growth factor/scatter factor:c-Met-driven models of leiomyosarcoma. Mol. Cancer. Ther. 2009, 8, 2803–2810. [Google Scholar] [CrossRef] [PubMed]
- Glodde, N.; Bald, T.; van den Boorn-Konijnenberg, D.; Nakamura, K.; O’Donnell, J.S.; Szczepanski, S.; Brandes, M.; Eickhoff, S.; Das, I.; Shridhar, N.; et al. Reactive neutrophil responses dependent on the receptor tyrosine kinase c-MET limit cancer immunotherapy. Immunuity. 2017, 47, 789–802. [Google Scholar] [CrossRef] [PubMed]
- Benkhoucha, M.; Molnarfi, N.; Kaya, G.; Belnoue, E.; Bjarnadóttir, K.; Dietrich, P.Y.; Walker, P.R.; Martinvalet, D.; Derouazi, M.; et al. Identification of a novel population of highly cytotoxic c-Met-expressing CD8+ T lymphocytes. EMBO. Rep. 2017, 18, 1545–1558. [Google Scholar] [CrossRef] [PubMed]
- Noh, J.; Kim, O.; Jung, Y.; Han, H.; Kim, J.; Kim, S.; Lee, S.; Park, J.; Jung, R.; Kim, S.; et al. High-throughput retrieval of physical DNA for NGS-identifiable clones in phage display library. bioRxiv 2018, 370809. [Google Scholar] [CrossRef]
- Bolotin, D.A.; Poslavsky, S.; Mitrophanov, I.; Shugay, M.; Mamedov, I.Z.; Putintseva, E.V.; Chudakov, D.M. MiXCR: Software for comprehensive adaptive immunity profiling. Nat. Methods 2015, 12, 380–381. [Google Scholar] [CrossRef] [PubMed]
- Miho, E.; Grieff, V.; Roskar, R.; Reddy, S.T. The fundamental principles of antibody repertoire architecture revealed by large-scale network analysis. bioRxiv 2017, 124578. [Google Scholar] [CrossRef]
- Madi, A.; Poran, A.; Shifrut, E.; Reich-Zelinger, S.; Greenstein, E.; Zaretsky, I.; Arnon, T.; Laethem, F.V.; Singer, A.; Lu, J.; et al. T cell receptor repertoires of mice and humans are clustered in similarity networks around conserved public CDR3 sequences. Elife 2017, 6, e22057. [Google Scholar] [CrossRef]
- Bashford-Rogers, R.J.M.; Palser, A.L.; Huntly, B.J.; Rance, R.; Vassiliou, G.S.; Follows, G.A.; Kellam, P. Network properties derived from deep sequencing of human B-cell receptor repertoires delineate B-cell populations. Genome Res. 2013, 23, 1874–1884. [Google Scholar] [CrossRef]
- Levenshtein, V.I. Binary codes capable of correcting deletions, insertions and reversals. Sov. Phys. Dokl. 1966, 4, 261–283. [Google Scholar]
- Newman, M. Networks: An Introduction, 1st ed.; Oxford University Press: Oxford, UK, 2010; ISBN 978-0-19-920665-0. [Google Scholar]
- Hershberg, U.; Uduman, M.; Shlomchik, M.J.; Kleinstein, S.H. Improved methods for detecting selection by mutation analysis of Ig V region sequences. Int. Immunol. 2008, 20, 683–694. [Google Scholar] [CrossRef]
- Ratcliffe, M.J. Antibodies, immunoglobulin genes and the bursa of Fabricius in chicken B cell development. Dev. Comp. Immunol. 2006, 30, 101–118. [Google Scholar] [CrossRef] [PubMed]
- Cormen, T.H.; Leiserson, C.E.; Rivest, R.L.; Stein, C. Introduction to Algorithms; MIT Press: Cambridge, MA, USA, 2001. [Google Scholar]
- Loisel, S.; Ohresser, M.; Pallardy, M.; Daydé, D.; Berthou, C.; Cartron, G.; Watier, H. Relevance, advantages and limitations of animal models used in the development of monoclonal antibodies for cancer treatment. Crit. Rev. Oncol. Hematol. 2007, 62, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Fukuzawa, T.; Sampei, Z.; Haraya, K.; Ruike, Y.; Shida-Kawazoe, M.; Shimizu, Y.; Gan, S.W.; Irie, M.; Tsuboi, Y.; Tai, H.; et al. Long lasting neutralization of C5 by SKY59, a novel recycling antibody, is a potential therapy for complement-mediated diseases. Sci. Rep. 2017, 7, 1080. [Google Scholar] [CrossRef] [PubMed]
- Koo, M.Y.; Park, J.; Lim, J.M.; Joo, S.Y.; Shin, S.P.; Shim, H.B.; Chung, J.; Kang, D.; Woo, H.A.; Rhee, S.G. Selective inhibition of the function of tyrosine-phosphorylated STAT3 with a phosphorylation site-specific intrabody. Proc. Natl. Acad. Sci. USA 2014, 111, 6269–6274. [Google Scholar] [CrossRef] [PubMed]
- Jo, D.H.; Kim, J.H.; Yang, W.; Kim, H.; Chang, S.; Kim, D.; Chang, M.; Lee, K.; Chung, J.; Kim, J.H. Anti-complement component 5 antibody targeting MG4 domain inhibits choroidal neovascularization. Oncotarget 2017, 8, 45506–45516. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Yoon, A.; Lee, S.; Kim, S.; Han, J.; Chung, J. Next-generation sequencing enables the discovery of more diverse positive clones from a phage-displayed antibody library. Exp. Mol. Med. 2017, 49, e308. [Google Scholar] [CrossRef] [PubMed]
- Christley, S.; Levin, M.K.; Toby, I.T.; Fonner, J.M.; Monson, N.L.; Rounds, W.H.; Rubelt, F.; Scarborough, W.; Scheuermann, R.H.; Cowell, L.G. VDJPipe: A pipelined tool for pre-processing immune repertoire sequencing data. BMC. Bioinform. 2017, 18, 448. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High throughput Sequence Data; Babraham Institute: Cambridge, UK, 2010. [Google Scholar]
- Csardi, G.; Nepusz, T. The igraph software package for complex network research. InterJ. Complex Syst. 2006, 1695. [Google Scholar]
- Lee, M.S.; Lee, J.C.; Choi, C.Y.; Chung, J. Production and characterization of monoclonal antibody to botulinum neurotoxin type B light chain by phage display. Hybridoma (Larchmt) 2008, 27, 18. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, H.; Chung, J. An antibody reactive to the Gly63-Lys68 epitope of NT-proBNP exhibits O-glycosylation-independent binding. Exp. Mol. Med. 2014, 46, e114. [Google Scholar] [CrossRef]
- Mathonet, P.; Ullman, C.G. The application of next generation sequencing to the understanding of antibody repertoires. Front. Immunol. 2013, 4, 265. [Google Scholar] [CrossRef] [PubMed]
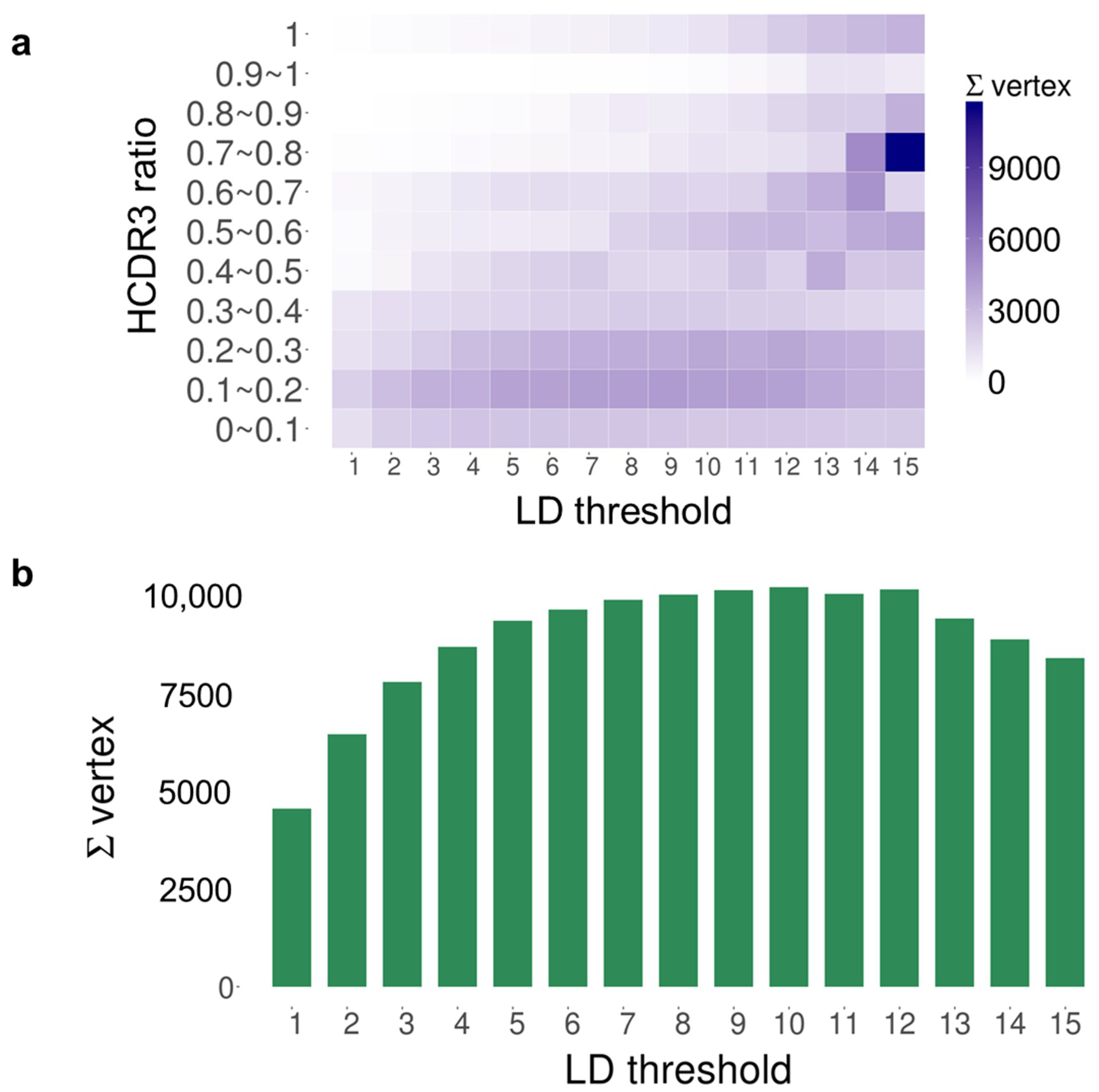
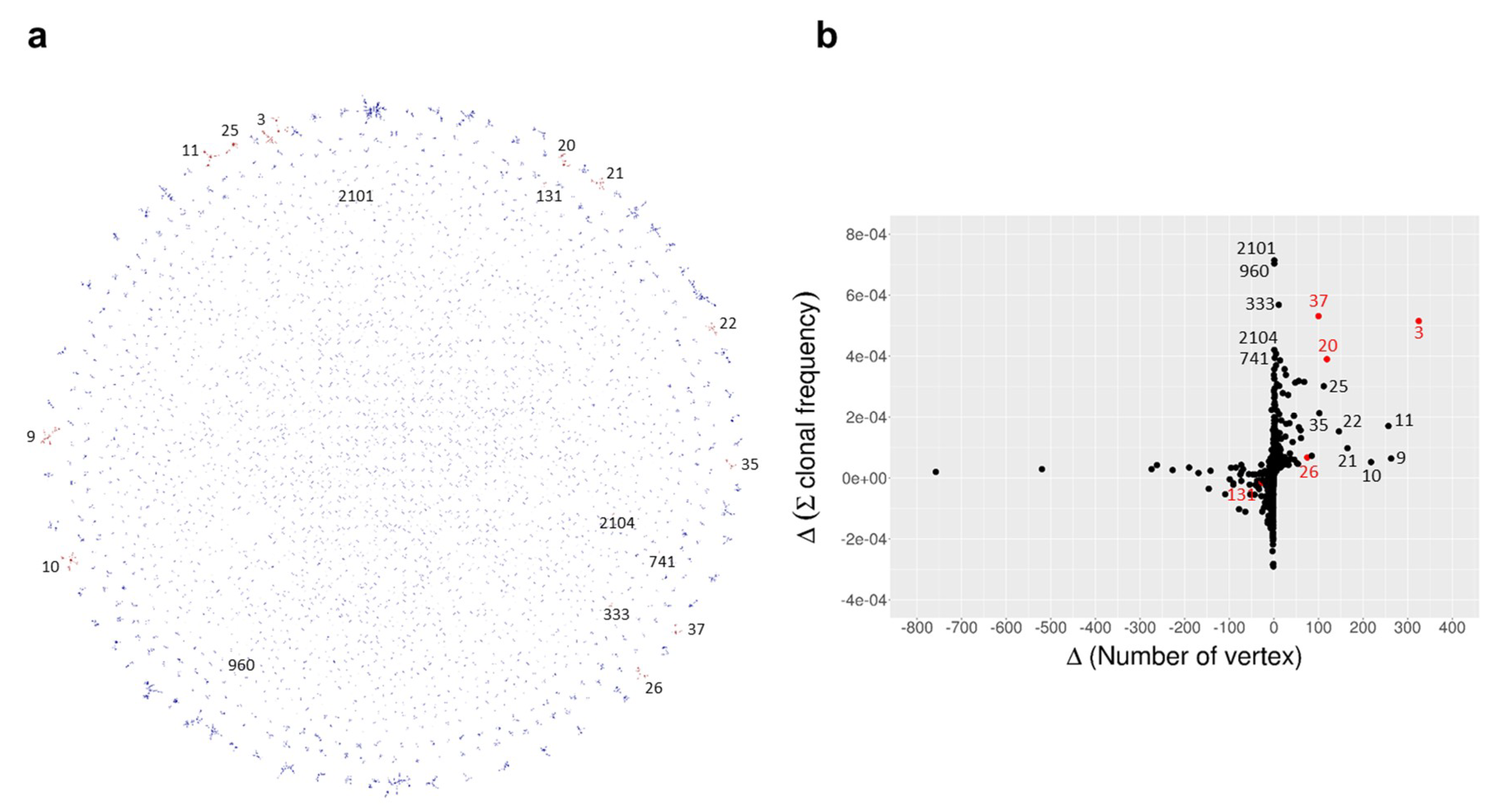
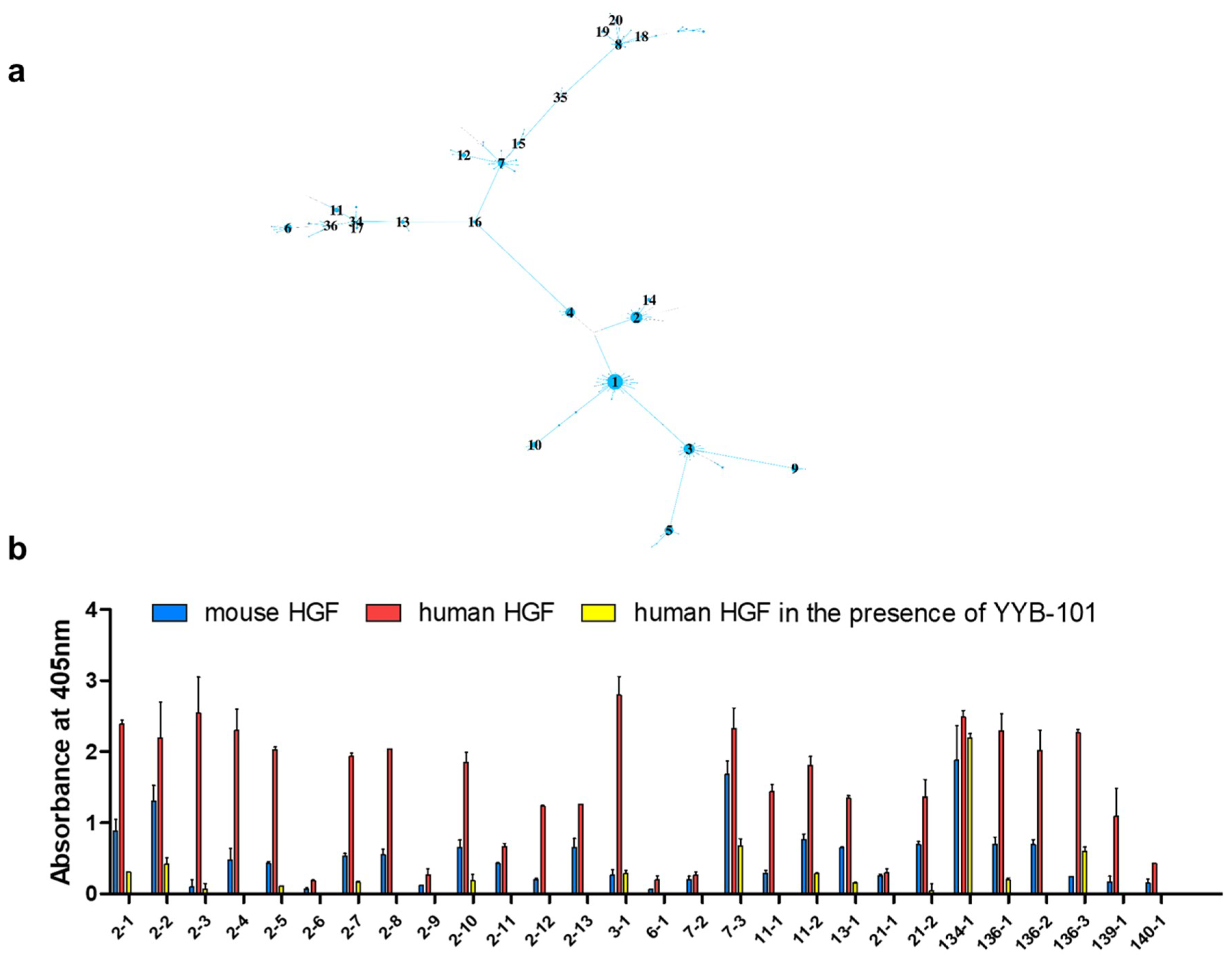
| Time Point | Blood Sampling | Immunization | NGS Reads | VH Gene |
|---|---|---|---|---|
| Week 0 | O | O | 347,331 | 84,693 |
| Week 2 | O | O | 213,534 | 21,556 |
| Week 4 | O | O | 94,925 | 17,213 |
| Week 5 | O | 95,293 | 11,309 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.; Lee, H.; Noh, J.; Lee, Y.; Han, H.; Yoo, D.K.; Kim, H.; Kwon, S.; Chung, J. Efficient Selection of Antibodies Reactive to Homologous Epitopes on Human and Mouse Hepatocyte Growth Factors by Next-Generation Sequencing-Based Analysis of the B Cell Repertoire. Int. J. Mol. Sci. 2019, 20, 417. https://doi.org/10.3390/ijms20020417
Kim S, Lee H, Noh J, Lee Y, Han H, Yoo DK, Kim H, Kwon S, Chung J. Efficient Selection of Antibodies Reactive to Homologous Epitopes on Human and Mouse Hepatocyte Growth Factors by Next-Generation Sequencing-Based Analysis of the B Cell Repertoire. International Journal of Molecular Sciences. 2019; 20(2):417. https://doi.org/10.3390/ijms20020417
Chicago/Turabian StyleKim, Soohyun, Hyunho Lee, Jinsung Noh, Yonghee Lee, Haejun Han, Duck Kyun Yoo, Hyori Kim, Sunghoon Kwon, and Junho Chung. 2019. "Efficient Selection of Antibodies Reactive to Homologous Epitopes on Human and Mouse Hepatocyte Growth Factors by Next-Generation Sequencing-Based Analysis of the B Cell Repertoire" International Journal of Molecular Sciences 20, no. 2: 417. https://doi.org/10.3390/ijms20020417
APA StyleKim, S., Lee, H., Noh, J., Lee, Y., Han, H., Yoo, D. K., Kim, H., Kwon, S., & Chung, J. (2019). Efficient Selection of Antibodies Reactive to Homologous Epitopes on Human and Mouse Hepatocyte Growth Factors by Next-Generation Sequencing-Based Analysis of the B Cell Repertoire. International Journal of Molecular Sciences, 20(2), 417. https://doi.org/10.3390/ijms20020417





