Active DNA Demethylation in Plants
Abstract
1. Introduction
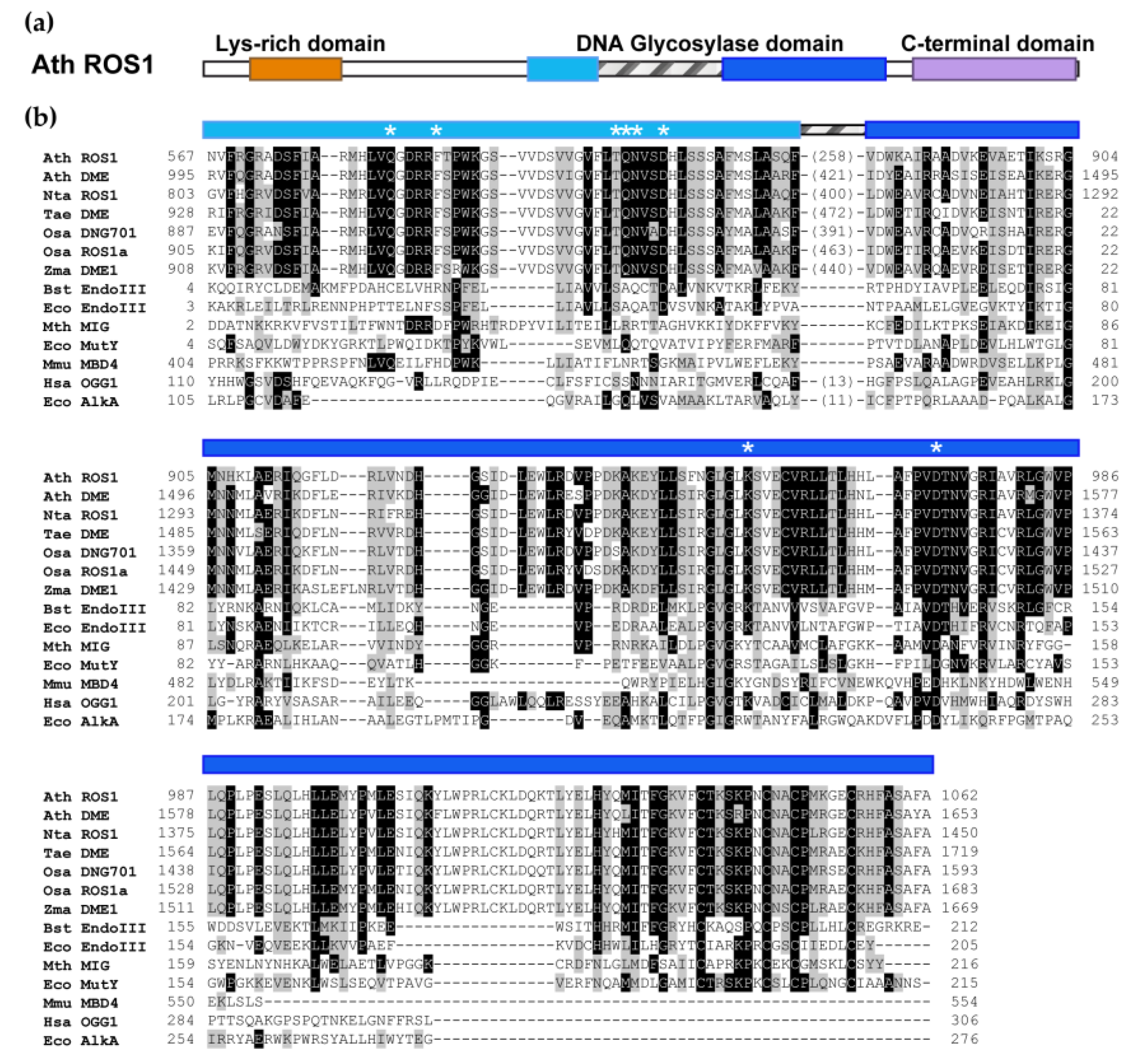
2. The DML Family: Plant-Specific DNA Glycosylases That Excise 5-meC
3. The Plant DNA Demethylation Pathway
3.1. Base Excision and Strand Incision
3.2. Gap Tailoring by 3´-End Cleaning
3.3. Gap Filling and DNA Ligation
3.4. Regulation of Active DNA Demethylation
4. Biological Roles of DNA Demethylation
4.1. Maintenance of Genome Stability by Preventing Hypermethylation
4.2. Methylome Reprogramming During Reproductive Development
4.3. Gene Regulation in Response to Biotic Stimuli
4.4. Gene Regulation in Response to Abiotic Stress
5. Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Law, J.A.; Jacobsen, S.E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010, 11, 204–220. [Google Scholar] [CrossRef] [PubMed]
- Goll, M.G.; Bestor, T.H. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005, 74, 481–514. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.W.; Henderson, I.R.; Jacobsen, S.E. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat. Rev. Genet. 2005, 6, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Zemach, A.; Kim, M.Y.; Hsieh, P.H.; Coleman-Derr, D.; Eshed-Williams, L.; Thao, K.; Harmer, S.L.; Zilberman, D. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 2013, 153, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, T.; Kobayashi, A.; Saze, H.; Kakutani, T. RNAi-independent de novo DNA methylation revealed in Arabidopsis mutants of chromatin remodeling gene DDM1. Plant J. 2012, 70, 750–758. [Google Scholar] [CrossRef] [PubMed]
- Kress, C.; Thomassin, H.; Grange, T. Local DNA demethylation in vertebrates: How could it be performed and targeted? FEBS Lett. 2001, 494, 135–140. [Google Scholar] [CrossRef]
- Ooi, S.K.; Bestor, T.H. The colorful history of active DNA demethylation. Cell 2008, 133, 1145–1148. [Google Scholar] [CrossRef]
- Wu, X.; Zhang, Y. TET-mediated active DNA demethylation: Mechanism, function and beyond. Nat. Rev. Genet. 2017, 18, 517–534. [Google Scholar] [CrossRef] [PubMed]
- Roldan-Arjona, T.; Ariza, R.R. DNA demethylation. In DNA and RNA modification Enzymes: Comparative Structure, Mechanism, Functions, Cellular Interactions and Evolution; Grosjean, H., Ed.; Landes Bioscience: Austin, TX, USA, 2009; pp. 149–161. [Google Scholar]
- Zhu, J.K. Active DNA demethylation mediated by DNA glycosylases. Annu. Rev. Genet. 2009, 43, 143–166. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Morales-Ruiz, T.; Ariza, R.R.; Roldan-Arjona, T.; David, L.; Zhu, J.K. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 2002, 111, 803–814. [Google Scholar] [CrossRef]
- Morales-Ruiz, T.; Ortega-Galisteo, A.P.; Ponferrada-Marin, M.I.; Martinez-Macias, M.I.; Ariza, R.R.; Roldan-Arjona, T. DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases. Proc. Natl. Acad. Sci. USA 2006, 103, 6853–6858. [Google Scholar] [CrossRef] [PubMed]
- Agius, F.; Kapoor, A.; Zhu, J.K. Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proc. Natl. Acad. Sci. USA 2006, 103, 11796–11801. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Gehring, M.; Johnson, L.; Hannon, M.; Harada, J.J.; Goldberg, R.B.; Jacobsen, S.E.; Fischer, R.L. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell 2002, 110, 33–42. [Google Scholar] [CrossRef]
- Penterman, J.; Zilberman, D.; Huh, J.H.; Ballinger, T.; Henikoff, S.; Fischer, R.L. DNA demethylation in the Arabidopsis genome. Proc. Natl. Acad. Sci. USA 2007, 104, 6752–6757. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Galisteo, A.P.; Morales-Ruiz, T.; Ariza, R.R.; Roldan-Arjona, T. Arabidopsis DEMETER-LIKE proteins DML2 and DML3 are required for appropriate distribution of DNA methylation marks. Plant Mol. Biol. 2008, 67, 671–681. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Kapoor, A.; Sridhar, V.V.; Agius, F.; Zhu, J.K. The DNA glycosylase/lyase ROS1 functions in pruning DNA methylation patterns in Arabidopsis. Curr. Biol. 2007, 17, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Gehring, M.; Huh, J.H.; Hsieh, T.F.; Penterman, J.; Choi, Y.; Harada, J.J.; Goldberg, R.B.; Fischer, R.L. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell 2006, 124, 495–506. [Google Scholar] [CrossRef]
- Calarco, J.P.; Borges, F.; Donoghue, M.T.; Van Ex, F.; Jullien, P.E.; Lopes, T.; Gardner, R.; Berger, F.; Feijo, J.A.; Becker, J.D.; et al. Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell 2012, 151, 194–205. [Google Scholar] [CrossRef]
- Ibarra, C.A.; Feng, X.; Schoft, V.K.; Hsieh, T.F.; Uzawa, R.; Rodrigues, J.A.; Zemach, A.; Chumak, N.; Machlicova, A.; Nishimura, T.; et al. Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science 2012, 337, 1360–1364. [Google Scholar] [CrossRef]
- Kawakatsu, T.; Nery, J.R.; Castanon, R.; Ecker, J.R. Dynamic DNA methylation reconfiguration during seed development and germination. Genome Biol. 2017, 18, 171. [Google Scholar] [CrossRef]
- Lang, Z.; Wang, Y.; Tang, K.; Tang, D.; Datsenka, T.; Cheng, J.; Zhang, Y.; Handa, A.K.; Zhu, J.K. Critical roles of DNA demethylation in the activation of ripening-induced genes and inhibition of ripening-repressed genes in tomato fruit. Proc. Natl. Acad. Sci. USA 2017, 114, 4511–4519. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; How-Kit, A.; Stammitti, L.; Teyssier, E.; Rolin, D.; Mortain-Bertrand, A.; Halle, S.; Liu, M.; Kong, J.; Wu, C.; et al. A DEMETER-like DNA demethylase governs tomato fruit ripening. Proc. Natl. Acad. Sci. USA 2015, 112, 10804–10809. [Google Scholar] [CrossRef] [PubMed]
- Pandey, G.; Sharma, N.; Sahu, P.P.; Prasad, M. Chromatin-Based Epigenetic Regulation of Plant Abiotic Stress Response. Curr. Genomics 2016, 17, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Yu, A.; Lepere, G.; Jay, F.; Wang, J.; Bapaume, L.; Wang, Y.; Abraham, A.L.; Penterman, J.; Fischer, R.L.; Voinnet, O.; et al. Dynamics and biological relevance of DNA demethylation in Arabidopsis antibacterial defense. Proc. Natl. Acad. Sci. USA 2013, 110, 2389–2394. [Google Scholar] [CrossRef] [PubMed]
- Nash, H.M.; Bruner, S.D.; Scharer, O.D.; Kawate, T.; Addona, T.A.; Spooner, E.; Lane, W.S.; Verdine, G.L. Cloning of a yeast 8-oxoguanine DNA glycosylase reveals the existence of a base-excision DNA-repair protein superfamily. Curr. Biol. 1996, 6, 968–980. [Google Scholar] [CrossRef]
- Ponferrada-Marín, M.I.; Parrilla-Doblas, J.T.; Roldán-Arjona, T.; Ariza, R.R. A discontinuous DNA glycosylase domain in a family of enzymes that excise 5-methylcytosine. Nucleic. Acids. Res. 2011, 39, 1473–1484. [Google Scholar] [CrossRef] [PubMed]
- Mok, Y.G.; Uzawa, R.; Lee, J.; Weiner, G.M.; Eichman, B.F.; Fischer, R.L.; Huh, J.H. Domain structure of the DEMETER 5-methylcytosine DNA glycosylase. Proc. Natl. Acad. Sci. USA 2010, 107, 19225–19230. [Google Scholar] [CrossRef]
- Krokan, H.E.; Standal, R.; Slupphaug, G. DNA glycosylases in the base excision repair of DNA. Biochem. J. 1997, 325, 1–16. [Google Scholar] [CrossRef]
- Parrilla-Doblas, J.T.; Ponferrada-Marin, M.I.; Roldan-Arjona, T.; Ariza, R.R. Early steps of active DNA demethylation initiated by ROS1 glycosylase require three putative helix-invading residues. Nucleic Acids Res. 2013, 41, 8654–8664. [Google Scholar] [CrossRef]
- Buzas, D.M. Emerging links between iron-sulfur clusters and 5-methylcytosine base excision repair in plants. Genes Genet. Syst. 2016, 91, 51–62. [Google Scholar] [CrossRef][Green Version]
- Ponferrada-Marín, M.I.; Martínez-Macías, M.I.; Morales-Ruiz, T.; Roldán-Arjona, T.; Ariza, R.R. Methylation-independent DNA binding modulates specificity of repressor of silencing 1 (ROS1) and facilitates demethylation in long substrates. J. Biol. Chem. 2010, 285, 23032–23039. [Google Scholar] [CrossRef] [PubMed]
- Ponferrada-Marín, M.I.; Roldán-Arjona, T.; Ariza, R.R. Demethylation initiated by ROS1 glycosylase involves random sliding along DNA. Nucleic Acids Res. 2012, 40, 11554–11562. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Hashimoto, H.; Kow, Y.W.; Zhang, X.; Cheng, X. The carboxy-terminal domain of ROS1 is essential for 5-methylcytosine DNA glycosylase activity. J. Mol. Biol. 2014, 426, 3703–3712. [Google Scholar] [CrossRef] [PubMed]
- Ponferrada-Marín, M.I.; Roldán-Arjona, T.; Ariza, R.R. ROS1 5-methylcytosine DNA glycosylase is a slow-turnover catalyst that initiates DNA demethylation in a distributive fashion. Nucleic Acids Res. 2009, 37, 4264–4274. [Google Scholar] [CrossRef] [PubMed]
- Dodson, M.L.; Michaels, M.L.; Lloyd, R.S. Unified catalytic mechanism for DNA glycosylases. J. Biol. Chem. 1994, 269, 32709–32712. [Google Scholar] [PubMed]
- Martínez-Macías, M.I.; Qian, W.; Miki, D.; Pontes, O.; Liu, Y.; Tang, K.; Liu, R.; Morales-Ruiz, T.; Ariza, R.R.; Roldán-Arjona, T.; et al. A DNA 3′ phosphatase functions in active DNA demethylation in Arabidopsis. Mol. Cell 2012, 45, 357–370. [Google Scholar] [CrossRef] [PubMed]
- Córdoba-Cañero, D.; Roldan-Arjona, T.; Ariza, R.R. Arabidopsis ZDP DNA 3′-phosphatase and ARP endonuclease function in 8-oxoG repair initiated by FPG and OGG1 DNA glycosylases. Plant J. 2014, 79, 824–834. [Google Scholar] [CrossRef] [PubMed]
- Barbado, C.; Córdoba-Cañero, D.; Ariza, R.R.; Roldan-Arjona, T. Nonenzymatic release of N7-methylguanine channels repair of abasic sites into an AP endonuclease-independent pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E916–E924. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liang, W.; Li, Y.; Qian, W. Apurinic/apyrimidinic endonuclease2 and zinc finger dna 3′-phosphoesterase play overlapping roles in the maintenance of epigenome and genome stability. Plant Cell 2018, 30, 1954–1970. [Google Scholar] [CrossRef]
- Suh, D.; Wilson, D.M., 3rd; Povirk, L.F. 3′-phosphodiesterase activity of human apurinic/apyrimidinic endonuclease at DNA double-strand break ends. Nucleic Acids Res. 1997, 25, 2495–2500. [Google Scholar] [CrossRef]
- Li, Y.; Córdoba-Cañero, D.; Qian, W.; Zhu, X.; Tang, K.; Zhang, H.; Ariza, R.R.; Roldán-Arjona, T.; Zhu, J.K. An AP endonuclease functions in active DNA demethylation and gene imprinting in Arabidopsis. PLoS Genet 2015, 11, e1004905. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Jang, H.; Shin, H.; Choi, W.L.; Mok, Y.G.; Huh, J.H. AP endonucleases process 5-methylcytosine excision intermediates during active DNA demethylation in Arabidopsis. Nucleic Acids Res. 2015, 42, 11408–11418. [Google Scholar] [CrossRef] [PubMed]
- Fortini, P.; Dogliotti, E. Base damage and single-strand break repair: Mechanisms and functional significance of short- and long-patch repair subpathways. DNA Repair (Amst) 2007, 6, 398–409. [Google Scholar] [CrossRef] [PubMed]
- Córdoba-Cañero, D.; Morales-Ruiz, T.; Roldán-Arjona, T.; Ariza, R.R. Single-nucleotide and long-patch base excision repair of DNA damage in plants. Plant J. 2009, 60, 716–728. [Google Scholar] [CrossRef] [PubMed]
- Kubota, Y.; Nash, R.A.; Klungland, A.; Schar, P.; Barnes, D.E.; Lindahl, T. Reconstitution of DNA base excision-repair with purified human proteins: Interaction between DNA polymerase b and the XRCC1 protein. EMBO J. 1996, 15, 6662–6670. [Google Scholar] [CrossRef]
- Podlutsky, A.J.; Dianova, II.; Podust, V.N.; Bohr, V.A.; Dianov, G.L. Human DNA polymerase beta initiates DNA synthesis during long-patch repair of reduced AP sites in DNA. Embo J. 2001, 20, 1477–1482. [Google Scholar] [CrossRef]
- Uchiyama, Y.; Kimura, S.; Yamamoto, T.; Ishibashi, T.; Sakaguchi, K. Plant DNA polymerase l, a DNA repair enzyme that functions in plant meristematic and meiotic tissues. Eur. J. Biochem. 2004, 271, 2799–2807. [Google Scholar] [CrossRef]
- Amoroso, A.; Concia, L.; Maggio, C.; Raynaud, C.; Bergounioux, C.; Crespan, E.; Cella, R.; Maga, G. Oxidative DNA damage bypass in Arabidopsis thaliana requires DNA Polymerase l and Proliferating Cell Nuclear Antigen 2. Plant Cell 2011, 23, 806–822. [Google Scholar] [CrossRef]
- Roy, S.; Choudhury, S.R.; Singh, S.K.; Das, K.P. AtPoll, a homolog of mammalian DNA polymerase l in Arabidopsis thaliana, is involved in the repair of UV-B induced DNA damage through the dark repair pathway. Plant Cell Physiol. 2011, 52, 448–467. [Google Scholar] [CrossRef]
- Balakrishnan, L.; Bambara, R.A. Flap endonuclease 1. Annu. Rev. Biochem. 2013, 82, 119–138. [Google Scholar] [CrossRef]
- Zhang, J.; Xie, S.; Zhu, J.K.; Gong, Z. Requirement for flap endonuclease 1 (FEN1) to maintain genomic stability and transcriptional gene silencing in Arabidopsis. Plant J. 2016, 87, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Cappelli, E.; Taylor, R.; Cevasco, M.; Abbondandolo, A.; Caldecott, K.; Frosina, G. Involvement of XRCC1 and DNA ligase III gene products in DNA base excision repair. J. Biol. Chem. 1997, 272, 23970–23975. [Google Scholar] [CrossRef] [PubMed]
- Sleeth, K.M.; Robson, R.L.; Dianov, G.L. Exchangeability of mammalian DNA ligases between base excision repair pathways. Biochemistry 2004, 43, 12924–12930. [Google Scholar] [CrossRef] [PubMed]
- Bonatto, D.; Brendel, M.; Henriques, J.A.P. A new group of plant-specific ATP-dependent DNA ligases identified by protein phylogeny, hydrophobic cluster analysis and 3-dimensional modelling. Funct. Plant Biol. 2005, 32, 161–174. [Google Scholar] [CrossRef]
- Waterworth, W.M.; Kozak, J.; Provost, C.M.; Bray, C.M.; Angelis, K.J.; West, C.E. DNA ligase 1 deficient plants display severe growth defects and delayed repair of both DNA single and double strand breaks. BMC Plant Biol. 2009, 9, 79. [Google Scholar] [CrossRef]
- Waterworth, W.M.; Masnavi, G.; Bhardwaj, R.M.; Jiang, Q.; Bray, C.M.; West, C.E. A higher plant DNA ligase is an important determinant of seed longevity. Plant J. 2010, 63, 848–860. [Google Scholar] [CrossRef] [PubMed]
- West, C.E.; Waterworth, W.M.; Jiang, Q.; Bray, C.M. Arabidopsis DNA ligase IV is induced by gamma-irradiation and interacts with an Arabidopsis homologue of the double strand break repair protein XRCC4. Plant J. 2000, 24, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Córdoba-Cañero, D.; Roldán-Arjona, T.; Ariza, R.R. Arabidopsis ARP endonuclease functions in a branched base excision DNA repair pathway completed by LIG1. Plant J. 2011, 68, 693–702. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Duan, C.G.; Zhu, X.; Qian, W.; Zhu, J.K. A DNA ligase required for active DNA demethylation and genomic imprinting in Arabidopsis. Cell Res. 2015, 25, 757–760. [Google Scholar] [CrossRef]
- Andreuzza, S.; Li, J.; Guitton, A.E.; Faure, J.E.; Casanova, S.; Park, J.S.; Choi, Y.; Chen, Z.; Berger, F. DNA LIGASE I exerts a maternal effect on seed development in Arabidopsis thaliana. Development 2009, 137, 73–81. [Google Scholar] [CrossRef]
- Martinez-Macías, M.I.; Cordoba-Cañero, D.; Ariza, R.R.; Roldan-Arjona, T. The DNA repair protein XRCC1 functions in the plant DNA demethylation pathway by stimulating cytosine methylation (5-meC) excision, gap tailoring, and DNA ligation. J. Biol. Chem. 2013, 288, 5496–5505. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.; Lang, Z.; Zhang, H.; Zhu, J.K. The DNA demethylase ROS1 targets genomic regions with distinct chromatin modifications. Nat. Plants 2016, 2, 16169. [Google Scholar] [CrossRef] [PubMed]
- Qian, W.; Miki, D.; Zhang, H.; Liu, Y.; Zhang, X.; Tang, K.; Kan, Y.; La, H.; Li, X.; Li, S.; et al. A histone acetyltransferase regulates active DNA demethylation in Arabidopsis. Science 2012, 336, 1445–1448. [Google Scholar] [CrossRef] [PubMed]
- Qian, W.; Miki, D.; Lei, M.; Zhu, X.; Zhang, H.; Liu, Y.; Li, Y.; Lang, Z.; Wang, J.; Tang, K.; et al. Regulation of active DNA demethylation by an alpha-crystallin domain protein in Arabidopsis. Mol. Cell 2014, 55, 361–371. [Google Scholar] [CrossRef] [PubMed]
- Lang, Z.; Lei, M.; Wang, X.; Tang, K.; Miki, D.; Zhang, H.; Mangrauthia, S.K.; Liu, W.; Nie, W.; Ma, G.; et al. The methyl-CpG-binding protein MBD7 facilitates active DNA demethylation to limit DNA hyper-methylation and transcriptional gene silencing. Mol. Cell 2015, 57, 971–983. [Google Scholar] [CrossRef] [PubMed]
- Duan, C.G.; Wang, X.; Xie, S.; Pan, L.; Miki, D.; Tang, K.; Hsu, C.C.; Lei, M.; Zhong, Y.; Hou, Y.J.; et al. A pair of transposon-derived proteins function in a histone acetyltransferase complex for active DNA demethylation. Cell Res. 2017, 27, 226–240. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Pontes, O.; Zhu, J.; Miki, D.; Zhang, F.; Li, W.X.; Iida, K.; Kapoor, A.; Pikaard, C.S.; Zhu, J.K. ROS3 is an RNA-binding protein required for DNA demethylation in Arabidopsis. Nature 2008, 455, 1259–1262. [Google Scholar] [CrossRef][Green Version]
- Lister, R.; O’Malley, R.C.; Tonti-Filippini, J.; Gregory, B.D.; Berry, C.C.; Millar, A.H.; Ecker, J.R. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 2008, 133, 523–536. [Google Scholar] [CrossRef]
- Huettel, B.; Kanno, T.; Daxinger, L.; Aufsatz, W.; Matzke, A.J.; Matzke, M. Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. EMBO J. 2006, 25, 2828–2836. [Google Scholar] [CrossRef]
- Mathieu, O.; Reinders, J.; Caikovski, M.; Smathajitt, C.; Paszkowski, J. Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell 2007, 130, 851–862. [Google Scholar] [CrossRef]
- Penterman, J.; Uzawa, R.; Fischer, R.L. Genetic interactions between DNA demethylation and methylation in Arabidopsis. Plant Physiol. 2007, 145, 1549–1557. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Liu, H.L.; Daxinger, L.; Pontes, O.; He, X.; Qian, W.; Lin, H.; Xie, M.; Lorkovic, Z.J.; Zhang, S.; et al. An RNA polymerase II- and AGO4-associated protein acts in RNA-directed DNA methylation. Nature 2010, 465, 106–109. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Qian, W.; Zhao, Y.; Wang, C.; Shen, J.; Zhu, J.K.; Gong, Z. Antisilencing role of the RNA-directed DNA methylation pathway and a histone acetyltransferase in Arabidopsis. Proc. Natl. Acad Sci. USA 2012, 109, 11425–11430. [Google Scholar] [CrossRef] [PubMed]
- Lei, M.; Zhang, H.; Julian, R.; Tang, K.; Xie, S.; Zhu, J.K. Regulatory link between DNA methylation and active demethylation in Arabidopsis. Proc. Natl. Acad Sci. USA 2015, 112, 3553–3557. [Google Scholar] [CrossRef] [PubMed]
- Williams, B.P.; Pignatta, D.; Henikoff, S.; Gehring, M. Methylation-sensitive expression of a DNA demethylase gene serves as an epigenetic rheostat. PLoS Genet. 2015, 11, e1005142. [Google Scholar] [CrossRef] [PubMed]
- Wittschieben, B.O.; Iwai, S.; Wood, R.D. DDB1-DDB2 (xeroderma pigmentosum group E) protein complex recognizes a cyclobutane pyrimidine dimer, mismatches, apurinic/apyrimidinic sites, and compound lesions in DNA. J. Biol. Chem. 2005, 280, 39982–39989. [Google Scholar] [CrossRef] [PubMed]
- Schalk, C.; Drevensek, S.; Kramdi, A.; Kassam, M.; Ahmed, I.; Cognat, V.; Graindorge, S.; Bergdoll, M.; Baumberger, N.; Heintz, D.; et al. DNA DAMAGE BINDING PROTEIN2 Shapes the DNA Methylation Landscape. Plant Cell 2016, 28, 2043–2059. [Google Scholar] [CrossRef]
- Córdoba-Cañero, D.; Cognat, V.; Ariza, R.R.; Roldan Arjona, T.; Molinier, J. Dual control of ROS1-mediated active DNA demethylation by DNA damage-binding protein 2 (DDB2). Plant J. 2017, 92, 1170–1181. [Google Scholar] [CrossRef]
- Luo, D.; Bernard, D.G.; Balk, J.; Hai, H.; Cui, X. The DUF59 family gene AE7 acts in the cytosolic iron-sulfur cluster assembly pathway to maintain nuclear genome integrity in Arabidopsis. Plant Cell 2012, 24, 4135–4148. [Google Scholar] [CrossRef]
- Nakamura, M.; Buzas, D.M.; Kato, A.; Fujita, M.; Kurata, N.; Kinoshita, T. The role of Arabidopsis thaliana NAR1, a cytosolic iron-sulfur cluster assembly component, in gametophytic gene expression and oxidative stress responses in vegetative tissue. New Phytol. 2013, 199, 925–935. [Google Scholar] [CrossRef]
- Buzas, D.M.; Nakamura, M.; Kinoshita, T. Epigenetic role for the conserved Fe-S cluster biogenesis protein AtDRE2 in Arabidopsis thaliana. Proc. Natl. Acad Sci. USA 2014, 111, 13565–13570. [Google Scholar] [CrossRef] [PubMed]
- Duan, C.G.; Wang, X.; Tang, K.; Zhang, H.; Mangrauthia, S.K.; Lei, M.; Hsu, C.C.; Hou, Y.J.; Wang, C.; Li, Y.; et al. MET18 Connects the cytosolic iron-sulfur cluster assembly pathway to active dna demethylation in arabidopsis. PLoS Genet. 2015, 11, e1005559. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.F.; Huang, H.W.; Li, L.; Cai, T.; Chen, S.; He, X.J. The cytosolic iron-sulfur cluster assembly protein mms19 regulates transcriptional gene silencing, dna repair, and flowering time in arabidopsis. PLoS ONE 2015, 10, e0129137. [Google Scholar] [CrossRef] [PubMed]
- Tran, R.K.; Zilberman, D.; de Bustos, C.; Ditt, R.F.; Henikoff, J.G.; Lindroth, A.M.; Delrow, J.; Boyle, T.; Kwong, S.; Bryson, T.D.; et al. Chromatin and siRNA pathways cooperate to maintain DNA methylation of small transposable elements in Arabidopsis. Genome Biol. 2005, 6, R90. [Google Scholar] [CrossRef] [PubMed]
- Stroud, H.; Do, T.; Du, J.; Zhong, X.; Feng, S.; Johnson, L.; Patel, D.J.; Jacobsen, S.E. Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat. Struct. Mol. Biol. 2014, 21, 64–72. [Google Scholar] [CrossRef] [PubMed]
- La, H.; Ding, B.; Mishra, G.P.; Zhou, B.; Yang, H.; Bellizzi Mdel, R.; Chen, S.; Meyers, B.C.; Peng, Z.; Zhu, J.K.; et al. A 5-methylcytosine DNA glycosylase/lyase demethylates the retrotransposon Tos17 and promotes its transposition in rice. Proc. Natl. Acad. Sci. USA 2011, 108, 15498–15503. [Google Scholar] [CrossRef] [PubMed]
- Yamamuro, C.; Miki, D.; Zheng, Z.; Ma, J.; Wang, J.; Yang, Z.; Dong, J.; Zhu, J.K. overproduction of stomatal lineage cells in arabidopsis mutants defective in active dna demethylation. Nat. Commun. 2014, 5, 4062. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wang, J.; Miki, D.; Xia, R.; Yu, W.; He, J.; Zheng, Z.; Zhu, J.K.; Gong, Z. DNA replication factor c1 mediates genomic stability and transcriptional gene silencing in Arabidopsis. Plant Cell 2010, 22, 2336–2352. [Google Scholar] [CrossRef] [PubMed]
- Huh, J.H.; Bauer, M.J.; Hsieh, T.F.; Fischer, R.L. Cellular programming of plant gene imprinting. Cell 2008, 132, 735–744. [Google Scholar] [CrossRef]
- Park, K.; Kim, M.Y.; Vickers, M.; Park, J.S.; Hyun, Y.; Okamoto, T.; Zilberman, D.; Fischer, R.L.; Feng, X.; Choi, Y.; et al. DNA demethylation is initiated in the central cells of Arabidopsis and rice. Proc. Natl. Acad. Sci. USA 2016, 113, 15138–15143. [Google Scholar] [CrossRef]
- Kinoshita, T.; Miura, A.; Choi, Y.; Kinoshita, Y.; Cao, X.; Jacobsen, S.E.; Fischer, R.L.; Kakutani, T. one-way control of FWA imprinting in arabidopsis endosperm by DNA methylation. Science 2004, 303, 521–523. [Google Scholar] [CrossRef] [PubMed]
- Jullien, P.E.; Kinoshita, T.; Ohad, N.; Berger, F. Maintenance of DNA methylation during the Arabidopsis life cycle is essential for parental imprinting. Plant Cell 2006, 18, 1360–1372. [Google Scholar] [CrossRef] [PubMed]
- Gehring, M.; Bubb, K.L.; Henikoff, S. Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science 2009, 324, 1447–1451. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, T.F.; Shin, J.; Uzawa, R.; Silva, P.; Cohen, S.; Bauer, M.J.; Hashimoto, M.; Kirkbride, R.C.; Harada, J.J.; Zilberman, D.; et al. Regulation of imprinted gene expression in Arabidopsis endosperm. Proc. Natl. Acad. Sci. USA 2011, 108, 1755–1762. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, T.F.; Ibarra, C.A.; Silva, P.; Zemach, A.; Eshed-Williams, L.; Fischer, R.L.; Zilberman, D. Genome-wide demethylation of Arabidopsis endosperm. Science 2009, 324, 1451–1454. [Google Scholar] [CrossRef]
- Schoft, V.K.; Chumak, N.; Choi, Y.; Hannon, M.; Garcia-Aguilar, M.; Machlicova, A.; Slusarz, L.; Mosiolek, M.; Park, J.S.; Park, G.T.; et al. Function of the DEMETER DNA glycosylase in the Arabidopsis thaliana male gametophyte. Proc. Natl. Acad. Sci. USA 2011, 108, 8042–8047. [Google Scholar] [CrossRef] [PubMed]
- Park, J.S.; Frost, J.M.; Park, K.; Ohr, H.; Park, G.T.; Kim, S.; Eom, H.; Lee, I.; Brooks, J.S.; Fischer, R.L.; et al. Control of DEMETER DNA demethylase gene transcription in male and female gamete companion cells in Arabidopsis thaliana. Pro. Natl. Acad. Sci. USA 2017, 114, 2078–2083. [Google Scholar] [CrossRef] [PubMed]
- Slotkin, R.K.; Vaughn, M.; Borges, F.; Tanurdzic, M.; Becker, J.D.; Feijo, J.A.; Martienssen, R.A. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 2009, 136, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Martinez, G.; Panda, K.; Kohler, C.; Slotkin, R.K. Silencing in sperm cells is directed by RNA movement from the surrounding nurse cell. Nat. Plants 2016, 2, 16030. [Google Scholar] [CrossRef] [PubMed]
- Zemach, A.; Kim, M.Y.; Silva, P.; Rodrigues, J.A.; Dotson, B.; Brooks, M.D.; Zilberman, D. Local DNA hypomethylation activates genes in rice endosperm. Proc. Natl. Acad. Sci. USA 2010, 107, 18729–18734. [Google Scholar] [CrossRef] [PubMed]
- Ono, A.; Yamaguchi, K.; Fukada-Tanaka, S.; Terada, R.; Mitsui, T.; Iida, S. A null mutation of ROS1a for DNA demethylation in rice is not transmittable to progeny. Plant J. 2012, 71, 564–574. [Google Scholar] [CrossRef]
- Kim, M.Y.; Ono, A.; Scholten, S.; Kinoshita, T.; Zilberman, D.; Okamoto, T.; Fischer, R.L. DNA demethylation by ROS1a in rice vegetative cells promotes methylation in sperm. Proc. Natl. Acad. Sci. USA 2019, 116, 9652–9657. [Google Scholar] [CrossRef]
- Wang, P.; Xia, H.; Zhang, Y.; Zhao, S.; Zhao, C.; Hou, L.; Li, C.; Li, A.; Ma, C.; Wang, X. Genome-wide high-resolution mapping of DNA methylation identifies epigenetic variation across embryo and endosperm in Maize (Zea may). BMC Genomics 2015, 16, 21. [Google Scholar] [CrossRef]
- Wen, S.; Wen, N.; Pang, J.; Langen, G.; Brew-Appiah, R.A.; Mejias, J.H.; Osorio, C.; Yang, M.; Gemini, R.; Moehs, C.P.; et al. Structural genes of wheat and barley 5-methylcytosine DNA glycosylases and their potential applications for human health. Proc. Natl. Acad. Sci. USA 2012, 109, 20543–20548. [Google Scholar] [CrossRef]
- Cheng, J.; Niu, Q.; Zhang, B.; Chen, K.; Yang, R.; Zhu, J.K.; Zhang, Y.; Lang, Z. Downregulation of RdDM during strawberry fruit ripening. Genome Biol. 2018, 19, 212. [Google Scholar] [CrossRef]
- Huang, H.; Liu, R.; Niu, Q.; Tang, K.; Zhang, B.; Zhang, H.; Chen, K.; Zhu, J.K.; Lang, Z. Global increase in DNA methylation during orange fruit development and ripening. Proc. Natl. Acad. Sci. USA 2019, 116, 1430–1436. [Google Scholar] [CrossRef]
- Satge, C.; Moreau, S.; Sallet, E.; Lefort, G.; Auriac, M.C.; Rembliere, C.; Cottret, L.; Gallardo, K.; Noirot, C.; Jardinaud, M.F.; et al. Reprogramming of DNA methylation is critical for nodule development in Medicago truncatula. Nat. Plants 2016, 2, 16166. [Google Scholar] [CrossRef]
- Rodriguez-Negrete, E.; Lozano-Duran, R.; Piedra-Aguilera, A.; Cruzado, L.; Bejarano, E.R.; Castillo, A.G. Geminivirus Rep protein interferes with the plant DNA methylation machinery and suppresses transcriptional gene silencing. New Phytol. 2013, 199, 464–475. [Google Scholar] [CrossRef]
- Le, T.N.; Schumann, U.; Smith, N.A.; Tiwari, S.; Au, P.; Zhu, Q.H.; Taylor, J.; Kazan, K.; Llewellyn, D.J.; Zhang, R.; et al. DNA demethylases target promoter transposable elements to positively regulate stress responsive genes in Arabidopsis. Genome Biol. 2014, 15, 458. [Google Scholar] [CrossRef]
- Schumann, U.; Lee, J.; Kazan, K.; Ayliffe, M.; Wang, M.B. DNA-Demethylase Regulated Genes Show Methylation-Independent Spatiotemporal Expression Patterns. Front Plant Sci. 2017, 8, 1449. [Google Scholar] [CrossRef]
- Lopez Sanchez, A.; Stassen, J.H.; Furci, L.; Smith, L.M.; Ton, J. The role of DNA (de)methylation in immune responsiveness of Arabidopsis. Plant J. 2016, 88, 361–374. [Google Scholar] [CrossRef]
- Chen, X.; Schonberger, B.; Menz, J.; Ludewig, U. Plasticity of DNA methylation and gene expression under zinc deficiency in Arabidopsis roots. Plant Cell Physiol. 2018, 59, 790–1802. [Google Scholar] [CrossRef]
- Mager, S.; Ludewig, U. Massive Loss of DNA Methylation in Nitrogen-, but Not in Phosphorus-Deficient Zea mays Roots Is Poorly Correlated With Gene Expression Differences. Front Plant Sci. 2018, 9, 497. [Google Scholar] [CrossRef]
- Yong-Villalobos, L.; Gonzalez-Morales, S.I.; Wrobel, K.; Gutierrez-Alanis, D.; Cervantes-Perez, S.A.; Hayano-Kanashiro, C.; Oropeza-Aburto, A.; Cruz-Ramirez, A.; Martinez, O.; Herrera-Estrella, L. Methylome analysis reveals an important role for epigenetic changes in the regulation of the Arabidopsis response to phosphate starvation. Proc. Natl. Acad. Sci. USA 2015, 112, 7293–7302. [Google Scholar] [CrossRef]
- Ferreira, L.J.; Azevedo, V.; Maroco, J.; Oliveira, M.M.; Santos, A.P. Salt tolerant and sensitive rice varieties display differential methylome flexibility under salt stress. PLoS ONE 2015, 10, e0124060. [Google Scholar] [CrossRef]
- Wibowo, A.; Becker, C.; Marconi, G.; Durr, J.; Price, J.; Hagmann, J.; Papareddy, R.; Putra, H.; Kageyama, J.; Becker, J.; et al. Hyperosmotic stress memory in Arabidopsis is mediated by distinct epigenetically labile sites in the genome and is restricted in the male germline by DNA glycosylase activity. Elife 2016, 5, E13546. [Google Scholar] [CrossRef]
- Kapazoglou, A.; Drosou, V.; Argiriou, A.; Tsaftaris, A.S. The study of a barley epigenetic regulator, HvDME, in seed development and under drought. BMC Plant Biol. 2013, 13, 172. [Google Scholar] [CrossRef]
- Chwialkowska, K.; Nowakowska, U.; Mroziewicz, A.; Szarejko, I.; Kwasniewski, M. Water-deficiency conditions differently modulate the methylome of roots and leaves in barley (Hordeum vulgare L.). J. Exp. Bot. 2016, 67, 1109–1121. [Google Scholar] [CrossRef]
- Xu, P.; Chen, H.; Jin, J.; Cai, W. Single-base resolution methylome analysis shows epigenetic changes in Arabidopsis seedlings exposed to microgravity spaceflight conditions on board the SJ-10 recoverable satellite. NPJ Microgravity 2018, 4, 12. [Google Scholar] [CrossRef]
- Naydenov, M.; Baev, V.; Apostolova, E.; Gospodinova, N.; Sablok, G.; Gozmanova, M.; Yahubyan, G. High-temperature effect on genes engaged in DNA methylation and affected by DNA methylation in Arabidopsis. Plant Physiol. Biochem. 2015, 87, 102–108. [Google Scholar] [CrossRef]
- Zhang, B.; Tieman, D.M.; Jiao, C.; Xu, Y.; Chen, K.; Fei, Z.; Giovannoni, J.J.; Klee, H.J. Chilling-induced tomato flavor loss is associated with altered volatile synthesis and transient changes in DNA methylation. Proc. Natl. Acad. Sci. USA 2016, 113, 12580–12585. [Google Scholar] [CrossRef]
- Liang, X.; Hou, X.; Li, J.; Han, Y.; Zhang, Y.; Feng, N.; Du, J.; Zhang, W.; Zheng, D.; Fang, S. High-resolution DNA methylome reveals that demethylation enhances adaptability to continuous cropping comprehensive stress in soybean. BMC Plant Biol. 2019, 19, 79. [Google Scholar] [CrossRef]
- Ou, X.; Zhang, Y.; Xu, C.; Lin, X.; Zang, Q.; Zhuang, T.; Jiang, L.; von Wettstein, D.; Liu, B. Transgenerational inheritance of modified DNA methylation patterns and enhanced tolerance induced by heavy metal stress in rice (Oryza sativa L.). PLoS ONE 2012, 7, e41143. [Google Scholar] [CrossRef]
- Xu, W.; Wang, T.; Xu, S.; Xu, S.; Wu, L.; Wu, Y.; Bian, P. Radiation-induced epigenetic bystander effects demonstrated in Arabidopsis thaliana. Radiat. Res. 2015, 183, 511–524. [Google Scholar] [CrossRef]
- Kim, J.S.; Lim, J.Y.; Shin, H.; Kim, B.G.; Yoo, S.D.; Kim, W.T.; Huh, J.H. ROS1-dependent DNA demethylation is required for ABA-inducible NIC3 expression. Plant Physiol. 2019, 179, 1810–1821. [Google Scholar] [CrossRef]
- Kim, J.Y.; Kwak, K.J.; Jung, H.J.; Lee, H.J.; Kang, H. MicroRNA402 affects seed germination of Arabidopsis thaliana under stress conditions via targeting DEMETER-LIKE Protein3 mRNA. Plant Cell Physiol. 2010, 51, 1079–1083. [Google Scholar] [CrossRef]
- Mok, Y.G.; Choi, K.Y.; Hong, S.H.; Huh, J.H. DEMETER plant DNA demethylase induces antiviral response by interferon signalling in animal cells. Sci. Rep. 2017, 7, 9160. [Google Scholar] [CrossRef]
- Morales-Ruiz, T.; Garcia-Ortiz, M.V.; Devesa-Guerra, I.; Raya-Ruiz, L.; Tejedor, J.R.; Bayon, G.F.; Sierra, M.I.; Fraga, M.F.; Ariza, R.R.; Roldan-Arjona, T. DNA methylation reprogramming of human cancer cells by expression of a plant 5-methylcytosine DNA glycosylase. Epigenetics 2018, 13, 95–107. [Google Scholar] [CrossRef]
- Parrilla-Doblas, J.T.; Ariza, R.R.; Roldan-Arjona, T. Targeted DNA demethylation in human cells by fusion of a plant 5-methylcytosine DNA glycosylase to a sequence-specific DNA binding domain. Epigenetics 2017, 12, 296–303. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parrilla-Doblas, J.T.; Roldán-Arjona, T.; Ariza, R.R.; Córdoba-Cañero, D. Active DNA Demethylation in Plants. Int. J. Mol. Sci. 2019, 20, 4683. https://doi.org/10.3390/ijms20194683
Parrilla-Doblas JT, Roldán-Arjona T, Ariza RR, Córdoba-Cañero D. Active DNA Demethylation in Plants. International Journal of Molecular Sciences. 2019; 20(19):4683. https://doi.org/10.3390/ijms20194683
Chicago/Turabian StyleParrilla-Doblas, Jara Teresa, Teresa Roldán-Arjona, Rafael R. Ariza, and Dolores Córdoba-Cañero. 2019. "Active DNA Demethylation in Plants" International Journal of Molecular Sciences 20, no. 19: 4683. https://doi.org/10.3390/ijms20194683
APA StyleParrilla-Doblas, J. T., Roldán-Arjona, T., Ariza, R. R., & Córdoba-Cañero, D. (2019). Active DNA Demethylation in Plants. International Journal of Molecular Sciences, 20(19), 4683. https://doi.org/10.3390/ijms20194683




