Genetic Dissection of Seed Storability and Validation of Candidate Gene Associated with Antioxidant Capability in Rice (Oryza sativa L.)
Abstract
1. Introduction
2. Results
2.1. Construction of High-Density Bin Map in BRILs
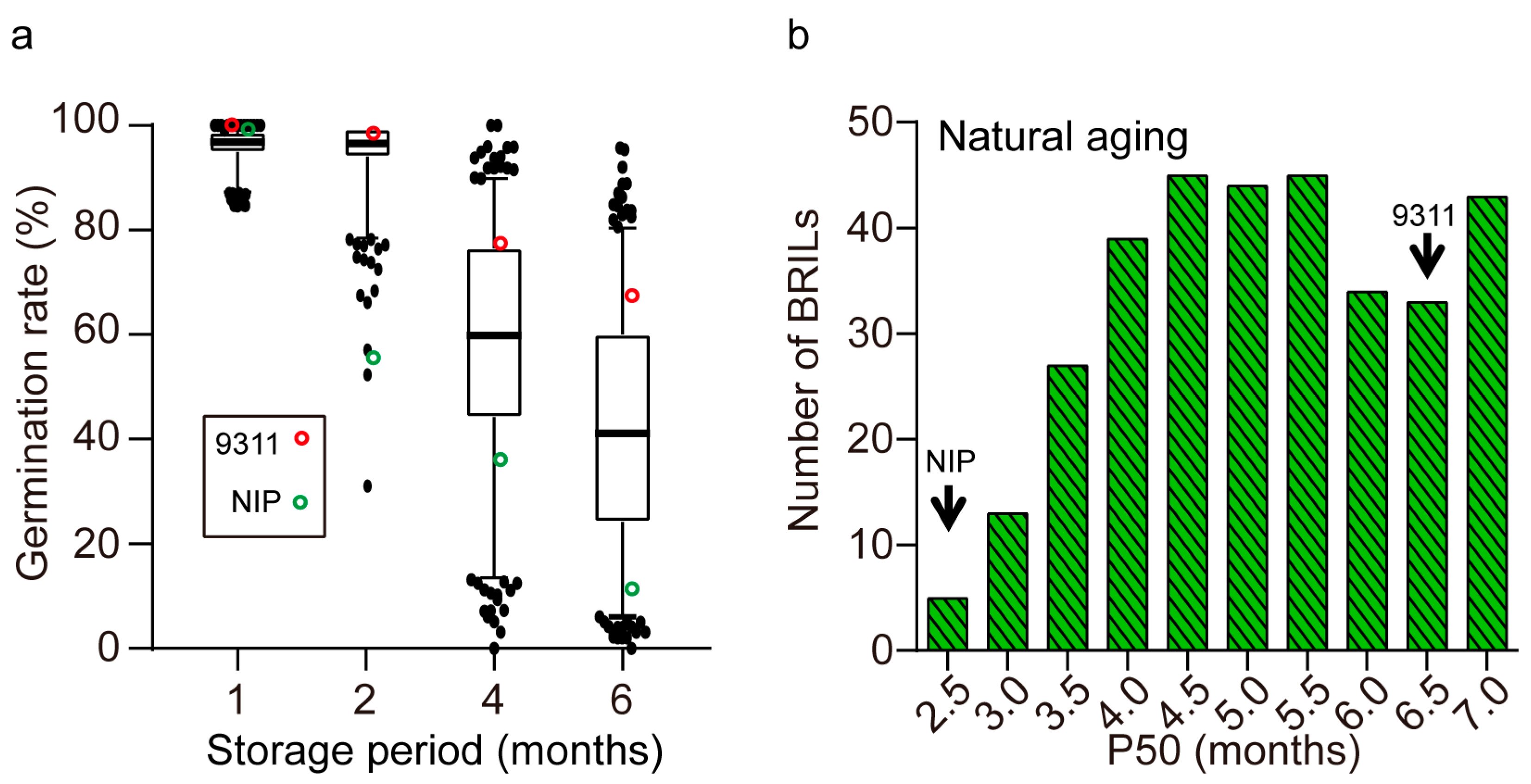
2.2. Seed Storability of the Parents and BRILs
2.3. QTLs for Seed Storability
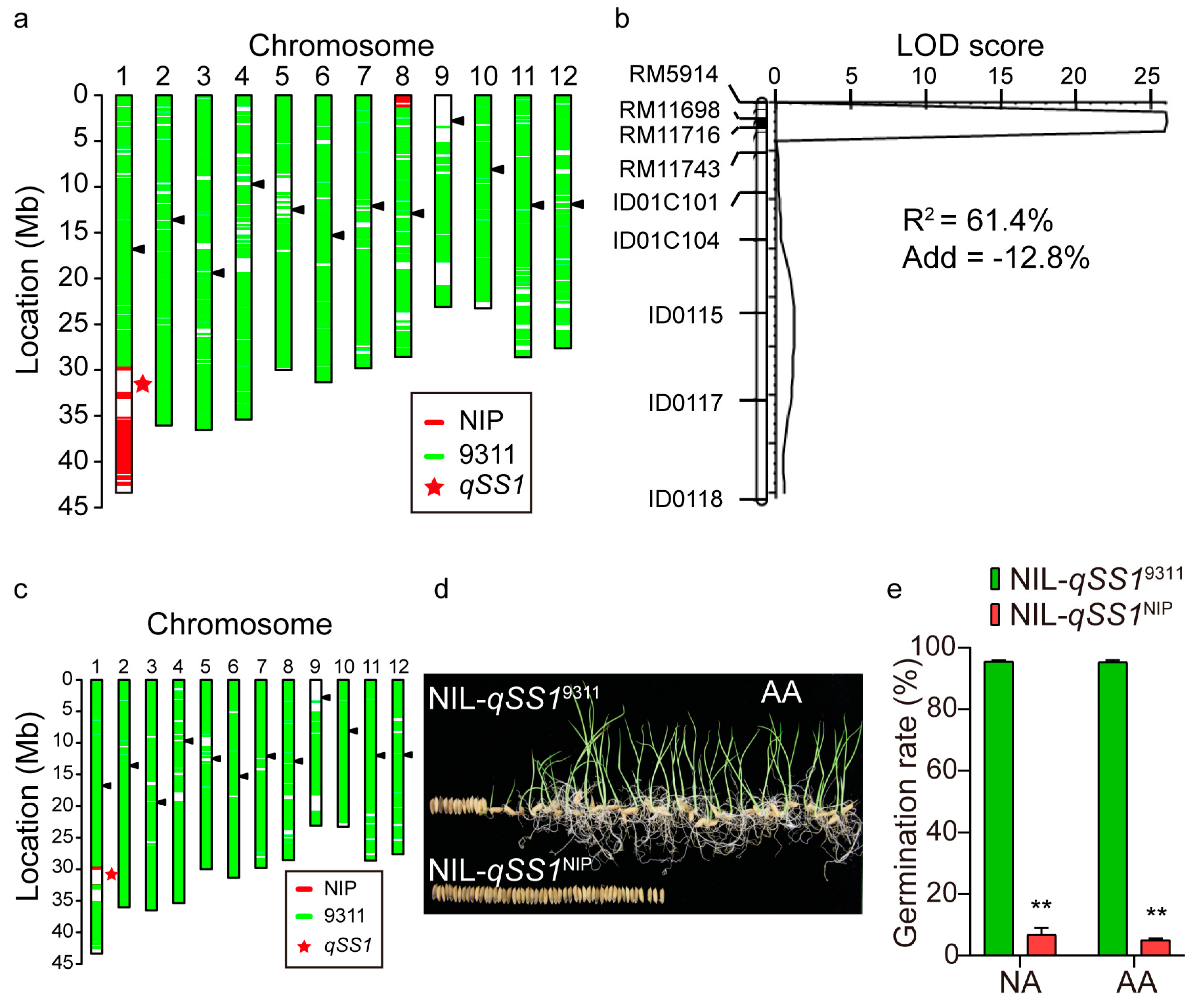
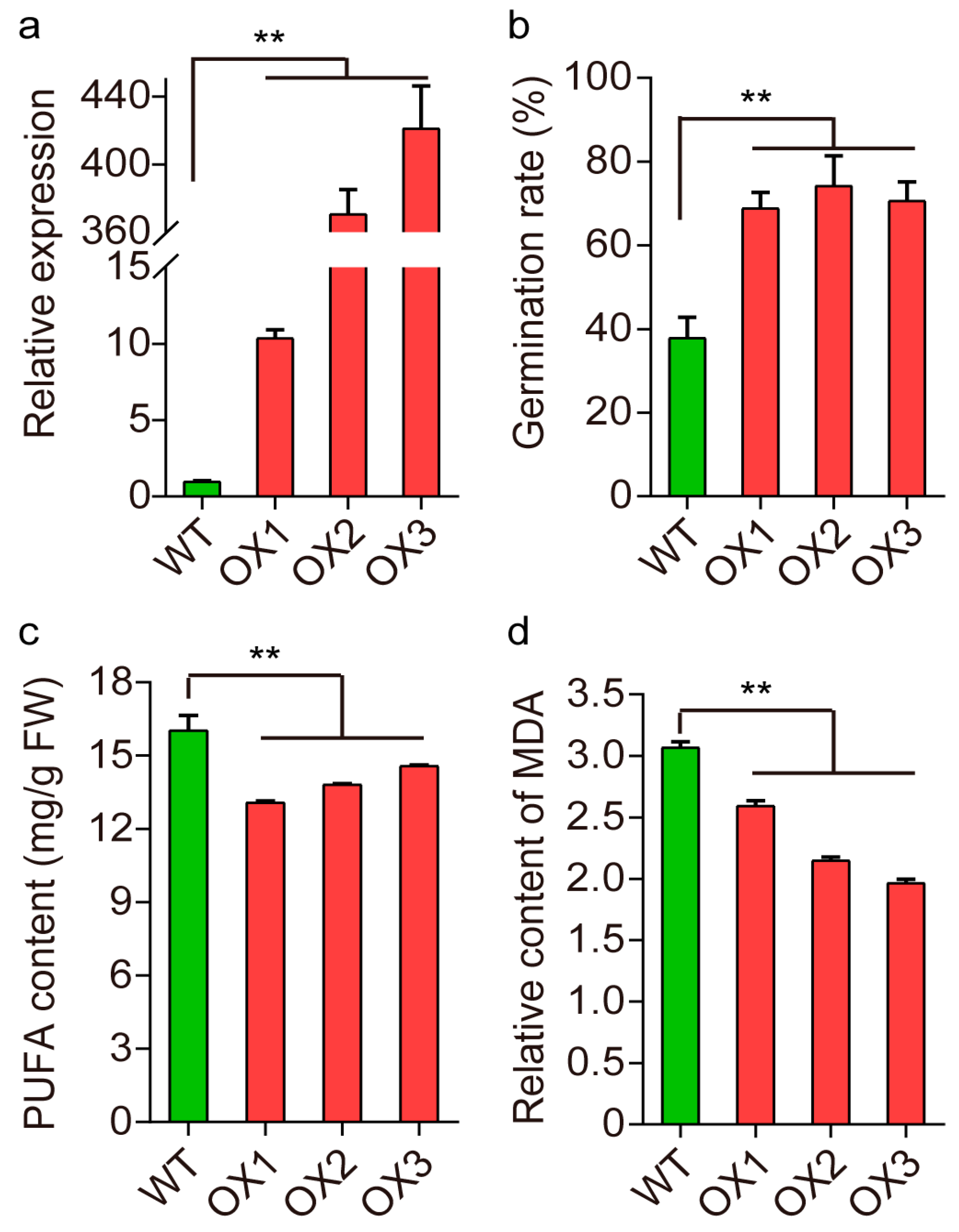
2.4. Validation of qSS1 for Seed Storability
2.5. QTLs for Germination Tolerance to Hydrogen Peroxide
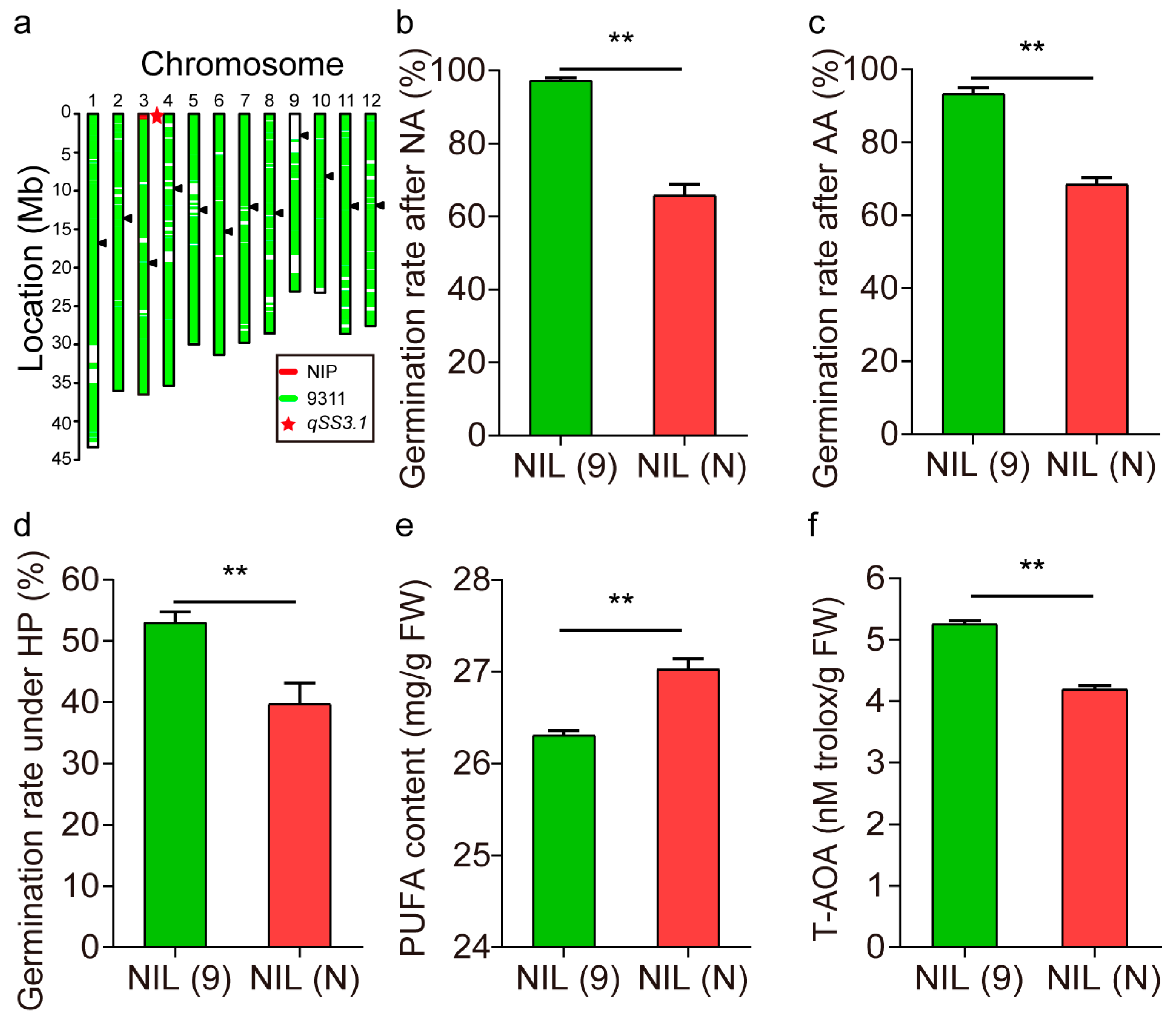
2.6. Validation of qSS3.1
2.7. Candidate Gene for qSS3.1
3. Discussion
3.1. High-Density Bin Map Developed in BRILs for QTL Detection
3.2. Seed Storability and Antioxidant Ability
4. Materials and Methods
4.1. Plant Materials
4.2. Seed Storability Evaluation
4.3. Determination of Total Antioxidant Activities
4.4. Measurement of Relative Electrolyte Leakage and MDA Content
4.5. Total PUFA Measurement Using the GC-MS Method
4.6. DNA Extraction and SNP Genotyping
4.7. QTL Analyses
4.8. Vector Construction and Rice Transformation
4.9. Quantitative Real-Time PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BRIL | Backcross Recombinant Inbred Line |
| AA | Artificial Aging |
| AOC | Antioxidant Capability |
| FAH | Fatty Acid Hydroxylase |
| GBS | Genotyping by Sequencing |
| LOX | Lipoxygenases |
| LOD | Logarithm of Odds |
| MDA | Malondialdehyde |
| NA | Natural Aging |
| NIL | Near-isogenic Line |
| PUFA | Polyunsaturated Fatty Acids |
| PVE | Phenotypic Variance Explained |
| QTLs | Quantitative Trait Loci |
| ROS | Reactive Oxygen Species |
| SNP | Single Nucleotide Polymorphism |
| SS | Seed Storability |
| T-AOA | Total Antioxidant Activity |
| TCP | Teosinte branched1/Cycloidea/Proliferating cell factor |
References
- Siddique, S.B.; Seshu, D.V.; Pardee, W.D. Rice cultivars variability in tolerance for accelerated aging of seed [Philippines]. IRRI Res. Pap. Ser. (Philipp.) 1988, 131, 2–7. [Google Scholar]
- Rajjou, L.; Debeaujon, I. Seed longevity: Survival and maintenance of high germination ability of dry seeds. C. R. Biol. 2008, 331, 796–805. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.B.; Wei, Y.D.; Zhu, Y.S.; Lian, L.; Xie, H.G.; Cai, Q.H.; Chen, Q.S.; Lin, Z.P.; Wang, Z.H.; Xie, H.A.; et al. Antisense suppression of LOX3 gene expression in rice endosperm enhances seed longevity. Plant Biotech. J. 2015, 13, 526–539. [Google Scholar] [CrossRef] [PubMed]
- Ellis, R.H.; Hong, T.D.; Jackson, M.T. Seed production environment, time of harvest, and the potential longevity of seeds of 3 cultivars of rice (Oryza sativa L). Ann. Bot. 1993, 72, 583–590. [Google Scholar] [CrossRef]
- Rao, N.K.; Jackson, M.T. Variation in seed longevity of rice cultivars belonging to different isozyme groups. Genet. Resour. Crop Evol. 1997, 44, 159–164. [Google Scholar]
- Miura, K.; Lin, S.Y.; Yano, M.; Nagamine, T. Mapping quantitative trait loci controlling seed longevity in rice (Oryza sativa L.). Theor. Appl. Genet. 2002, 104, 981–986. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, K.; Fukuta, Y.; Sato, T. Mapping of quantitative trait loci controlling seed longevity of rice (Oryza sativa L.) after various periods of seed storage. Plant Breed. 2005, 124, 361–366. [Google Scholar] [CrossRef]
- Zeng, D.L.; Guo, L.B.; Xu, Y.B.; Yasukumi, K.; Zhu, L.H.; Qian, Q. QTL analysis of seed storability in rice. Plant Breed. 2006, 125, 57–60. [Google Scholar] [CrossRef]
- Hang, N.T.; Lin, Q.Y.; Liu, L.L.; Liu, X.; Liu, S.J.; Wang, W.Y.; Li, L.F.; He, N.Q.; Liu, Z.; Jiang, L. Mapping QTLs related to rice seed storability under natural and artificial aging storage conditions. Euphytica 2015, 203, 673–681. [Google Scholar] [CrossRef]
- Li, C.S.; Shao, G.S.; Wang, L.; Wang, Z.F.; Mao, Y.J.; Wang, X.Q.; Zhang, X.H.; Liu, S.T.; Zhang, H.S. QTL identification and fine mapping for seed storability in rice (Oryza sativa L.). Euphytica 2017, 213, 12. [Google Scholar] [CrossRef]
- Li, L.F.; Lin, Q.Y.; Liu, S.J.; Liu, X.; Wang, W.Y.; Hang, N.T.; Liu, F.; Zhao, Z.G.; Jiang, L.; Wan, J.M. Identification of quantitative trait loci for seed storability in rice (Oryza sativa L.). Plant Breed. 2012, 131, 739–743. [Google Scholar] [CrossRef]
- Lin, Q.Y.; Wang, W.Y.; Ren, Y.K.; Jiang, Y.M.; Sun, A.L.; Qian, Y.; Zhang, Y.F.; He, N.Q.; Hang, N.T.; Liu, Z.; et al. Genetic dissection of seed storability using two different populations with a same parent rice cultivar N22. Breed. Sci. 2015, 65, 411–419. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Zhang, S.Q.; Yao, Q.H.; Peng, R.H.; Xiong, A.S.; Li, X.; Zhu, W.M.; Zhu, Y.Y.; Zha, D.S. Identification of quantitative trait loci for seed storability in rice (Oryza sativa L.). Euphytica 2008, 164, 739–744. [Google Scholar] [CrossRef]
- Sasaki, K.; Takeuchi, Y.; Miura, K.; Yamaguchi, T.; Ando, T.; Ebitani, T.; Higashitani, A.; Yamaya, T.; Yano, M.; Sato, T. Fine mapping of a major quantitative trait locus, qLG-9, that controls seed longevity in rice (Oryza sativa L.). Theor. Appl. Genet. 2015, 128, 769–778. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.Y.; Jiang, Y.M.; Sun, A.L.; Cao, P.H.; Li, L.F.; Liu, X.; Tian, Y.L.; He, J.; Liu, S.J.; Chen, L.M. Fine mapping of qSS-9, a major and stable quantitative trait locus, for seed storability in rice (Oryza sativa L.). Plant Breed. 2015, 134, 293–299. [Google Scholar] [CrossRef]
- Kumar, S.P.J.; Prasad, S.R.; Banerjee, R.; Thammineni, C. Seed birth to death: Dual functions of reactive oxygen species in seed physiology. Ann. Bot. 2015, 116, 663–668. [Google Scholar] [CrossRef] [PubMed]
- McDonald, M.B. Seed deterioration: Physiology, repair and assessment. Seed Sci. Technol. 1999, 27, 177–237. [Google Scholar]
- Moller, I.M.; Jensen, P.E.; Hansson, A. Oxidative modifications to cellular components in plants. Annu. Rev. Plant Biol. 2007, 58, 459–481. [Google Scholar] [CrossRef]
- Xin, X.; Tian, Q.; Yin, G.; Chen, X.; Zhang, J.; Ng, S.; Lu, X. Reduced mitochondrial and ascorbate-glutathione activity after artificial ageing in soybean seed. J. Plant Physiol. 2014, 171, 140–147. [Google Scholar] [CrossRef]
- Yin, G.; Xin, X.; Song, C.; Chen, X.; Zhang, J.; Wu, S.; Li, R.; Liu, X.; Lu, X. Activity levels and expression of antioxidant enzymes in the ascorbate-glutathione cycle in artificially aged rice seed. Plant Physiol. Biochem. 2014, 80, 1–9. [Google Scholar] [CrossRef]
- Liavonchanka, A.; Feussner, N. Lipoxygenases: Occurrence, functions and catalysis. J Plant Physiol. 2006, 163, 348–357. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Miura, K.; Shigemune, A.; Sasahara, H.; Ohta, H.; Uehara, Y.; Ishikawa, T.; Hamada, S.; Shirasawa, K. Marker-assisted breeding of a LOX-3-null rice line with improved storability and resistance to preharvest sprouting. Theor. Appl. Genet. 2015, 128, 1421–1430. [Google Scholar] [CrossRef] [PubMed]
- Long, Q.Z.; Zhang, W.W.; Wang, P.; Shen, W.B.; Zhou, T.; Liu, N.N.; Wang, R.; Jiang, L.; Huang, J.X.; Wang, Y.H.; et al. Molecular genetic characterization of rice seed lipoxygenase 3 and assessment of its effects on seed longevity. J. Plant Biol. 2013, 56, 232–242. [Google Scholar] [CrossRef]
- Kim, K.R.; Oh, D.K. Production of hydroxy fatty acids by microbial fatty acid-hydroxylation enzymes. Biotechnol. Adv. 2013, 31, 1473–1485. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Matsuda, M.; Hatanaka, S.; Kanauchi, M.; Kasahara, S.; Shimoyamada, M. Cloning and Sequence Analysis of Fatty Acid Hydroxylase Gene in Lactobacillus sakei Y-20 Strain and Characteristics of Fatty Acid Hydroxylase. J. Am. Soc. Brew. Chem. 2016, 74, 77–84. [Google Scholar] [CrossRef]
- Greco, M.; Chiappetta, A.; Bruno, L.; Bitonti, M.B. In Posidonia oceanica cadmium induces changes in DNA methylation and chromatin patterning. J. Exp. Bot. 2012, 63, 695–709. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, M.; Cubas, P. TCP factors: New kids on the signaling block. Curr. Opin. Plant Biol. 2016, 33, 33–41. [Google Scholar] [CrossRef]
- Resentini, F.; Felipo-Benavent, A.; Colombo, L.; Blazquez, M.A.; Alabadi, D.; Masiero, S. TCP14 and TCP15 mediate the promotion of seed germination by gibberellins in Arabidopsis thaliana. Mol. Plant 2015, 8, 482–485. [Google Scholar] [CrossRef]
- Hu, Z.; Yamauchi, T.; Yang, J.; Jikumaru, Y.; Tsuchida-Mayama, T.; Ichikawa, H.; Takamure, I.; Nagamura, Y.; Tsutsumi, N.; Yamaguchi, S.; et al. Strigolactone and cytokinin act antagonistically in regulating rice mesocotyl elongation in darkness. Plant Cell Physiol. 2014, 55, 30–41. [Google Scholar] [CrossRef]
- Zhang, L.X.; Wang, S.F.; Jiang, L.; Wan, J.M. QTL analysis of cold tolerance at the bud bursting period in rice (Oryza sativa L.) by using recombinant inbred lines. J. Nanjing Agric. Univ. 2007, 30, 1–5. [Google Scholar]
- Wang, P.; Zhou, G.L.; Yu, H.H.; Yu, S.B. Fine mapping a major QTL for flag leaf size and yield-related traits in rice. Theor. Appl. Genet. 2011, 123, 1319–1330. [Google Scholar] [PubMed]
- Zhang, G.H.; Li, S.Y.; Wang, L.; Ye, W.J.; Zeng, D.L.; Rao, Y.C.; Peng, Y.L.; Hu, J.; Yang, Y.L.; Xu, J.; et al. LSCHL4 from japonica cultivar, which Is allelic to NAL1, Increases yield of indica super rice 93–11. Mol. Plant 2014, 7, 1350–1364. [Google Scholar] [CrossRef] [PubMed]
- Sano, N.; Rajjou, L.; North, H.M.; Debeaujon, I.; Marion-Poll, A.; Seo, M. Staying alive: Molecular aspects of seed longevity. Plant Cell Physiol. 2016, 57, 660–674. [Google Scholar] [CrossRef] [PubMed]
- Rajjou, L.; Lovigny, Y.; Groot, S.P.; Belghazi, M.; Job, C.; Job, D. Proteome-wide characterization of seed aging in Arabidopsis: A comparison between artificial and natural aging protocols. Plant Physiol. 2008, 148, 620–641. [Google Scholar] [CrossRef] [PubMed]
- Joosen, R.V.L.; Kodde, J.; Willems, L.A.J.; Ligterink, W.; van der Plas, L.H.W.; Hilhorst, H.W.M. GERMINATOR: A software package for high-throughput scoring and curve fitting of Arabidopsis seed germination. Plant J. 2010, 62, 148–159. [Google Scholar] [CrossRef]
- Yang, Q.M.; Pan, X.H.; Kong, W.B.; Yang, H.; Su, Y.D.; Zhang, L.; Zhang, Y.N.; Yang, Y.L.; Ding, L.; Liu, G.A. Antioxidant activities of malt extract from barley (Hordeum vulgare L.) toward various oxidative stress in vitro and in vivo. Food Chem. 2010, 118, 84–89. [Google Scholar]
- Lv, Y.; Guo, Z.L.; Li, X.K.; Ye, H.Y.; Li, X.H.; Xiong, L.Z. New insights into the genetic basis of natural chilling and cold shock tolerance in rice by genome-wide association analysis. Plant Cell Environ. 2016, 39, 556–570. [Google Scholar] [CrossRef]
- Cui, M.Y.; Lin, Y.C.; Zu, Y.G.; Efferth, T.; Li, D.W.; Tang, Z.H. Ethylene increases accumulation of compatible solutes and decreases oxidative stress to improve plant tolerance to water stress in Arabidopsis. J. Plant Biol. 2015, 58, 193–201. [Google Scholar] [CrossRef]
- Lu, S.P.; Yao, S.B.; Wang, G.L.; Guo, L.; Zhou, Y.M.; Hong, Y.Y.; Wang, X.M. Phospholipase Dε enhances Braasca napus growth and seed production in response to nitrogen availability. Plant. Biotech. J. 2016, 14, 926–937. [Google Scholar] [CrossRef]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 2011, 6, 10. [Google Scholar] [CrossRef] [PubMed]
- Li, R.Q.; Yu, C.; Li, Y.R.; Lam, T.W.; Yiu, S.M.; Kristiansen, K.; Wang, J. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 2009, 25, 1966–1967. [Google Scholar] [CrossRef] [PubMed]
- Paran, I.; Zamir, D. Quantitative traits in plants: Beyond the QTL. Trends Genet. 2003, 19, 303–306. [Google Scholar] [CrossRef]
- Xie, W.; Feng, Q.; Yu, H.; Huang, X.; Zhao, Q.; Xing, Y.; Yu, S.; Han, B.; Zhang, Q. Parent-independent genotyping for constructing an ultrahigh-density linkage map based on population sequencing. Proc. Natl. Acad. Sci. USA 2010, 107, 10578–10583. [Google Scholar] [CrossRef] [PubMed]
- Broman, K.W.; Wu, H.; Sen, S.; Churchill, G.A. R/qtl: QTL mapping in experimental crosses. Bioinformatics 2003, 19, 889–890. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Basten, C.J.; Zeng, Z.B. Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC. 2012. Available online: http://statgen.ncsu.edu/qtlcart/winqtl cart.htm. (accessed on 1 August 2012).
- Zhou, Y.; Cai, H.; Xiao, J.; Li, X.; Zhang, Q.; Lian, X. Over-expression of aspartate aminotransferase genes in rice resulted in altered nitrogen metabolism and increased amino acid content in seeds. Theor. Appl. Genet. 2009, 118, 1381–1390. [Google Scholar] [CrossRef] [PubMed]
- Hiei, Y.; Ohta, S.; Komari, T.; Kumashiro, T. Efficient transformation of rice (Oryza sativa L.) mediated by agrobacterium and sequence-analysis of the boundaries of the T-DNA. Plant J. 1994, 6, 271–282. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆Ct method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
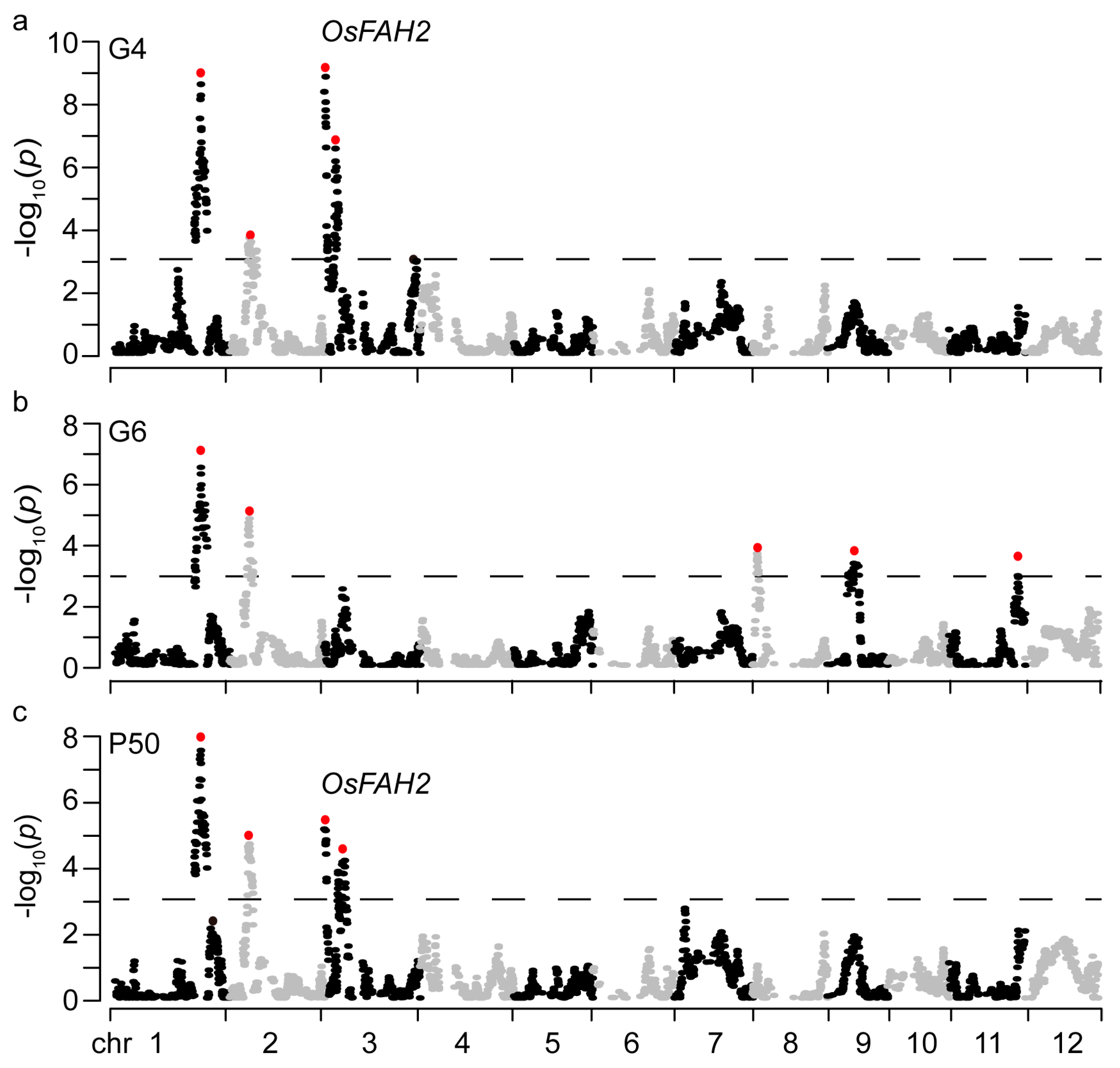
| Trait | QTL | Chr | Bin | Start (Mb) | Bin Size (kb) | Add | LOD | PVE (%) | Candidate Gene |
|---|---|---|---|---|---|---|---|---|---|
| G4 | qG4S1 | 1 | B01C270 | 32.09 | 23.93 | -0.09 | 8.7 | 9.0 | |
| qG4S2 | 2 | B02C083 | 7.32 | 47.8 | -0.06 | 3.6 | 3.6 | ||
| qG4S3.1 | 3 | B03C003 | 0.48 | 26.21 | -0.09 | 8.7 | 9.0 | OsFAH2 | |
| qG4S3.2 | 3 | B03C058 | 4.25 | 62.7 | 0.08 | 6.5 | 6.7 | ||
| G6 | qG6S1 | 1 | B01C270 | 32.09 | 23.93 | -0.08 | 7.0 | 8.5 | |
| qG6S2 | 2 | B02C083 | 7.32 | 47.8 | -0.08 | 5.0 | 5.9 | ||
| qG6S8 | 8 | B08C021 | 1.73 | 114.23 | 0.07 | 3.7 | 4.3 | ||
| qG6S9 | 9 | B09C054 | 9.97 | 153.97 | -0.07 | 3.7 | 4.4 | OsTPP7 | |
| qG6S11 | 11 | B11C210 | 25.84 | 19.6 | 0.06 | 3.5 | 4.2 | ||
| P50 | qSS1 | 1 | B01C270 | 32.09 | 23.93 | -0.39 | 7.9 | 8.7 | |
| qSS2 | 2 | B02C078 | 7.03 | 78.64 | -0.34 | 4.9 | 5.2 | ||
| qSS3.1 | 3 | B03C003 | 0.48 | 26.21 | -0.33 | 5.3 | 5.7 | OsFAH2 | |
| qSS3.2 | 3 | B03C094 | 6.84 | 22.69 | 0.37 | 4.5 | 4.8 | ||
| AOC | qAOC1 | 1 | B01C345 | 38.37 | 41.05 | -0.06 | 6.7 | 6.8 | SD1 |
| qAOC3.1 | 3 | B03C003 | 0.48 | 26.21 | -0.08 | 8.9 | 9.0 | OsFAH2 | |
| qAOC3.2 | 3 | B03C265 | 34.31 | 32.75 | 0.06 | 3.9 | 4.0 | ||
| qAOC4 | 4 | B04C113 | 20.11 | 49.21 | -0.04 | 3.0 | 3.0 | ||
| qAOC11 | 11 | B11C106 | 10.91 | 36.49 | -0.05 | 3.5 | 3.4 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, Z.; Fan, K.; Xia, L.; Ding, X.; Tian, L.; Sun, W.; He, H.; Yu, S. Genetic Dissection of Seed Storability and Validation of Candidate Gene Associated with Antioxidant Capability in Rice (Oryza sativa L.). Int. J. Mol. Sci. 2019, 20, 4442. https://doi.org/10.3390/ijms20184442
Yuan Z, Fan K, Xia L, Ding X, Tian L, Sun W, He H, Yu S. Genetic Dissection of Seed Storability and Validation of Candidate Gene Associated with Antioxidant Capability in Rice (Oryza sativa L.). International Journal of Molecular Sciences. 2019; 20(18):4442. https://doi.org/10.3390/ijms20184442
Chicago/Turabian StyleYuan, Zhiyang, Kai Fan, Laifu Xia, Xiali Ding, Li Tian, Wenqiang Sun, Hanzi He, and Sibin Yu. 2019. "Genetic Dissection of Seed Storability and Validation of Candidate Gene Associated with Antioxidant Capability in Rice (Oryza sativa L.)" International Journal of Molecular Sciences 20, no. 18: 4442. https://doi.org/10.3390/ijms20184442
APA StyleYuan, Z., Fan, K., Xia, L., Ding, X., Tian, L., Sun, W., He, H., & Yu, S. (2019). Genetic Dissection of Seed Storability and Validation of Candidate Gene Associated with Antioxidant Capability in Rice (Oryza sativa L.). International Journal of Molecular Sciences, 20(18), 4442. https://doi.org/10.3390/ijms20184442





