Changes in the Substrate Source Reveal Novel Interactions in the Sediment-Derived Methanogenic Microbial Community
Abstract
1. Introduction
2. Results
2.1. Methane Production
2.2. Physicochemical Conditions
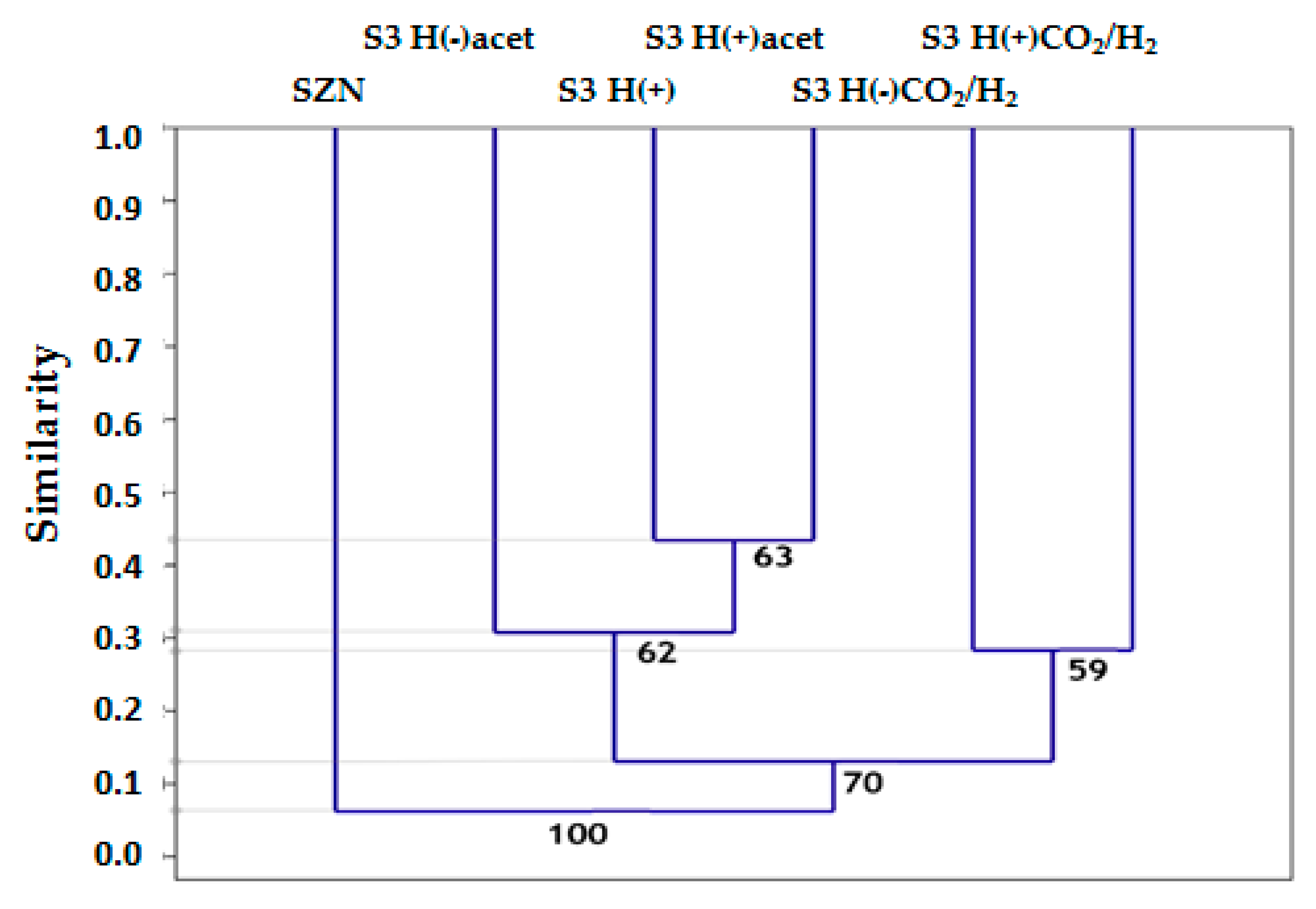
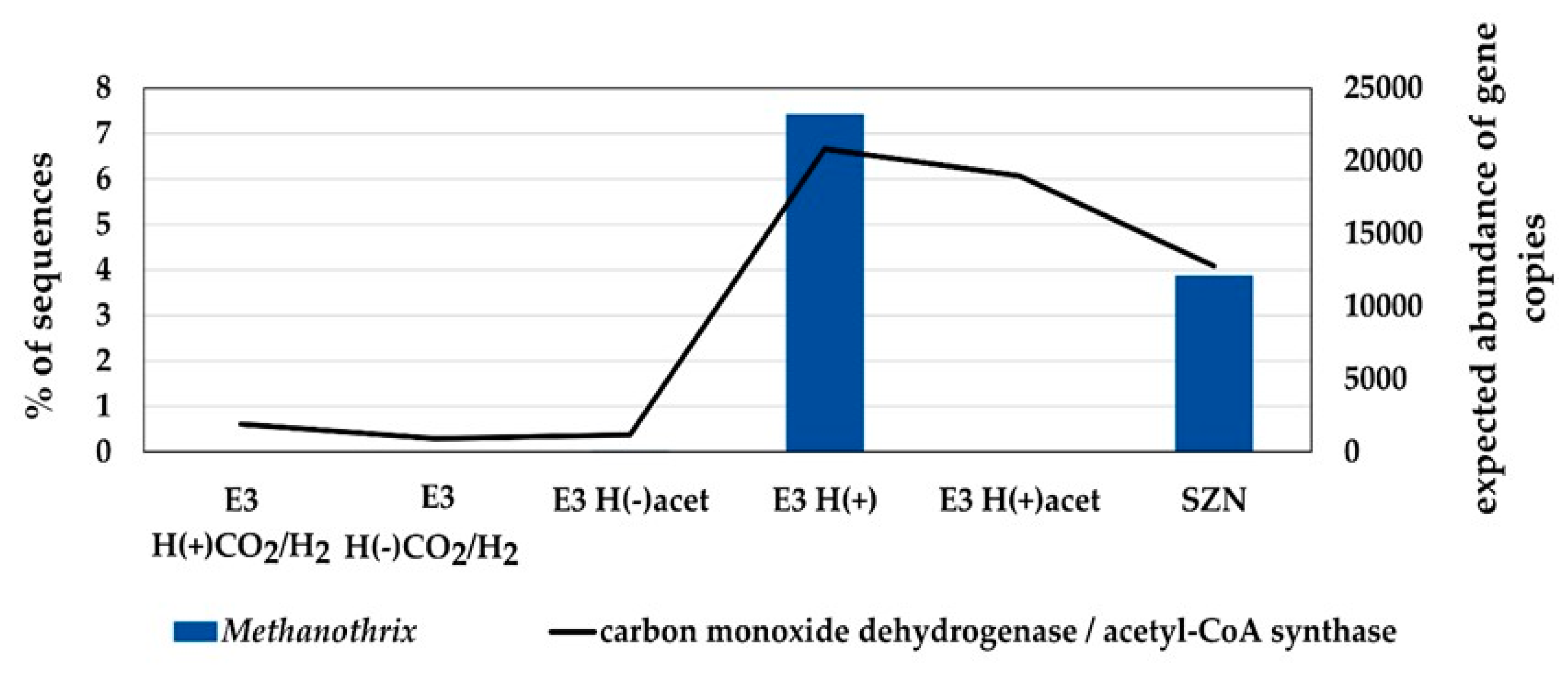
2.3. Microbial Community Structure Across Sediment and Media Combinations
2.4. Microbial Biodiversity
3. Discussion
3.1. Lake Sediment
3.2. Enrichment Cultures
3.3. Microbial Biodiversity
4. Materials and Methods
4.1. Physical and Chemical Analysis
4.2. Incubation Experiments
4.2.1. Assessment of the Methane Production Rate in Sediment
4.2.2. Cultivation Conditions of the Enrichment Cultures
4.2.3. Chromatographic Analyses and Calculation of the Methane Production
4.3. DNA Extraction and NGS Procedure
4.4. Bioinformatic Analysis
4.5. Metabolic Pathways Prediction Using PICRUSt Software
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| PICRUSt | Phylogenetic Investigation of Communities by Reconstruction of Unobserved States, software |
References
- Boughner, L.A.; Singh, P. Microbial Ecology: Where are we now? Postdoc. J. 2016, 4, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Eckert, I.M.K.; Littlefair, J.E.; Zhang, G.K.; Chain, F.J.J.; Crease, T.J.; Cristescu, M.E. Bioinformatics for biomonitoring: Species detection and diversity estimates across Next-Generation Sequencing Platforms. Adv. Ecol. Res. 2018, 59, 1–32. [Google Scholar] [CrossRef]
- Costessi, A.; van den Bogert, B.; May, A.; Ver Loren van Themaat, E.; Roubos, J.A.; Kolkman, M.A.B.; Butler, D.; Pirovano, W. Novel sequencing technologies to support industrial biotechnology. FEMS Microbiol. Lett. 2018, 365, fny103. [Google Scholar] [CrossRef] [PubMed]
- Detman, A.; Mielecki, D.; Pleśniak, Ł.; Bucha, M.; Janiga, M.; Matyasik, I.; Chojnacka, A.; Jędrysek, M.-O.; Błaszczyk, B.K.; Sikora, A. Methane-yielding microbial communities processing lactate-rich substrates: A piece of the anaerobic digestion puzzle. Biotechnol. Biofuels 2018, 11, 116. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Hou, X.; Su, H. Exploration of the relationship between biogas production and microbial community under high salinity conditions. Sci. Rep. 2017, 7, 1149. [Google Scholar] [CrossRef] [PubMed]
- Wojcieszak, M.; Pyzik, A.; Poszytek, K.; Krawczyk, P.S.; Sobczak, A.; Lipinski, L.; Roubinek, O.; Palige, J.; Sklodowska, A.; Drewniak, L. Adaptation of methanogenic inocula to anaerobic digestion of maize silage. Front. Microbiol. 2017, 8, 1881. [Google Scholar] [CrossRef] [PubMed]
- Fuertez, J.; Boakye, R.; McLennan, J.; Adamsc, D.J.; Sparks, T.D.; Gottschalke, A. Developing methanogenic microbial consortia from diverse coal sources and environments. J. Nat. Gas Sci. Eng. 2017, 46, 637–650. [Google Scholar] [CrossRef]
- Green, M.S.; Flanegan, K.C.; Gilcrease, P.C. Characterization of a methanogenic consortium enriched from a coalbed methane well in the Powder River Basin, U.S.A. Int. J. Coal Geol. 2008, 76, 34–45. [Google Scholar] [CrossRef]
- Plugge, C.M. Biogas. Microb. Biotechnol. 2017, 10, 1128–1130. [Google Scholar] [CrossRef]
- Rapheal, S.V.; Swaminathan, K.R.; Lalitha, K. Metabolic characteristics of an aerobe isolated from a methylotrophic methanogenic enrichment culture. J. Biosci. 2003, 28, 235–242. [Google Scholar] [CrossRef]
- Enzmann, F.; Mayer, F.; Rother, M.; Holtmann, D. Methanogens: Biochemical background and biotechnological applications. AMB Expr. 2018, 8, 1. [Google Scholar] [CrossRef] [PubMed]
- Evans, P.N.; Parks, D.H.; Chadwick, G.L.; Robbins, S.J.; Orphan, W.J.; Golding, S.D.; Tyson, G.W. Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science 2015, 350, 434–438. [Google Scholar] [CrossRef] [PubMed]
- Vanwonterghem, I.; Evans, P.N.; Parks, D.H.; Jensen, P.D.; Woodcroft, B.J.; Hugenholtz, P.; Tyson, G.W. Methylotrophic methanogenesis discovered in the archaeal phylum Verstraetearchaeota. Nat. Microbiol. 2016, 1, 16170. [Google Scholar] [CrossRef] [PubMed]
- Purwantini, E.; Torto-Alalibo, T.; Lomax, J.; Setubal, J.C.; Tyler, B.M.; Mukhopadhyay, B. Genetic resources for methane production from biomass described with the Gene Ontology. Front. Microbiol. 2014, 5, 634. [Google Scholar] [CrossRef] [PubMed]
- Conrad, R.; Klose, M.; Claus, P.; Enrich-Prast, A. Methanogenic pathway, C-13 isotope fractionation, and archaeal community composition in the sediment of two clear-water lakes of Amazonia. Limnol. Oceanogr. 2010, 55, 689–702. [Google Scholar] [CrossRef]
- Lovley, D.R.; Klug, M.J. Model for the distribution of sulfate reduction and methanogenesis in freshwater sediments. Geochim. Cosmochim. Acta 1986, 50, 11–18. [Google Scholar] [CrossRef]
- Esteves, A.I.S.; Hardoim, C.C.P.; Xavier, J.R.; Gonçalves, J.M.; Costa, R. Molecular richness and biotechnological potential of bacteria cultured from Irciniidae sponges in the north-east Atlantic. FEMS Microbiol. Ecol. 2013, 85, 519–536. [Google Scholar] [CrossRef] [PubMed]
- Zurita, G.; Pe’er, G.; Bellocq, M.I.; Hansbauer, M.M. Edge effects and their influence on habitat suitability calculations: A continuous approach applied to birds of the Atlantic forest. J Appl. Ecol. 2012, 49, 503–512. [Google Scholar] [CrossRef]
- Kark, S. Effects of ecotones on biodiversity. In Encyclopedia of Biodiversity; Levin, S.A., Ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2007; pp. 1–10. [Google Scholar]
- Dąbrowska-Prot, E.; Wasiłowska, A. Ecological importance of meadow patches in protected forest area: Floristic diversity and the dynamics of insect communities. Pol. J. Ecol. 2010, 58, 741–758. [Google Scholar]
- Lachavanne, J.-B.; Juge, R. Biodiversity in Land–Inland Water Ecotones; Taylor & Francis: Abingdon, UK, 1997; p. 326. [Google Scholar]
- Shen, C.; Shi, Y.; Ni, Y.; Deng, Y.; Van Nostrand, J.D.; He, Z.; Zhou, J.; Chu, H. Dramatic increases of soil microbial functional gene diversity at the treeline ecotone of Changbai Mountain. Front. Microbiol. 2016, 7, 1184. [Google Scholar] [CrossRef]
- Mieczan, T.; Adamczuk, M.; Tarkowska-Kukuryk, M.; Nawrot, D. Effect of water chemistry on zooplanktonic and microbial communities across freshwater ecotones in different macrophyte-dominated shallow lakes. J. Limnol. 2016, 75, 262–274. [Google Scholar] [CrossRef]
- Jiang, H.; Dong, H.; Zhang, G.; Yu, B.; Chapman, L.R.; Fields, M.W. Microbial diversity in water and sediment of Lake Chaka, an athalassohaline lake in northwestern China. Appl. Environ. Microbiol. 2006, 72, 3832–3845. [Google Scholar] [CrossRef] [PubMed]
- Laskar, F.; Das Purkayastha, S.; Sen, A.; Bhattacharya, M.K.; Misra, B.B. Diversity of methanogenic Archaea in freshwater sediments of lacustrine ecosystems. J. Basic Microbiol. 2018, 58, 101–119. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.S. Methane production in river and lake sediments in Taiwan. Environ. Geochem. Health 1998, 20, 245–249. [Google Scholar] [CrossRef]
- Schwarz, J.I.K.; Eckert, W.; Conrad, R. Response of the methanogenic microbial community of a profundal lake sediment (Lake Kinneret, Israel) to algal deposition. Limnol. Oceanogr. 2008, 53, 113–121. [Google Scholar] [CrossRef]
- Chan, O.C.; Wolf, M.; Hepperle, D.; Casper, P. Methanogenic archaeal community in the sediment of an artificially partitioned acidic bog lake. FEMS Microbiol. Ecol. 2002, 42, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Dai, Y.; Li, N.; Li, B.; Xie, S.; Liu, Y. Temporal and spatial dynamics of sediment anaerobic ammonium oxidation (Anammox) bacteria in freshwater lakes. Microb. Ecol. 2017, 73, 285–295. [Google Scholar] [CrossRef]
- Rojas, P.; Rodríguez, N.; de la Fuente, V.; Sánchez-Mata, D.; Amils, R.; Sanz, J.L. Microbial diversity associated with the anaerobic sediments of a soda lake (Mono Lake, California, USA). Can. J. Microbiol. 2018, 64, 385–392. [Google Scholar] [CrossRef]
- Ji, Y.; Angel, R.; Klose, M.; Claus, P.; Marotta, H.; Pinho, L.; Enrich-Prast, A.; Conrad, R. Structure and function of methanogenic microbial communities in sediments of Amazonian lakes with different water types. Environ. Microbiol. 2016, 18, 5082–5100. [Google Scholar] [CrossRef]
- Huang, W.; Chen, X.; Jiang, X.; Zheng, B. Characterization of sediment bacterial communities in plain lakes with different trophic statuses. MicrobiologyOpen 2017, 6, e00503. [Google Scholar] [CrossRef]
- Xing, L.; Yang, S.; Yin, Q.; Xie, S.; Strong, P.J.; Wu, G. Effects of carbon source on methanogenic activities and pathways incorporating metagenomic analysis of microbial community. Bioresour. Technol. 2017, 244, 982–988. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, L.; Osman, O.A.; Bertilsson, S.; Eiler, A. Microbial community composition and diversity via 16S rRNA gene amplicons: Evaluating the illumina platform. PLoS ONE 2015, 10, e0116955. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Ruan, X.; Zhang, Y.; Li, R. Illumina sequencing-based analysis of sediment bacteria community in different trophic status freshwater lakes. MicrobiologyOpen 2017, 6, e00450. [Google Scholar] [CrossRef] [PubMed]
- Lan, G.; Li, Y.; Jatoi, M.T.; Tan, Z.; Wu, Z.; Xie, G. Change in soil microbial community compositions and diversity following the conversion of tropical forest to rubber plantations in Xishuangbanan, Southwest China. Trop. Conserv. Sci. 2017, 10, 1–14. [Google Scholar] [CrossRef]
- Wu, X.; Xu, H.; Liu, G.; Ma, X.; Mu, C.; Zhao, L. Bacterial communities in the upper soil layers in the permafrost regions on the Qinghai-Tibetan plateau. Appl. Soil Ecol. 2017, 120, 81–88. [Google Scholar] [CrossRef]
- Chen, Y.; Dai, Y.; Wang, Y.; Wu, Z.; Xie, S.; Liu, Y. Distribution of bacterial communities across plateau freshwater lake and upslope soils. J. Environ. Sci. 2016, 43, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Parulekar, N.N.; Kolekar, P.; Jenkins, A.; Kleiven, S.; Utkilen, H.; Johansen, A.; Sawant, S.; Kulkarni-Kale, U.; Kale, M.; Sæbø, M. Characterization of bacterial community associated with phytoplankton bloom in a eutrophic lake in South Norway using 16S rRNA gene amplicon sequence analysis. PLoS ONE 2017, 12, e0173408. [Google Scholar] [CrossRef] [PubMed]
- Jetten, M.S.M.; Stams, A.J.M.; Zehnder, A.J.B. Methanogenesis from acetate: A comparison of the acetate metabolism in Methanothrix soehngenii and Methanosarcina spp. FEMS Microbiol. Lett. 1992, 88, 181–197. [Google Scholar] [CrossRef]
- Hubalek, V.; Buck, M.; Tan, B.; Foght, J.; Wendeberg, A.; Berry, D.; Bertilsson, S.; Eiler, A. Vitamin and Amino Acid Auxotrophy in Anaerobic Consortia Operating under Methanogenic Conditions. Msystems 2017, 2, e00038-17. [Google Scholar] [CrossRef]
- Fournier, G.P.; Gogarten, J.P. Evolution of acetoclastic methanogenesis in Methanosarcina via horizontal gene transfer from cellulolytic Clostridia. J. Bacteriol. 2008, 190, 1124–1127. [Google Scholar] [CrossRef]
- Walker, C.B.; Redding-Johanson, A.M.; Baidoo, E.E.; Rajeev, L.; Hendrickson, E.L.; Joachimiak, M.P.; Stolyar, S.; Arkin, A.P.; Leigh, J.A.; Zhou, J.A.; et al. Functional responses of methanogenic archaea to syntrophic growth. ISME J. 2012, 6, 2045–2055. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Wang, D.I. Elucidation of growth inhibition and acetic acid production by Clostridium thermoaceticum. Appl. Environ. Microbiol. 1984, 47, 294–298. [Google Scholar] [PubMed]
- Hugenholtz, P.; Pitulle, C.; Hershberger, K.L.; Pace, N.R. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 1998, 180, 366–376. [Google Scholar] [PubMed]
- Mori, K.; Yamaguchi, K.; Sakiyama, Y.; Urabe, T.; Suzuki, K. Caldisericum exile gen. nov., sp. nov., an anaerobic, thermophilic, filamentous bacterium of a novel bacterial phylum, Caldiserica phyl. nov., originally called the candidate phylum OP5, and description of Caldisericaceae fam. nov., Caldisericales ord. no. Int. J. Syst. Evol. Microbiol. 2009, 59, 2894–2898. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.-T.; Liu, L.-Y.; Rui, J.-P.; Yuan, Q.; Feng, D.-S.; Zhou, Z.; Dai, L.-R.; Zena, W.-Q.; Zhang, H.; Cheng, L. Coexistence and competition of sulfate-reducing and methanogenic populations in an anaerobic hexadecane-degrading culture. Biotechnol. Biofuels. 2017, 10, 207. [Google Scholar] [CrossRef] [PubMed]
- Finster, K. Microbiological disproportionation of inorganic sulfur compounds. J. Sulfur. Chem. 2008, 29, 281–292. [Google Scholar] [CrossRef]
- Loka Bharathi, P.A. Sulfur cycle. In Encyclopedia of Ecology; Jørgensen, S.E., Fath, B.D., Eds.; Academic Press: Oxford, UK, 2008; pp. 3424–3431. [Google Scholar]
- Mori, K.; Sunamura, M.; Yanagawa, K.; Ishibashi, J.; Miyoshi, Y.; Iino, T.; Suzuki, K.; Urabe, T. First Cultivation and ecological investigation of a bacterium affiliated with the candidate phylum OP5 from hot springs. Appl. Environ. Microbiol. 2008, 74, 6223–6229. [Google Scholar] [CrossRef] [PubMed]
- Stott, M.B.; Saito, J.A.; Crowe, M.A.; Dunfield, P.F.; Hou, S.; Nakasone, E.; Daughney, C.J.; Smirnova, A.V.; Mountain, B.W.; Takai, K.; et al. Culture-independent characterization of a novel microbial community at a hydrothermal vent at Brothers volcano, Kermadec arc, New Zealand. J. Geophys. Res. Solid Earth 2008, 113, B08S06. [Google Scholar] [CrossRef]
- Monteux, S.; Weedon, J.T.; Blume-Werry, G.; Gavazov, K.; Jassey, E.J.; Johansson, M.; Keuper, F.; Olid, C.; Dorrepaal, E. Long-term in situ permafrost thaw effects on bacterial communities and potential aerobic respiration. ISME J. 2018, 12, 2129–2141. [Google Scholar] [CrossRef]
- Martinez, M.A.; Woodcroft, B.J.; Ignacio Espinoza, J.C.; Zayed, A.A.; Singleton, C.M.; Boyd, J.A.; Li, Y.F.; Purvine, S.; Maughan, H.; Hodgkins, S.B.; et al. Discovery and ecogenomic context of a global Caldiserica-related phylum active in thawing permafrost, Candidatus Cryosericota phylum nov., Ca. Cryosericia class nov., Ca. Cryosericales ord. nov., Ca. Cryosericaceae fam. nov., comprising the four species Cryosericum septentrionale gen. nov. sp. nov., Ca. C. hinesii sp. nov., Ca. C. odellii sp. nov., Ca. C. terrychapinii sp. nov. Syst. Appl. Microbiol. 2019, 42, 54–56. [Google Scholar] [CrossRef]
- Tiodjio, R.E.; Sakatoku, A.; Nakamura, A.; Tanaka, D.; Fantong, W.Y.; Tchakam, K.B.; Tanyileke, G.; Ohba, T.; Hell, V.J.; Kusakaba, M.; et al. Bacterial and archaeal communities in Lake Nyos (Cameroon, Central Africa). Sci. Rep. 2014, 4, 6151. [Google Scholar] [CrossRef] [PubMed]
- Kurilkina, M.I.; Zakharova, Y.R.; Galachyants, Y.P.; Petrova, D.P.; Bukin, Y.S.; Domysheva, V.M.; Blinov, W.; Likhoshway, Y.V. Bacterial community composition in the water column of the deepest freshwater Lake Baikal as determined by next-generation sequencing. FEMS Microbiol. Ecol. 2016, 92, fiw094. [Google Scholar] [CrossRef] [PubMed]
- Dridi, B.; Fardeau, M.-L.; Ollivier, B.; Raoult, D.; Drancourt, M. Methanomassiliicoccus luminyensis gen. nov., sp. nov., a methanogenic archaeon isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2012, 62, 1902–1907. [Google Scholar] [CrossRef] [PubMed]
- Borrel, G.; Parisot, N.; Harris, H.M.B.; Peyretaillade, E.; Gaci, N.; Tottey, W.; Bardot, O.; Raymann, K.; Gribaldo, S.; Peyret, P.; et al. Comparative genomics highlights the unique biology of Methanomassiliicoccales, a Thermoplasmatales-related seventh order of methanogenic archaea that encodes pyrrolysine. BMC Genomics 2014, 15, 679. [Google Scholar] [CrossRef] [PubMed]
- Chojnacka, A.; Szczęsny, P.; Błaszczyk, M.K.; Zielenkiewicz, U.; Detman, A.; Salamon, A.; Sikora, A. Noteworthy facts about a methane-producing microbial community processing acidic effluent from sugar beet molasses fermentation. PLoS ONE 2015, 10, e0128008. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Li, H.; Zhang, Y.; Si, D.; Chen, Q. Evolution of microbial community along with increasing solid concentration during high-solids anaerobic digestion of sewage sludge. Bioresour. Technol. 2016, 216, 87–94. [Google Scholar] [CrossRef]
- Brugère, J.-F.; Borrel, G.; Gaci, N.; Tottey, W.; O’Toole, P.W.; Malpuech-Brugère, C. Archaebiotics: Proposed therapeutic use of archaea to prevent trimethylaminuria and cardiovascular disease. Gut Microbes 2014, 5, 5–10. [Google Scholar] [CrossRef]
- Dridi, B.; Henry, M.; Richet, H.; Raoult, H.; Drancourt, M. Age-related prevalence of Methanomassiliicoccus luminyensis in the human gut microbiome. Apmis 2012, 120, 773–777. [Google Scholar] [CrossRef]
- Bang, C.; Vierbuchen, T.; Gutsmann, T.; Heine, H.; Schmitz, R.A. Immunogenic properties of the human gut-associated archaeon Methanomassiliicoccus luminyensis and its susceptibility to antimicrobial peptides. PLoS ONE 2017, 12, e0185919. [Google Scholar] [CrossRef]
- Kröninger, L.; Gottschling, J.; Deppenmeier, U. Growth characteristics of Methanomassiliicoccus luminyensis and expression of methyltransferase encoding genes. Archaea 2017, 2017, 2756573. [Google Scholar] [CrossRef]
- Gorlas, A.; Robert, C.; Gimenez, G.; Drancourt, M.; Raoult, D. Complete genome sequence of Methanomassiliicoccus luminyensis, the largest genome of a human-associated Archaea species. J. Bacteriol. 2012, 194, 4745. [Google Scholar] [CrossRef] [PubMed]
- Paul, K.; Nonoh, J.O.; Mikulski, L.; Brune, A. ‘Methanoplasmatales,’ Thermoplasmatales-related archaea in termite guts and other environments, are the seventh order of methanogens. Appl. Environ. Microbiol. 2012, 78, 8245–8253. [Google Scholar] [CrossRef] [PubMed]
- Bowen De León, K.; Gerlach, R.; Peyton, B.; Fields, M. Archaeal and bacterial communities in three alkaline hot springs in Heart Lake Geyser Basin, Yellowstone National Park. Front. Microbiol. 2013, 4, 330. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.; Zhou, L.; Wang, L.-Y.; Liu, J.-F.; Gu, J.-D.; Mu, B.-Z.; Yang, S.-Z. Selective inhibition of methanogenesis by sulfate in enrichment culture with production water from low-temperature oil reservoir. Int. Biodeterior. Biodegrad. 2016, 108, 133–141. [Google Scholar] [CrossRef]
- Söllinger, A.; Schwab, C.; Weinmaier, T.; Loy, A.; Tveit, A.T.; Schleper, C.; Urich, T. Phylogenetic and genomic analysis of Methanomassiliicoccales in wetlands and animal intestinal tracts reveals clade-specific habitat preferences. FEMS Microbiol. Ecol. 2016, 92, fiv149. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Xing, P. Differences in the composition of archaeal communities in sediments from sontrasting zones of Lake Taihu. Front. Microbiol. 2016, 7, 1510. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Zhao, W.; Xia, D. The diversity of hydrogen-producing bacteria and methanogens within an in situ coal seam. Biotechnol. Biofuels 2018, 11, 245. [Google Scholar] [CrossRef] [PubMed]
- LWB. Integrated Report of the Lubelski Węgiel BOGDANKA Capital Group for the Year 2016; Lubelski Węgiel Bogdanka SA: Lublin Voivodeship, Poland, 2017; pp. 122–134. [Google Scholar]
- Ciosmak, M. The anthropogenic inundated area ”Szczecin” in the zone of active underground exploitation of hard coal in the Lublin coal basin. J. Ecol. Eng. 2016, 17, 70–80. [Google Scholar] [CrossRef]
- Sawicki, B. Geological and mining conditions of sublevel operation. In Proceedings of the 12th International Scientific and Technical Conference—Mining Natural Hazards, Katowice, Poland, 21–24 November 2005; The Central Mining Institute: Katowice, Poland, 2005; pp. 75–79. (In Polish). [Google Scholar]
- Szafranek-Nakonieczna, A.; Stępniewska, Z. The influence of the aeration status (ODR, Eh) of peat soils on their ability to produce methane. Wetl. Ecol. Manag. 2015, 23, 665–676. [Google Scholar] [CrossRef]
- Banach, A.M.; Banach, K.; Visser, E.J.W.; Stępniewska, Z.; Smits, A.J.M.; Roelofs, J.G.M.; Lamers, L.P.M. Effects of summer flooding on floodplain biogeochemistry in Poland; implications for increased flooding frequency. Biogeochemistry 2009, 92, 247–262. [Google Scholar] [CrossRef]
- Horn, M.A.; Matthies, C.; Küsel, K.; Schramm, A.; Drake, H.L. Hydrogenotrophic methanogenesis by moderately acid-tolerant methanogens of a methane-emitting acidic peat. Appl. Environ. Microbiol. 2003, 69, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Balch, W.E.; Fox, G.E.; Magrum, L.J.; Woese, C.R.; Wolfe, R.S. Methanogens: Reevaluation of a unique biological group. Microbiol. Rev. 1979, 43, 260–296. [Google Scholar] [PubMed]
- Wolfe, R.S. Techniques for cultivating methanogens. Methods Enzymol. 2011, 494, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Stępniewska, Z.; Goraj, W.; Kuźniar, A.; Łopacka, N.; Małysza, M. Enrichment culture and identification of endophytic methanotrophs isolated from peatland plants. Folia Microbiol. (Praha) 2017, 62, 381–391. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Szafranek-Nakonieczna, A.; Bennicelli, R.P. Ability of peat soil to oxidize methane and affect of temperature and layer deposition. Pol. J. Environ. Stud. 2010, 19, 805–810. [Google Scholar]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Development Core Team. A Language and Environment for Statistical Computing In R Foundation for Statistical Computing; R Development Core Team: Vienna, Austria, 2018; Available online: http: //www.R-project.org (accessed on 10 June 2018).
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, C.E.; Thurber, R.L.V.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. QIIME 2: Reproducible, interactive, scalable, and extensible microbiome data science. PeerJ Preprints 2018. [Google Scholar] [CrossRef]
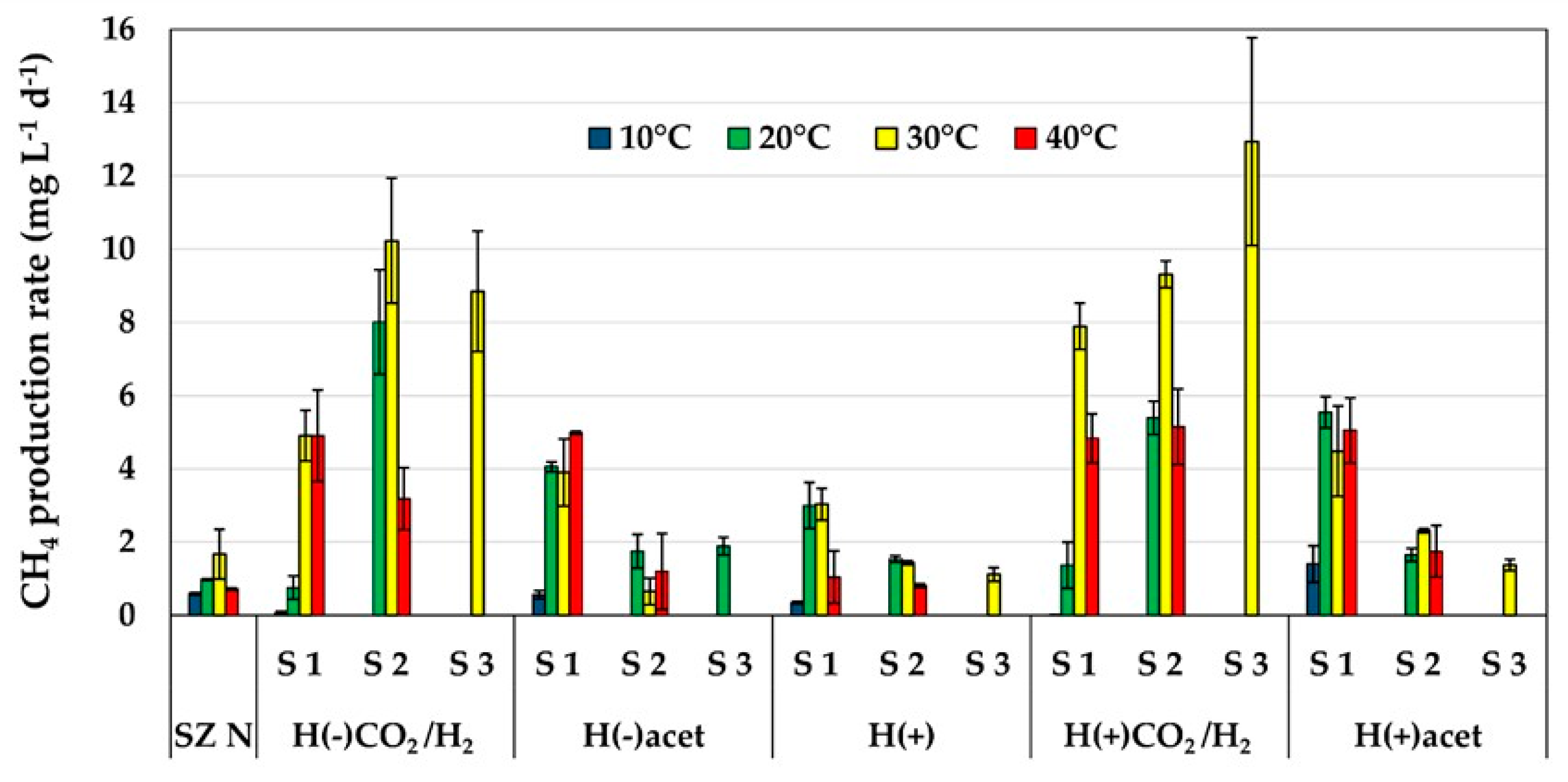
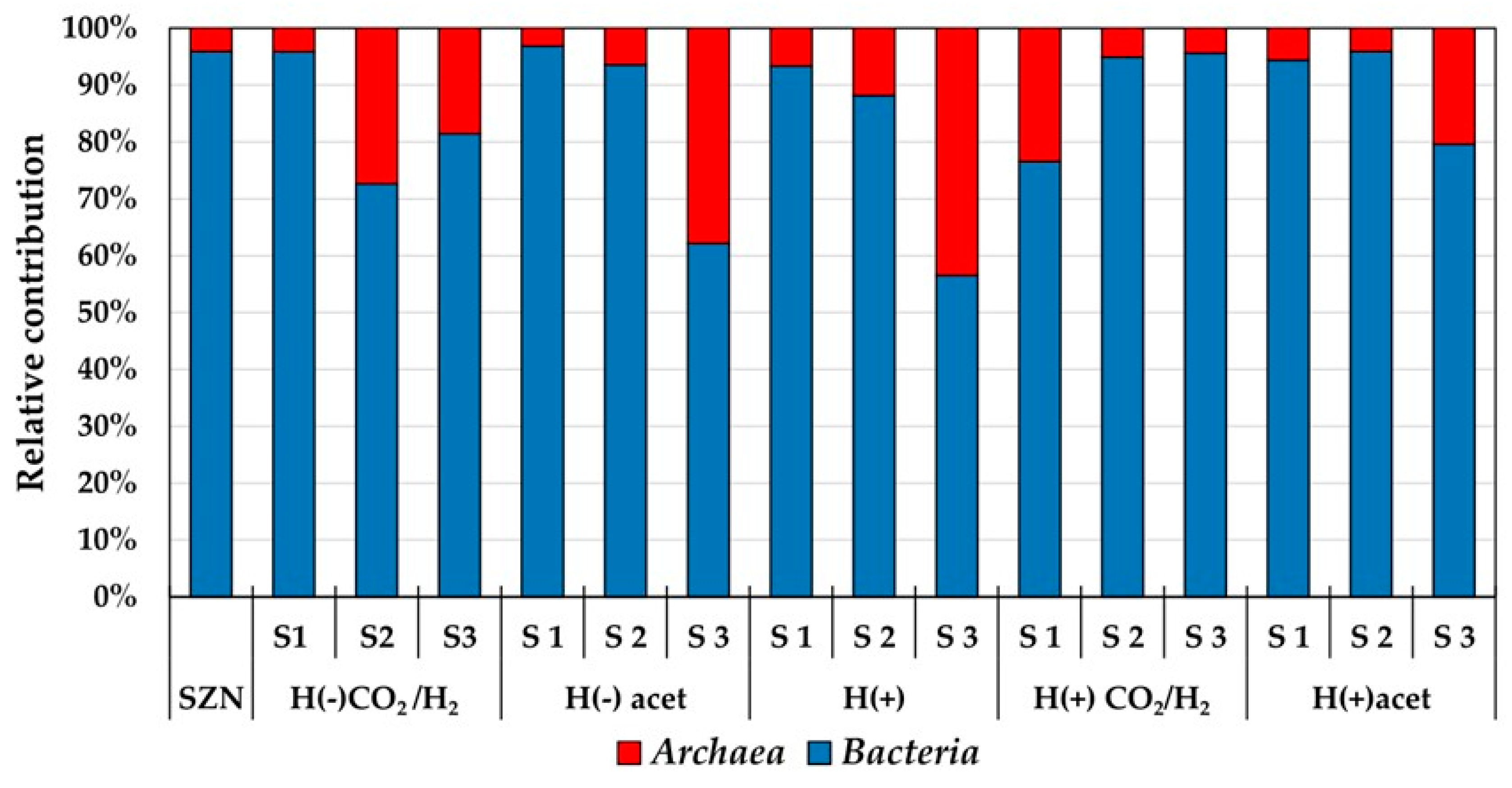
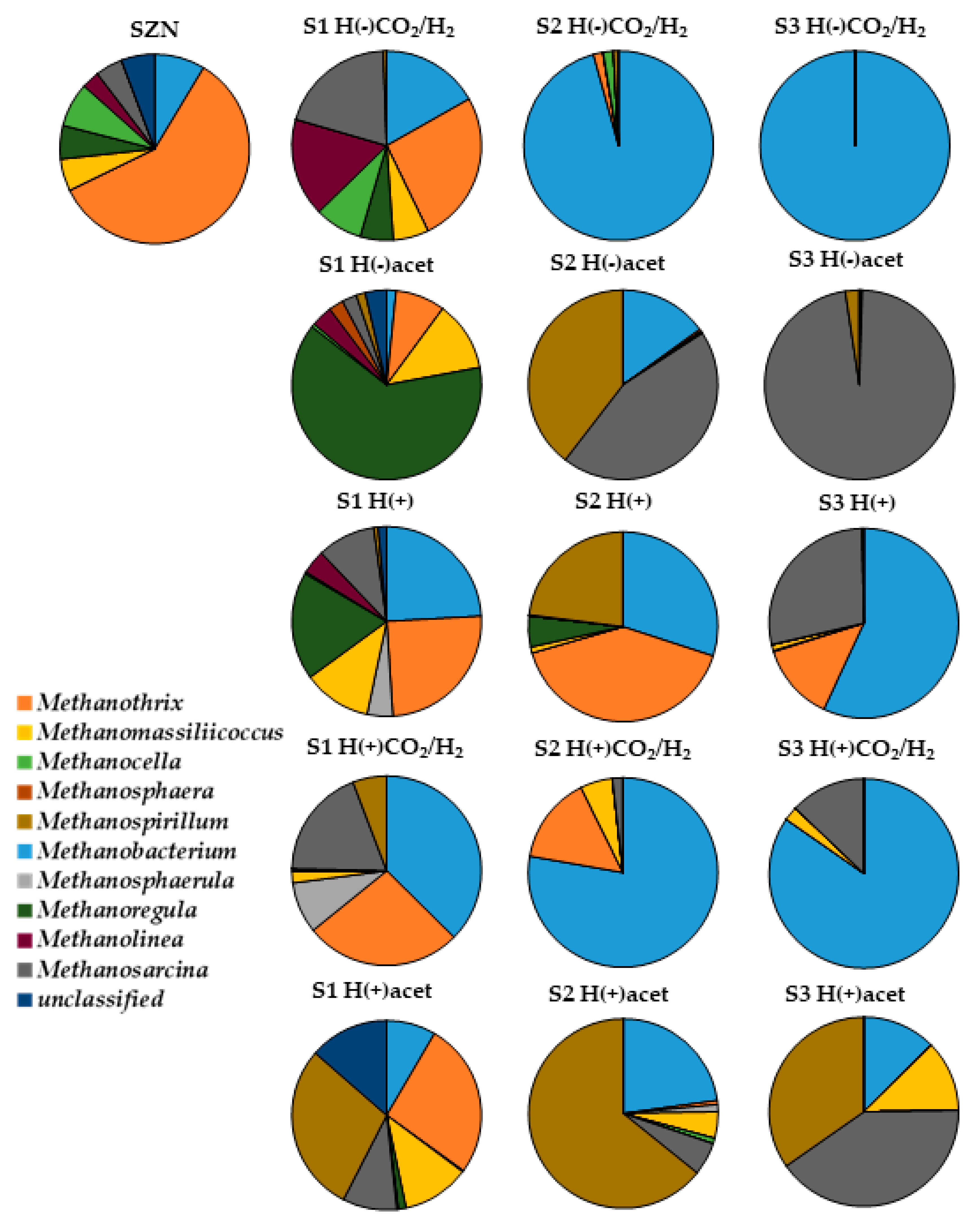
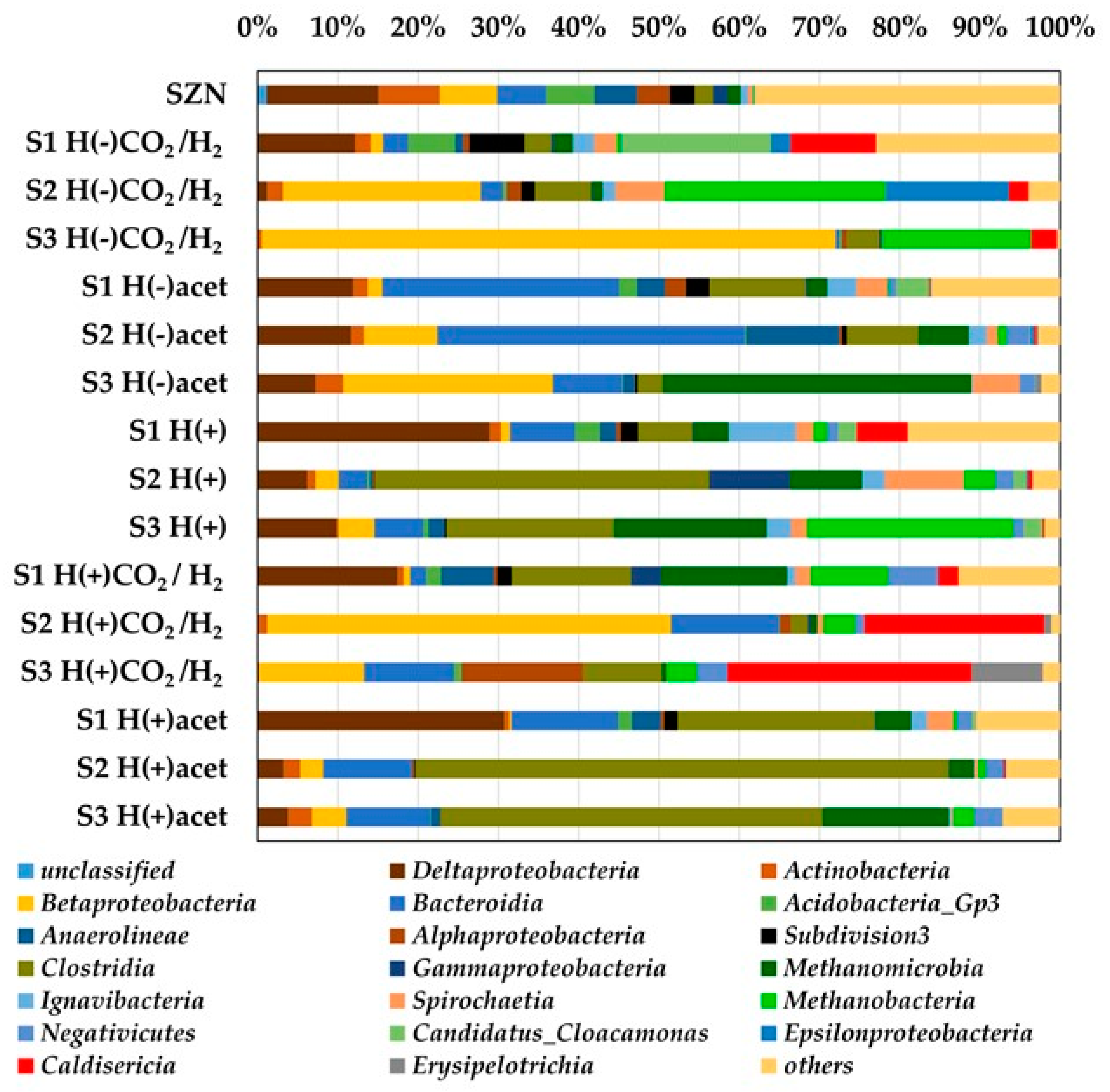
| N-NO2 | N-NO3 | N-NH4 | P-PO4 | TC | IC | TOC | Moisture | pH | Eh |
|---|---|---|---|---|---|---|---|---|---|
| mg kg d.w.−1 | g kg d.w.−1 | % | mV | ||||||
| 0.11 ± 0.01 | 0.30 ± 0.04 | 28.21 ± 4.88 | 10.05 ± 0.25 | 48.0 ± 0.8 | 0.00 | 48.0 ± 0.8 | 58.71 ± 2.28 | 5.46 ± 0.02 | −265.63 ± 2.89 |
| Stages of Bacterial Cultivation | Temperature (°C) | Lag Phase (d) | ||||
|---|---|---|---|---|---|---|
| H(−)CO2/H2 | H(−)acet | H(+) | H(+)CO2/H2 | H(+)acet | ||
| S1 | 10 | 14 | 31 | 14 | 14 | 31 |
| 20 | 10 | 21 | 10 | 10 | 10 | |
| 30 | 1 | 10 | 1 | 1 | 1 | |
| 40 | 1 | 7 | 1 | 1 | 1 | |
| S2 | 20 | 6 | 6 | 6 | 1 | 6 |
| 30 | 1 | 7 | 1 | 1 | 1 | |
| 40 | 1 | 6 | 1 | 1 | 1 | |
| S3 | 20 | - | 4 | - | - | - |
| 30 | 1 | - | 1 | 1 | 1 | |
| Stages of Bacterial Cultivation/Parameters | Medium Variants | |||||
|---|---|---|---|---|---|---|
| H(−)CO2/H2 | H(−)acet | H(+) | H(+)CO2/H2 | H(+)acet | ||
| Δ (start to end) | ||||||
| S1 | pH | −0.5 ± 0.01 | −0.24 ± 0.03 | −0.13 ± 0.02 | −0.21 ± 0.00 | −0.14 ± 0.01 |
| Eh (mV) | −383.20 ± 0.32 | −264.44 ± 0.06 | −337.27 ± 0.40 | −368.20 ± 0.10 | −303.87 ± 0.25 | |
| OD | na | na | na | na | na | |
| S2 | pH | −0.24 ± 0.01 | 1.07 ± 0.00 | 0.37 ± 0.00 | −0.36 ± 0.01 | 0.79 ± 0.02 |
| Eh (mV) | −330.60 ± 0.10 | −463.17 ± 0.30 | −347.77 ± 0.49 | −358.20 ± 0.10 | −467.53 ± 0.84 | |
| OD | 0.09 ± 0.02 | 0.08 ± 0.02 | 0.15 ± 0.01 | 0.09 ± 0.02 | 0.14 ± 0.01 | |
| S3 | pH | −1.18 ± 0.02 | 1.49 ± 0.00 | 0.16 ± 0.04 | −1.56 ± 0.04 | 0.76 ± 0.01 |
| Eh (mV) | −405.03 ± 0.49 | −466.33 ± 0.15 | −400.50 ± 1.42 | −465.80 ± 0.21 | −433.16 ± 1.05 | |
| OD | 0.09 ± 0.01 | 0.07 ± 0.02 | 0.16 ± 0.01 | 0.16 ± 0.02 | 0.16 ± 0.01 | |
| Treatment | Stage | Observed | Chao1 | ACE | Shannon | Simpson | InvSimpson | Fisher |
|---|---|---|---|---|---|---|---|---|
| SZN | - | 2536 | 2585.96 | 2565.61 | 7.06 | 1.00 | 422.83 | 559.06 |
| H(−)CO2/H2 | S1 | 1227 | 1232.69 | 1229.26 | 5.45 | 0.98 | 48.19 | 207.52 |
| S2 | 230 | 230.00 | 230.00 | 3.25 | 0.91 | 11.27 | 28.11 | |
| S3 | 96 | 96.00 | 96.00 | 1.40 | 0.53 | 2.11 | 10.55 | |
| H(−)acet | S1 | 1206 | 1206.56 | 1206.80 | 5.41 | 0.98 | 64.49 | 202.84 |
| S2 | 317 | 317.00 | 317.14 | 3.64 | 0.94 | 15.39 | 41.54 | |
| S3 | 184 | 184.00 | 184.20 | 2.59 | 0.80 | 5.00 | 22.77 | |
| H(+) | S1 | 996 | 996.30 | 996.43 | 5.10 | 0.97 | 33.28 | 158.44 |
| S2 | 353 | 353.00 | 353.14 | 4.07 | 0.96 | 25.17 | 48.10 | |
| S3 | 238 | 238.00 | 238.00 | 3.38 | 0.92 | 11.96 | 30.61 | |
| H(+)CO2/H2 | S1 | 574 | 574.00 | 574.14 | 4.93 | 0.98 | 66.40 | 85.08 |
| S2 | 121 | 121.00 | 121.00 | 1.89 | 0.69 | 3.27 | 13.60 | |
| S3 | 97 | 97.00 | 97.00 | 2.67 | 0.87 | 7.63 | 10.80 | |
| H(+)acet | S1 | 530 | 530.00 | 530.00 | 4.72 | 0.97 | 39.57 | 69.79 |
| S2 | 335 | 335.00 | 335.14 | 3.54 | 0.91 | 10.83 | 44.04 | |
| S3 | 241 | 241.00 | 241.28 | 3.83 | 0.95 | 20.53 | 31.56 |
| Media code | Tryptone (g L−1) | Yeast Extract (g L−1) | CH3COONa (g L−1) | Under Gases |
|---|---|---|---|---|
| SZN | Natural sediment with no additives | N2 | ||
| H(−)CO2/H2 | 0 | 0 | 0 | CO2/H2 (20:80 v/v) |
| H(−)acet | 0 | 0 | 2 | N2 |
| H(+) | 0.5 | 0.5 | 0 | N2 |
| H(+)CO2/H2 | 0.5 | 0.5 | 0 | CO2/H2 (20:80 v/v) |
| H(+)acet | 0.5 | 0.5 | 2 | N2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szafranek-Nakonieczna, A.; Pytlak, A.; Grządziel, J.; Kubaczyński, A.; Banach, A.; Górski, A.; Goraj, W.; Kuźniar, A.; Gałązka, A.; Stępniewska, Z. Changes in the Substrate Source Reveal Novel Interactions in the Sediment-Derived Methanogenic Microbial Community. Int. J. Mol. Sci. 2019, 20, 4415. https://doi.org/10.3390/ijms20184415
Szafranek-Nakonieczna A, Pytlak A, Grządziel J, Kubaczyński A, Banach A, Górski A, Goraj W, Kuźniar A, Gałązka A, Stępniewska Z. Changes in the Substrate Source Reveal Novel Interactions in the Sediment-Derived Methanogenic Microbial Community. International Journal of Molecular Sciences. 2019; 20(18):4415. https://doi.org/10.3390/ijms20184415
Chicago/Turabian StyleSzafranek-Nakonieczna, Anna, Anna Pytlak, Jarosław Grządziel, Adam Kubaczyński, Artur Banach, Andrzej Górski, Weronika Goraj, Agnieszka Kuźniar, Anna Gałązka, and Zofia Stępniewska. 2019. "Changes in the Substrate Source Reveal Novel Interactions in the Sediment-Derived Methanogenic Microbial Community" International Journal of Molecular Sciences 20, no. 18: 4415. https://doi.org/10.3390/ijms20184415
APA StyleSzafranek-Nakonieczna, A., Pytlak, A., Grządziel, J., Kubaczyński, A., Banach, A., Górski, A., Goraj, W., Kuźniar, A., Gałązka, A., & Stępniewska, Z. (2019). Changes in the Substrate Source Reveal Novel Interactions in the Sediment-Derived Methanogenic Microbial Community. International Journal of Molecular Sciences, 20(18), 4415. https://doi.org/10.3390/ijms20184415







