A Yersinia ruckeri TIR Domain-Containing Protein (STIR-2) Mediates Immune Evasion by Targeting the MyD88 Adaptor
Abstract
1. Introduction
2. Results
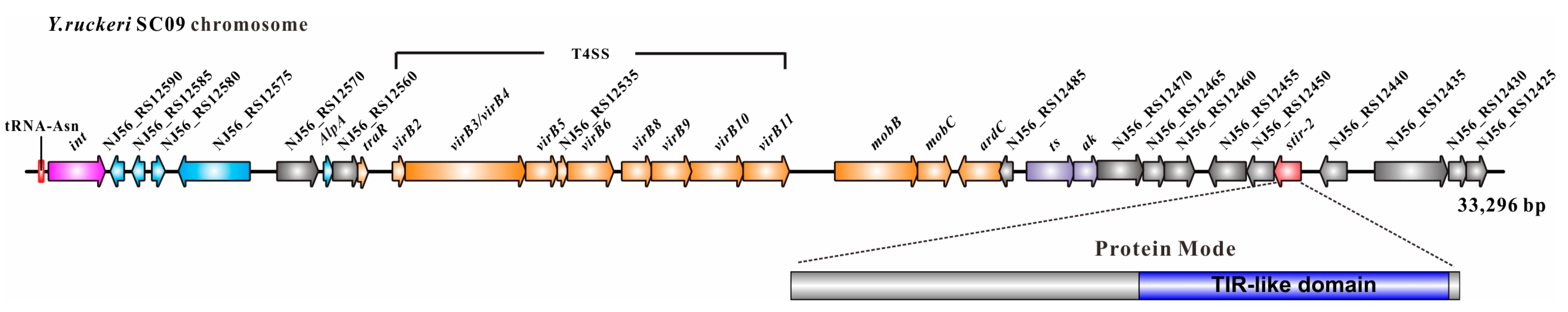
2.1. Genomic Location of Stir-2 in Yersinia ruckeri
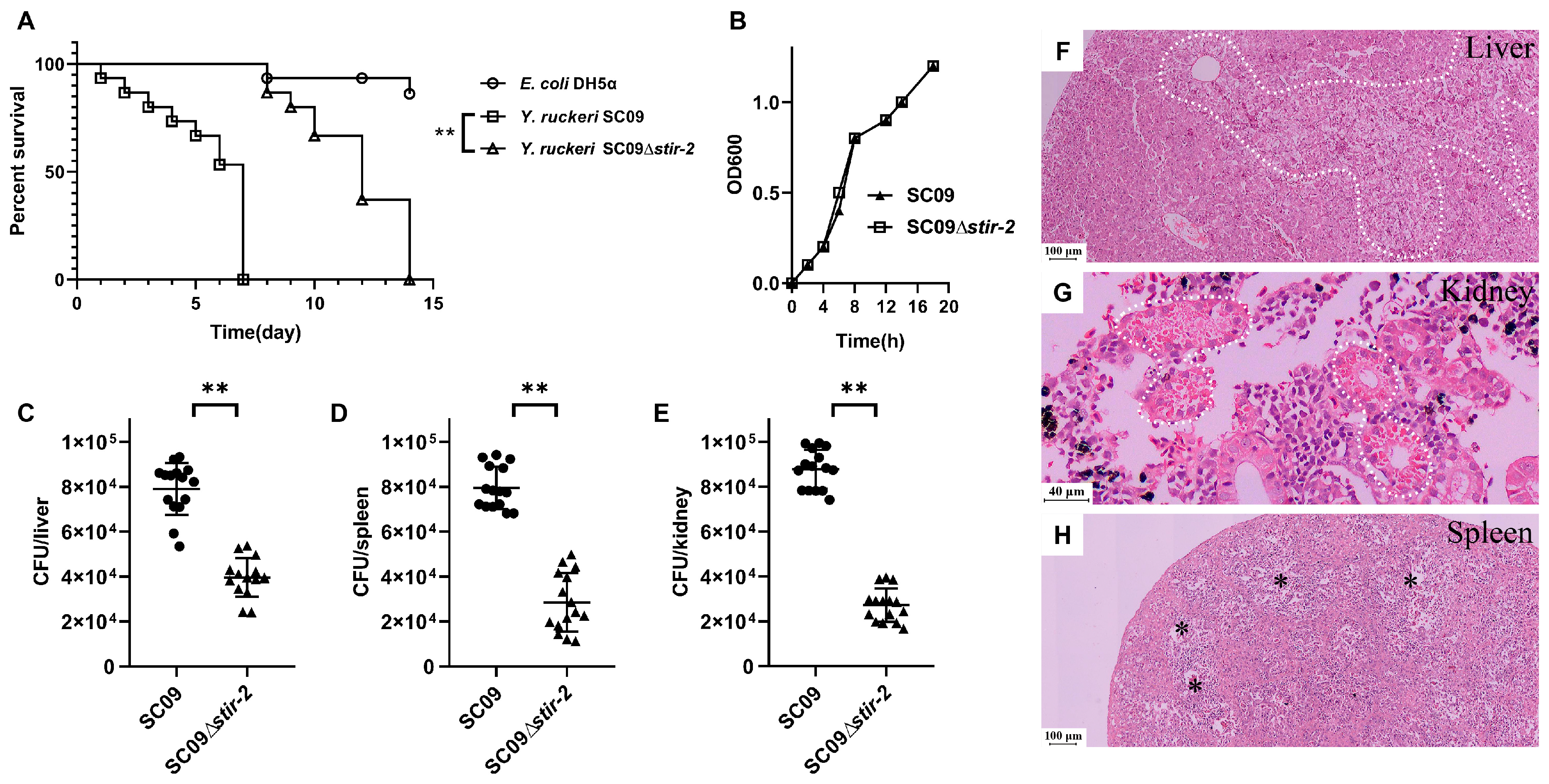
2.2. STIR-2 is Required for Yersinia ruckeri SC09 Infection in Vivo and in Vitro
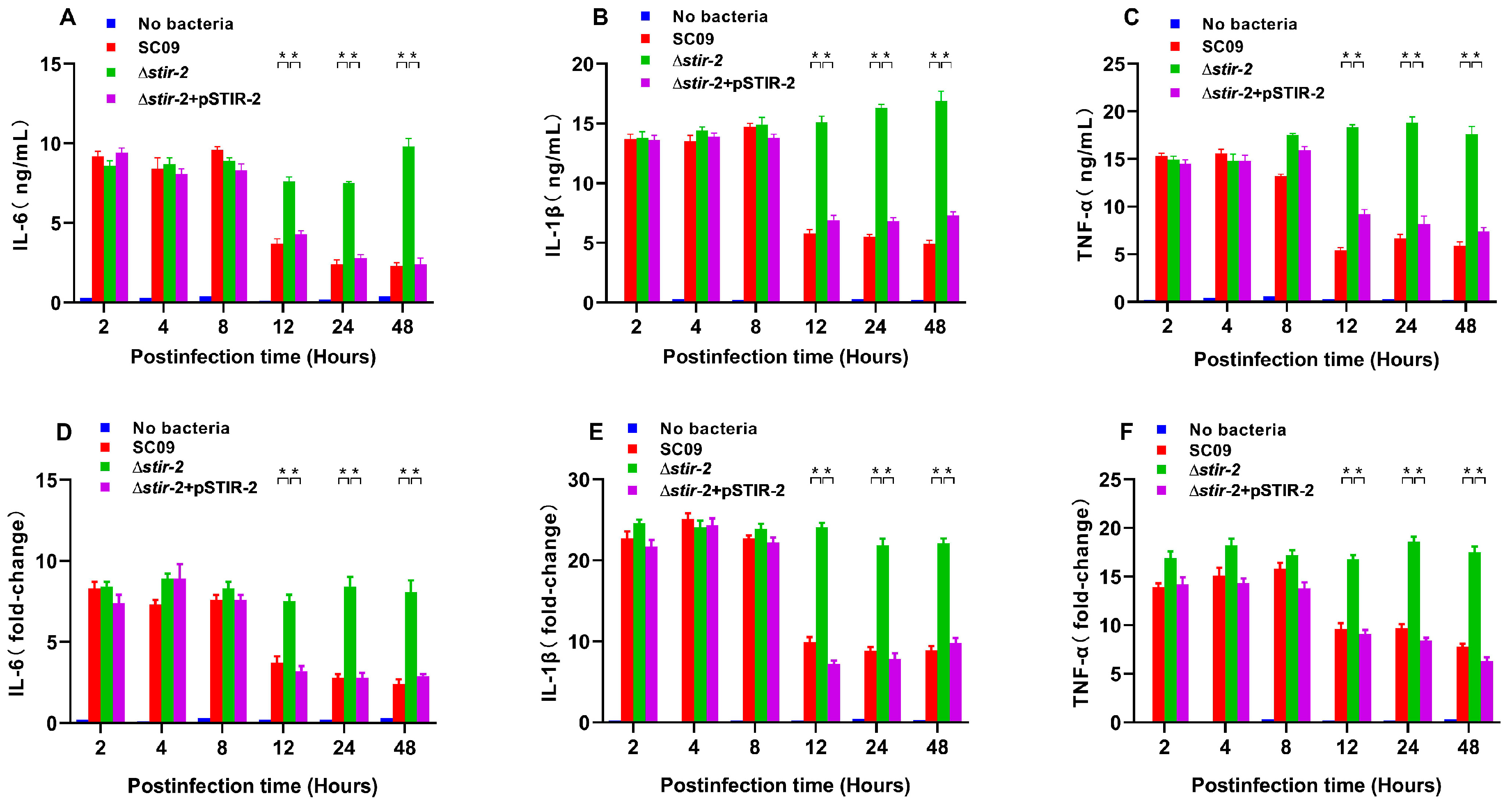
2.3. STIR-2 Decreases the Secretion and Transcription of Proinflammatory Cytokines
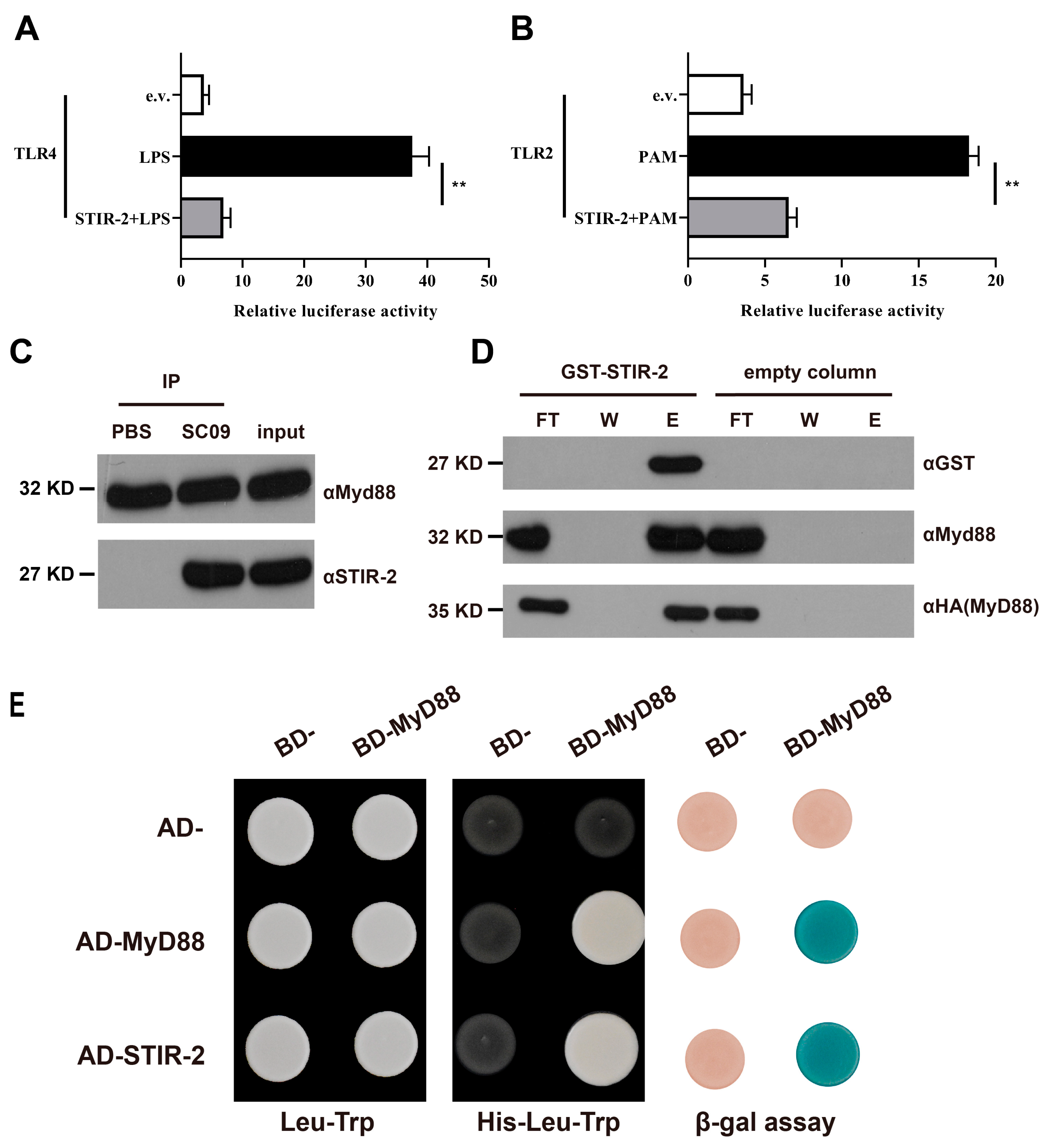
2.4. STIR-2 Affects Host TLR Signaling
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains
4.2. Construction of Y. ruckeri ∆stir-2 Mutant and Complemented Strains
4.3. Fish Infection Model
4.4. Bacterial Adherence, Invasion, and Intracellular Survival Assays
4.5. Enzyme-Linked Immunosorbent Assays and qPCR
4.6. Construction of Prokaryotic Expression Vectors
4.7. Construction of Eukaryotic Expression Vectors
4.8. Transfection and NF-κB-Dependent Luciferase Reporter Assay
4.9. Co-Immunoprecipitations (CO-IP)
4.10. Pulldowns from Cell Extracts
4.11. Yeast Two-Hybrid Assay
Author Contributions
Funding
Conflicts of Interest
References
- Rivers, J.K.; Mistry, B.D. Soft-Tissue Infection Caused by Streptococcus anginosus After Intramucosal Hyaluronidase Injection: A Rare Complication Related to Dermal Filler Injection. Derm. Surg. 2018, 44, S51–S53. [Google Scholar] [CrossRef] [PubMed]
- Ibberson, C.B.; Jones, C.L.; Singh, S.; Wise, M.C.; Hart, M.E.; Zurawski, D.V.; Horswill, A.R. Staphylococcus aureus hyaluronidase is a CodY-regulated virulence factor. Infect. Immun. 2014, 82, 4253–4264. [Google Scholar] [CrossRef] [PubMed]
- Zukaite, V.; Biziulevicius, G.A. Acceleration of hyaluronidase production in the course of batch cultivation of Clostridium perfringens can be achieved with bacteriolytic enzymes. Lett. Appl. Microbiol. 2000, 30, 203–206. [Google Scholar] [CrossRef] [PubMed]
- Matsuka, Y.V.; Pillai, S.; Gubba, S.; Musser, J.M.; Olmsted, S.B. Fibrinogen cleavage by the Streptococcus pyogenes extracellular cysteine protease and generation of antibodies that inhibit enzyme proteolytic activity. Infect. Immun. 1999, 67, 4326–4333. [Google Scholar] [PubMed]
- Wiemels, R.E.; Cech, S.M.; Meyer, N.M.; Burke, C.A.; Weiss, A.; Parks, A.R.; Shaw, L.N.; Carroll, R.K. An Intracellular Peptidyl-Prolyl cis/trans Isomerase Is Required for Folding and Activity of the Staphylococcus aureus Secreted Virulence Factor Nuclease. J. Bacteriol. 2017, 199, e00453–e00516. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Xiong, N.; Zhang, Y.; Rayner, S.; Chen, S. Functional characterization of lipase in the pathogenesis of Staphylococcus aureus. Biochem. Biophys. Res. Commun. 2012, 419, 617–620. [Google Scholar] [CrossRef] [PubMed]
- Thakur, A.; Mikkelsen, H.; Jungersen, G. Intracellular Pathogens: Host Immunity and Microbial Persistence Strategies. J. Immunol. Res. 2019, 2019, 1356540. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Suzuki, Y.; Guan, L.L. Dissect the mode of action of probiotics in affecting host-microbial interactions and immunity in food producing animals. Vet. Immunol. Immunopathol. 2018, 205, 35–48. [Google Scholar] [CrossRef]
- Ohtani, M.; Villumsen, K.R.; Strøm, H.K.; Lauritsen, A.H.; Aalbæk, B.; Dalsgaard, I.; Nowak, B.; Raida, M.K.; Bojesen, A.M. Effects of fish size and route of infection on virulence of a Danish Yersinia ruckeri O1 biotype 2 strain in rainbow trout (Oncorhynchus mykiss). Aquaculture 2019, 503, 519–526. [Google Scholar] [CrossRef]
- Kumar, G.; Menanteau-Ledouble, S.; Saleh, M.; El-Matbouli, M. Yersinia ruckeri, the causative agent of enteric redmouth disease in fish. Vet. Res. 2015, 46, 103. [Google Scholar] [CrossRef]
- Altinok, I.; Capkin, E.; Boran, H. Comparison of molecular and biochemical heterogeneity of Yersinia ruckeri strains isolated from Turkey and the USA. Aquaculture 2016, 450, 80–88. [Google Scholar] [CrossRef]
- Cascales, D.; Guijarro, J.A.; García-Torrico, A.I.; Méndez, J. Comparative genome analysis reveals important genetic differences among serotype O1 and serotype O2 strains of Y. ruckeri and provides insights into host adaptation and virulence. MicrobiologyOpen 2017, 6, e00460. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, L.; Marquez, I.; Guijarro, J.A. Identification of specific in vivo-induced (ivi) genes in Yersinia ruckeri and analysis of ruckerbactin, a catecholate siderophore iron acquisition system. Appl. Env. Microbiol. 2004, 70, 5199–5207. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, L.; Prieto, M.; Guijarro, J.A. The iron- and temperature-regulated haemolysin YhlA is a virulence factor of Yersinia ruckeri. Microbiology 2007, 153, 483–489. [Google Scholar] [CrossRef] [PubMed]
- Evenhuis, J.P.; Lapatra, S.E.; Verner-Jeffreys, D.W.; Dalsgaard, I.; Welch, T.J. Identification of flagellar motility genes in Yersinia ruckeri by transposon mutagenesis. Appl. Env. Microbiol. 2009, 75, 6630–6633. [Google Scholar] [CrossRef]
- Dahiya, I.; Stevenson, R.M. The UvrY response regulator of the BarA-UvrY two-component system contributes to Yersinia ruckeri infection of rainbow trout (Oncorhynchus mykiss). Arch. Microbiol. 2010, 192, 541–547. [Google Scholar] [CrossRef]
- Liu, T.; Wang, K.Y.; Wang, J.; Chen, D.F.; Huang, X.L.; Ouyang, P.; Geng, Y.; He, Y.; Zhou, Y.; Min, J. Genome Sequence of the Fish Pathogen Yersinia ruckeri SC09 Provides Insights into Niche Adaptation and Pathogenic Mechanism. Int. J. Mol. Sci. 2016, 17, 557. [Google Scholar] [CrossRef]
- Delavat, F.; Miyazaki, R.; Carraro, N.; Pradervand, N.; van der Meer, J.R. The hidden life of integrative and conjugative elements. Fems. Microbiol. Rev. 2017, 41, 512–537. [Google Scholar] [CrossRef]
- Bellanger, X.; Payot, S.; Leblond-Bourget, N.; Guedon, G. Conjugative and mobilizable genomic islands in bacteria: Evolution and diversity. Fems. Microbiol. Rev. 2014, 38, 720–760. [Google Scholar] [CrossRef]
- Seth-Smith, H.M.; Fookes, M.C.; Okoro, C.K.; Baker, S.; Harris, S.R.; Scott, P.; Pickard, D.; Quail, M.A.; Churcher, C.; Sanders, M.; et al. Structure, diversity, and mobility of the Salmonella pathogenicity island 7 family of integrative and conjugative elements within Enterobacteriaceae. J. Bacteriol. 2012, 194, 1494–1504. [Google Scholar] [CrossRef]
- Palti, Y. Toll-like receptors in bony fish: From genomics to function. Dev. Comp. Immunol. 2011, 35, 1263–1272. [Google Scholar] [CrossRef] [PubMed]
- Quiniou, S.M.; Boudinot, P.; Bengten, E. Comprehensive survey and genomic characterization of Toll-like receptors (TLRs) in channel catfish, Ictalurus punctatus: Identification of novel fish TLRs. Immunogenetics 2013, 65, 511–530. [Google Scholar] [CrossRef] [PubMed]
- Ryckaert, J.; Bossier, P.; D’Herde, K.; Diez-Fraile, A.; Sorgeloos, P.; Haesebrouck, F.; Pasmans, F. Persistence of Yersinia ruckeri in trout macrophages. Fish. Shellfish Immunol. 2010, 29, 648–655. [Google Scholar] [CrossRef] [PubMed]
- Skippington, E.; Ragan, M.A. Lateral genetic transfer and the construction of genetic exchange communities. Fems. Microbiol. Rev. 2011, 35, 707–735. [Google Scholar] [CrossRef] [PubMed]
- Del Grosso, M.; Camilli, R.; Rizzi, E.; Pietrelli, A.; De Bellis, G.; Pantosti, A. ICESpy009, a Conjugative Genetic Element Carrying mef(E) in Streptococcus pyogenes. Antimicrob. Agents Chemother. 2016, 60, 3906–3912. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Miyazaki, R.; Minoia, M.; Pradervand, N.; Sulser, S.; Reinhard, F.; van der Meer, J.R. Cellular variability of RpoS expression underlies subpopulation activation of an integrative and conjugative element. PLoS Genet. 2012, 8, e1002818. [Google Scholar] [CrossRef]
- Kaplan-Turkoz, B.; Koelblen, T.; Felix, C.; Candusso, M.P.; O’Callaghan, D.; Vergunst, A.C.; Terradot, L. Structure of the Toll/interleukin 1 receptor (TIR) domain of the immunosuppressive Brucella effector BtpA/Btp1/TcpB. Febs Letters 2013, 587, 3412–3416. [Google Scholar] [CrossRef] [PubMed]
- Rana, R.R.; Zhang, M.; Spear, A.M.; Atkins, H.S.; Byrne, B. Bacterial TIR-containing proteins and host innate immune system evasion. Med. Microbiol. Immunol. 2013, 202, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Zhang, M. Genetic diversity and antigenicity analysis of Streptococcus pneumoniae pneumolysin isolated from children with pneumococcal infection. Int. J. Infect. Dis. 2019, e1002818. [Google Scholar] [CrossRef] [PubMed]
- Martins Gomes, S.F.; Westermann, A.J.; Sauerwein, T.; Hertlein, T.; Forstner, K.U.; Ohlsen, K.; Metzger, M.; Shusta, E.V.; Kim, B.J.; Appelt-Menzel, A.; et al. Induced Pluripotent Stem Cell-Derived Brain Endothelial Cells as a Cellular Model to Study Neisseria meningitidis Infection. Front. Microbiol. 2019, 10, 1181. [Google Scholar] [CrossRef]
- Shen, J.; Liang, Q.; Su, G.; Zhang, Y.; Wang, Z.; Baudouin, C.; Labbe, A. In Vitro Effect of Toluidine Blue Antimicrobial Photodynamic Chemotherapy on Staphylococcus epidermidis and Staphylococcus aureus Isolated from Ocular Surface Infection. Transl. Vis. Sci. Technol. 2019, 8, 45. [Google Scholar] [CrossRef]
- Kao, P.H.N.; Kline, K.A. Dr. Jekyll and Mr. Hide: How Enterococcus faecalis Subverts the Host Immune Response to Cause Infection. J. Mol. Biol. 2019, 431, 2932–2945. [Google Scholar] [CrossRef] [PubMed]
- Khan, R.; Petersen, F.C.; Shekhar, S. Commensal Bacteria: An Emerging Player in Defense Against Respiratory Pathogens. Front. Immunol. 2019, 10, 1203. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Kautto, L.; Meyer, W.; Chen, S.C.; Nevalainen, H. Effect of proteases secreted by the opportunistic pathogen Scedosporium aurantiacum on human epithelial cells. Can. J. Microbiol. 2019, 1, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Keller, L.E.; Rueff, A.S.; Kurushima, J.; Veening, J.W. Three New Integration Vectors and Fluorescent Proteins for Use in the Opportunistic Human Pathogen Streptococcus pneumoniae. Genes (Basel) 2019, 10, 394. [Google Scholar] [CrossRef] [PubMed]
- Raffa, N.; Keller, N.P. A call to arms: Mustering secondary metabolites for success and survival of an opportunistic pathogen. PLoS Pathog 2019, 15, e1007606. [Google Scholar] [CrossRef] [PubMed]
- Kirchhoff, L.; Olsowski, M.; Rath, P.M.; Steinmann, J. Exophiala dermatitidis: Key issues of an opportunistic fungal pathogen. Virulence 2019, 1–15. [Google Scholar] [CrossRef]
- Kruppa, M.D.; Jacobs, J.; King-Hook, K.; Galloway, K.; Berry, A.; Kintner, J.; Whittimore, J.D.; Fritz, R.; Schoborg, R.V.; Hall, J.V. Binding of Elementary Bodies by the Opportunistic Fungal Pathogen Candida albicans or Soluble beta-Glucan, Laminarin, Inhibits Chlamydia trachomatis Infectivity. Front. Microbiol. 2018, 9, 3270. [Google Scholar] [CrossRef] [PubMed]
- Bertin, C.; Pau-Roblot, C.; Courtois, J.; Manso-Silvan, L.; Tardy, F.; Poumarat, F.; Citti, C.; Sirand-Pugnet, P.; Gaurivaud, P.; Thiaucourt, F. Highly dynamic genomic loci drive the synthesis of two types of capsular or secreted polysaccharides within the Mycoplasma mycoides cluster. Appl. Env. Microbiol. 2015, 81, 676–687. [Google Scholar] [CrossRef]
- Delannoy, S.; Beutin, L.; Mariani-Kurkdjian, P.; Fleiss, A.; Bonacorsi, S.; Fach, P. The Escherichia coli Serogroup O1 and O2 Lipopolysaccharides Are Encoded by Multiple O-antigen Gene Clusters. Front. Cell Infect. Microbiol. 2017, 7, 30. [Google Scholar] [CrossRef]
- Matsuda, S.; Haneda, T.; Saito, H.; Miki, T.; Okada, N. Salmonella effectors SifA, SpvB, SseF, SseJ, and SteA contribute to T3SS-1-independent inflammation in a streptomycin-pretreated mouse model of colitis. Infect. Immun. 2019. [Google Scholar] [CrossRef]
- Frost, L.S.; Koraimann, G. Regulation of bacterial conjugation: Balancing opportunity with adversity. Future Microbiol. 2010, 5, 1057–1071. [Google Scholar] [CrossRef] [PubMed]
- Bi, D.; Xu, Z.; Harrison, E.M.; Tai, C.; Wei, Y.; He, X.; Jia, S.; Deng, Z.; Rajakumar, K.; Ou, H.Y. ICEberg: A web-based resource for integrative and conjugative elements found in Bacteria. Nucleic Acids Res. 2012, 40, D621–D626. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.P.; Mullany, P. A modular master on the move: The Tn916 family of mobile genetic elements. Trends Microbiol. 2009, 17, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Patterson, N.J.; Werling, D. To con protection: TIR-domain containing proteins (Tcp) and innate immune evasion. Vet. Immunol. Immunopathol. 2013, 155, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Cirl, C.; Wieser, A.; Yadav, M.; Duerr, S.; Schubert, S.; Fischer, H.; Stappert, D.; Wantia, N.; Rodriguez, N.; Wagner, H.; et al. Subversion of Toll-like receptor signaling by a unique family of bacterial Toll/interleukin-1 receptor domain-containing proteins. Nat. Med. 2008, 14, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Salcedo, S.P.; Marchesini, M.I.; Degos, C.; Terwagne, M.; Von Bargen, K.; Lepidi, H.; Herrmann, C.K.; Santos Lacerda, T.L.; Imbert, P.R.C.; Pierre, P.; et al. BtpB, a novel Brucella TIR-containing effector protein with immune modulatory functions. Front. Cell. Infect. Microbiol. 2013, 3, 1–13. [Google Scholar] [CrossRef]
- Spear, A.M.; Rana, R.R.; Jenner, D.C.; Flick-Smith, H.C.; Oyston, P.C.; Simpson, P.; Matthews, S.J.; Byrne, B.; Atkins, H.S. A Toll/interleukin (IL)-1 receptor domain protein from Yersinia pestis interacts with mammalian IL-1/Toll-like receptor pathways but does not play a central role in the virulence of Y. pestis in a mouse model of bubonic plague. Microbiology 2012, 158, 1593–1606. [Google Scholar] [CrossRef] [PubMed]
- Imbert, P.R.; Louche, A.; Luizet, J.B.; Grandjean, T.; Bigot, S.; Wood, T.E.; Gagne, S.; Blanco, A.; Wunderley, L.; Terradot, L.; et al. A Pseudomonas aeruginosa TIR effector mediates immune evasion by targeting UBAP1 and TLR adaptors. Embo J. 2017, 36, 1869–1887. [Google Scholar] [CrossRef]
- Patot, S.; Imbert, P.R.; Baude, J.; Martins Simoes, P.; Campergue, J.B.; Louche, A.; Nijland, R.; Bes, M.; Tristan, A.; Laurent, F.; et al. The TIR Homologue Lies near Resistance Genes in Staphylococcus aureus, Coupling Modulation of Virulence and Antimicrobial Susceptibility. PLoS Pathog. 2017, 13, e1006092. [Google Scholar] [CrossRef]
- Spear, A.M.; Loman, N.J.; Atkins, H.S.; Pallen, M.J. Microbial TIR domains: Not necessarily agents of subversion? Trends Microbiol. 2009, 17, 393–398. [Google Scholar] [CrossRef]
- Wu, Y.; Guo, M.; Hua, X.; Duan, K.; Lian, G.; Sun, L.; Tang, L.; Xu, Y.; Liu, M.; Li, Y. The role of infectious hematopoietic necrosis virus (IHNV) proteins in the modulation of NF-kappaB pathway during IHNV infection. Fish. Shellfish Immunol. 2017, 63, 500–506. [Google Scholar] [CrossRef] [PubMed]
- Luo, P.; He, X.; Liu, Q.; Hu, C. Developing Universal Genetic Tools for Rapid and Efficient Deletion Mutation in Vibrio Species Based on Suicide T-Vectors Carrying a Novel Counterselectable Marker, vmi480. PLoS ONE 2015, 10, e0144465. [Google Scholar] [CrossRef] [PubMed]
- Carraro, N.; Matteau, D.; Luo, P.; Rodrigue, S.; Burrus, V. The master activator of IncA/C conjugative plasmids stimulates genomic islands and multidrug resistance dissemination. PLoS Genet. 2014, 10, e1004714. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Zhao, W.; Xiong, Q.; Wang, K.; He, Y.; Wang, J.; Chen, D.; Geng, Y.; Huang, X.; Ouyang, P.; et al. Immune responses of channel catfish following the stimulation of three recombinant flagellins of Yersinia ruckeri in vitro and in vivo. Dev. Comp. Immunol. 2017, 73, 61–71. [Google Scholar] [CrossRef] [PubMed]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, T.; Wei, W.-Y.; Wang, K.-Y.; Wang, E.-L.; Yang, Q. A Yersinia ruckeri TIR Domain-Containing Protein (STIR-2) Mediates Immune Evasion by Targeting the MyD88 Adaptor. Int. J. Mol. Sci. 2019, 20, 4409. https://doi.org/10.3390/ijms20184409
Liu T, Wei W-Y, Wang K-Y, Wang E-L, Yang Q. A Yersinia ruckeri TIR Domain-Containing Protein (STIR-2) Mediates Immune Evasion by Targeting the MyD88 Adaptor. International Journal of Molecular Sciences. 2019; 20(18):4409. https://doi.org/10.3390/ijms20184409
Chicago/Turabian StyleLiu, Tao, Wen-Yan Wei, Kai-Yu Wang, Er-Long Wang, and Qian Yang. 2019. "A Yersinia ruckeri TIR Domain-Containing Protein (STIR-2) Mediates Immune Evasion by Targeting the MyD88 Adaptor" International Journal of Molecular Sciences 20, no. 18: 4409. https://doi.org/10.3390/ijms20184409
APA StyleLiu, T., Wei, W.-Y., Wang, K.-Y., Wang, E.-L., & Yang, Q. (2019). A Yersinia ruckeri TIR Domain-Containing Protein (STIR-2) Mediates Immune Evasion by Targeting the MyD88 Adaptor. International Journal of Molecular Sciences, 20(18), 4409. https://doi.org/10.3390/ijms20184409




