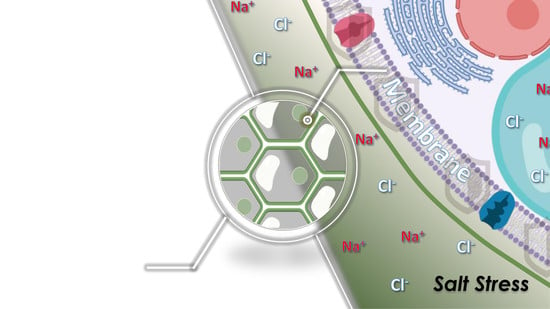
Membrane Lipid Remodeling in Response to Salinity
Abstract
1. Introduction
2. Plant Lipids
3. Alterations in Membrane Lipids in Response to Salt Stress
3.1. Total Membrane Lipid Content Changes under Salt Stress
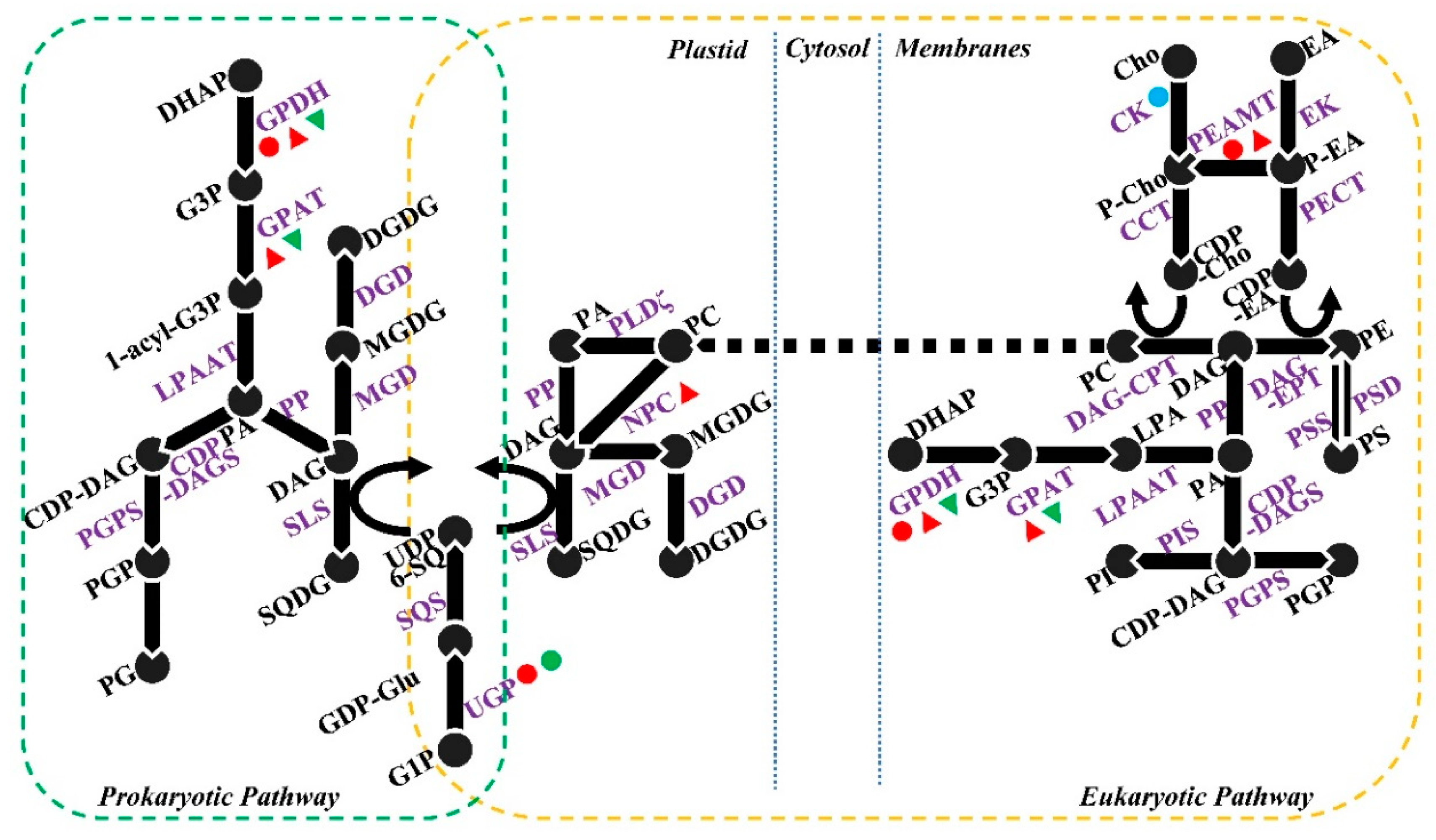
3.2. Changes in Membrane Phospholipids in Response to Salt Stress
3.3. Changes in Membrane Glycolipids in Response to Salt Stress
3.4. Changes in Membrane Sterols in Response to Salt Stress
3.5. Changes in Membrane Lipid Fatty Acids in Response to Salt Stress
3.6. Changes in Membrane Neutral Lipids in Response to Salt Stress
3.7. Membrane Lipids as Signaling Molecules in Plant Salt Tolerance
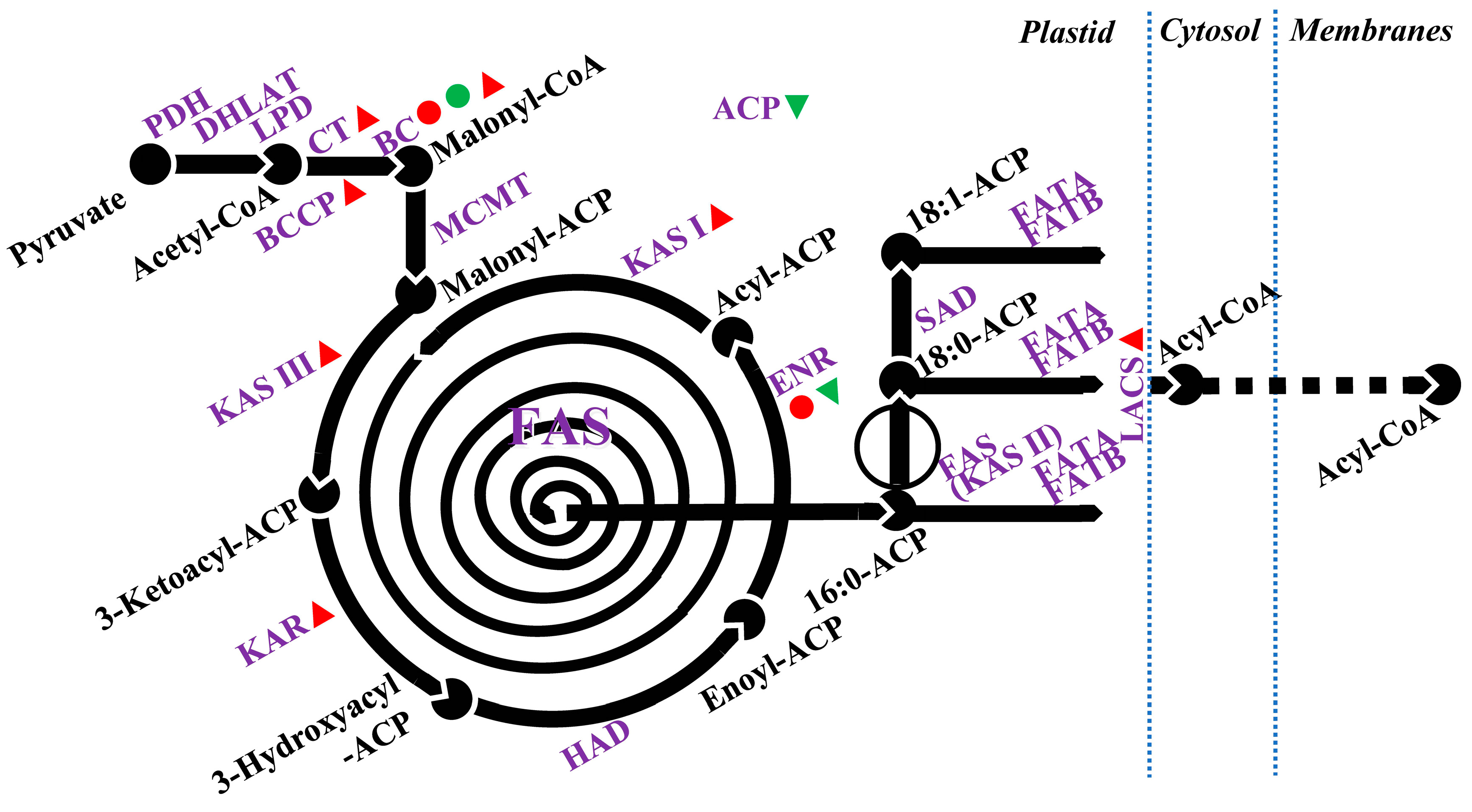
4. Salt-Induced Changes to Lipid Metabolism
5. Challenges and Perspectives
Acknowledgments
Conflicts of Interest
Appendix A
| Gene | Organism | Mutant/Overexpression | Salt Tolerance (Compared to WT) | Cellular Changes (Compared to WT) | Ref. |
|---|---|---|---|---|---|
| FAD2 | Arabidopsis thaliana | fad2 mutant | more sensitive during seed gemination and early seedling growth | lower polyunsaturation reduced Na+/H+ exchange activity higher Na+ accumulation in the cytoplasm of root cells | [206] |
| FAD3 | Lycopersicon esculentum | sense overexpression | enhanced tolerance | increased 18:3 FA decreased 18:2 FA improved maintenance of membrane integrity higher SOD and APX activity in the chloroplast | [207] |
| antisense overexpression | reduced tolerance | increased 18:2 FA decreased 18:3 FA | [207] | ||
| FAD6 | Arabidopsis thaliana | fad6 mutant | reduced tolerance | greater Na+ accumulation lower K+ accumulation increased electrolyte leakage rate and malondialdehyde production (more severe oxidative damage) decreased activities of anti-oxidative enzymes | [208] |
References
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Boil. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Allison, G.; Cook, P.; Barnett, S.; Walker, G.; Jolly, I.; Hughes, M. Land clearance and river salinisation in the western Murray Basin, Australia. J. Hydrol. 1990, 119, 1–20. [Google Scholar] [CrossRef]
- DeHaan, R.; Taylor, G. Field-derived spectra of salinized soils and vegetation as indicators of irrigation-induced soil salinization. Remote. Sens. Environ. 2002, 80, 406–417. [Google Scholar] [CrossRef]
- Darwish, T.; Atallah, T.; El Moujabber, M.; Khatib, N. Salinity evolution and crop response to secondary soil salinity in two agro-climatic zones in Lebanon. Agric. Water Manag. 2005, 78, 152–164. [Google Scholar] [CrossRef]
- Daliakopoulos, I.; Tsanis, I.; Koutroulis, A.; Kourgialas, N.; Varouchakis, A.; Karatzas, G.; Ritsema, C. The threat of soil salinity: A European scale review. Sci. Total. Environ. 2016, 573, 727–739. [Google Scholar] [CrossRef] [PubMed]
- Çullu, M.A. Estimation of the effect of soil salinity on crop yield using remote sensing and geographic information system. Turk. J. Agric. For. 2003, 27, 23–28. [Google Scholar]
- Katerji, N.; Van Hoorn, J.; Hamdy, A.; Mastrorilli, M. Salinity effect on crop development and yield, analysis of salt tolerance according to several classification methods. Agric. Water Manag. 2003, 62, 37–66. [Google Scholar] [CrossRef]
- Zeng, L.; Shannon, M.C. Salinity effects on seedling growth and yield components of rice. Crop. Sci. 2000, 40, 996. [Google Scholar] [CrossRef]
- Skaggs, R.; Van Schilfgaarde, J.; Maas, E.V.; Grattan, S.R. Crop Yields as Affected by Salinity. In Sorghum: State of the Art and Future Perspectives; American Society of Agronomy: Madison, WI, USA, 1999; pp. 55–108. [Google Scholar]
- Zhu, J.-K. Plant salt tolerance. Trends Plant Sci. 2001, 6, 66–71. [Google Scholar] [CrossRef]
- Acosta-Motos, J.R.; Ortuño, M.F.; Bernal-Vicente, A.; Diaz-Vivancos, P.; Sanchez-Blanco, M.J.; Hernandez, J.A. Plant responses to salt stress: Adaptive mechanisms. Agronomy 2017, 7, 18. [Google Scholar] [CrossRef]
- Deinlein, U.; Stephan, A.B.; Horie, T.; Luo, W.; Xu, G.; Schroeder, J.I. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014, 19, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Flowers, T.J.; Colmer, T.D. Plant salt tolerance: Adaptations in halophytes. Ann. Bot. 2015, 115, 327–331. [Google Scholar] [CrossRef] [PubMed]
- Isayenkov, S.V.; Maathuis, F.J.M. Plant salinity stress: Many unanswered questions remain. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, M.; Ali, Q. Relative membrane permeability and activities of some antioxidant enzymes as the key determinants of salt tolerance in canola (Brassica napus L.). Environ. Exp. Bot. 2008, 63, 266–273. [Google Scholar] [CrossRef]
- Magdy, M.; Mansour, F.; Lee-Stadelmann, O.Y.; Stadelmann, E.J. Salinity stress and cytoplasmic factors. A comparison of cell permeability and lipid partiality in salt sensitive and salt resistant cultivars and lines of Triticum aestivum and Hordeum vulgare. Physiol. Plant. 1993, 88, 141–148. [Google Scholar] [CrossRef]
- Mansour, M.M.F. Plasma membrane permeability as an indicator of salt tolerance in plants. Biol. Plantarum 2013, 57, 1–10. [Google Scholar] [CrossRef]
- Hulbert, A.J. Metabolism and longevity: Is there a role for membrane fatty acids? Integr. Comp. Boil. 2010, 50, 808–817. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, Y.; Saito, K. Roles of lipids as signaling molecules and mitigators during stress response in plants. Plant J. 2014, 79, 584–596. [Google Scholar] [CrossRef] [PubMed]
- Sévin, D.C.; Sauer, U. Ubiquinone accumulation improves osmotic-stress tolerance in Escherichia coli. Nat. Methods 2014, 10, 266–272. [Google Scholar] [CrossRef] [PubMed]
- Sévin, D.C.; Stählin, J.N.; Pollak, G.R.; Kuehne, A.; Sauer, U. Global metabolic responses to salt stress in fifteen species. PLoS ONE 2016, 11, e0148888. [Google Scholar] [CrossRef]
- Barkla, B.J.; Vera-Estrella, R. Single cell-type comparative metabolomics of epidermal bladder cells from the halophyte Mesembryanthemum crystallinum. Front. Plant Sci. 2015, 6, 435. [Google Scholar] [CrossRef]
- Surjus, A.; Durand, M. Lipid changes in soybean root membranes in response to salt treatment. J. Exp. Bot. 1996, 47, 17–23. [Google Scholar] [CrossRef]
- Elkahoui, S.; Smaoui, A.; Zarrouk, M.; Ghrir, R.; Limam, F. Salt-induced lipid changes in Catharanthus roseus cultured cell suspensions. Phytochemistry 2004, 65, 1911–1917. [Google Scholar] [CrossRef]
- Chalbi, N.; Hessini, K.; Gandour, M.; Mohamed, S.N.; Smaoui, A.; Abdelly, C.; Ben Youssef, N. Are changes in membrane lipids and fatty acid composition related to salt-stress resistance in wild and cultivated barley? J. Plant Nutr. Soil Sci. 2013, 176, 138–147. [Google Scholar] [CrossRef]
- Ben Hamed, K.; Ben Youssef, N.; Ranieri, A.; Zarrouk, M.; Abdelly, C. Changes in content and fatty acid profiles of total lipids and sulfolipids in the halophyte Crithmum maritimum under salt stress. J. Plant Physiol. 2005, 162, 599–602. [Google Scholar] [CrossRef]
- Magdy, M.; Mansour, F.; Hasselt, P.R.; Kuiper, P.J. Plasma membrane lipid alterations induced by NaCl in winter wheat roots. Physiol. Plant. 1994, 92, 473–478. [Google Scholar] [CrossRef]
- Lin, H.; Wu, L. Effects of salt stress on root plasma membrane characteristics of salt-tolerant and salt-sensitive buffalograss clones. Environ. Exp. Bot. 1996, 36, 239–254. [Google Scholar] [CrossRef]
- Alvarez-Pizarro, J.C.; Gomes-Filho, E.; de Lacerda, C.F.; Alencar, N.L.M.; Prisco, J.T. Salt-induced changes on H+-ATPase activity, sterol and phospholipid content and lipid peroxidation of root plasma membrane from dwarf-cashew (Anacardium occidentale L.) seedlings. Plant Growth Regul. 2009, 59, 125–135. [Google Scholar] [CrossRef]
- Salama, K.H.A.; Mansour, M.M.F.; Ali, F.Z.M.; Abou-Hadid, A.F. NaCl-induced changes in plasma membrane lipids and proteins of Zea mays L. cultivars differing in their response to salinity. Acta Physiol. Plant. 2007, 29, 351–359. [Google Scholar] [CrossRef]
- Salama, K.H.A.; Mansour, M.M.F. Choline priming-induced plasma membrane lipid alterations contributed to improved wheat salt tolerance. Acta Physiol. Plant. 2015, 37, 1–7. [Google Scholar] [CrossRef]
- Kerkeb, L.; Donaire, J.P.; Venema, K.; Rodríguez-Rosales, M.P. Tolerance to NaCl induces changes in plasma membrane lipid composition, fluidity and H+-ATPase activity of tomato calli. Physiol. Plantarum 2001, 113, 217–224. [Google Scholar] [CrossRef]
- Blits, K.; Gallagher, J. Effect of NaCl on lipid content of plasma membranes isolated from roots and cell suspension cultures of the dicot halophyte Kosteletzkya virginica (L.) Presl. Plant Cell Rep. 1990, 9, 156–159. [Google Scholar] [CrossRef]
- Barkla, B.J.; Garibay-Hernández, A.; Melzer, M.; Rupasinghe, T.W.; Roessner, U. Single cell-type analysis of cellular lipid remodelling in response to salinity in the epidermal bladder cells of the model halophyte Mesembryanthemum crystallinum. Plant Cell Environ. 2018, 41, 2390–2403. [Google Scholar] [CrossRef]
- Wu, J.; Seliskar, D.M.; Gallagher, J.L. The response of plasma membrane lipid composition in callus of the halophyte Spartina patens (Poaceae) to salinity stress. Am. J. Bot. 2005, 92, 852–858. [Google Scholar] [CrossRef]
- Sui, N.; Han, G. Salt-induced photoinhibition of PSII is alleviated in halophyte Thellungiella halophila by increases of unsaturated fatty acids in membrane lipids. Acta Physiol. Plant. 2014, 36, 983–992. [Google Scholar] [CrossRef]
- Zamani, S.; Bybordi, A.; Khorshidi, M.B.; Nezami, T. Effects of NaCl salinity levels on lipids and proteins of canola (Brassica napus L.) cultivars. Adv. Environ. Biol. 2010, 4, 397–403. [Google Scholar]
- Omoto, E.; Iwasaki, Y.; Miyake, H.; Taniguchi, M. Salinity induces membrane structure and lipid changes in maize mesophyll and bundle sheath chloroplasts. Physiol. Plant. 2016, 157, 13–23. [Google Scholar] [CrossRef]
- Bejaoui, F.; Salas, J.J.; Nouairi, I.; Smaoui, A.; Abdelly, C.; Force, E.M.; Ben Youssef, N. Changes in chloroplast lipid contents and chloroplast ultrastructure in Sulla carnosa and Sulla coronaria leaves under salt stress. J. Plant Physiol. 2016, 198, 32–38. [Google Scholar] [CrossRef]
- Ramani, B.; Zorn, H.; Papenbrock, J. Quantification and fatty acid profiles of sulfolipids in two halophytes and a glycophyte grown under different salt concentrations. Zeitschrift für Naturforschung C 2004, 59, 835–842. [Google Scholar] [CrossRef]
- Harayama, T.; Riezman, H. Understanding the diversity of membrane lipid composition. Nat. Rev. Mol. Cell Boil. 2018, 19, 281–296. [Google Scholar] [CrossRef]
- Van Meer, G. Cellular lipidomics. EMBO J. 2005, 24, 3159–3165. [Google Scholar] [CrossRef]
- Furt, F.; Simon-Plas, F.; Mongrand, S. Lipids of the plant plasma membrane. In The Plant Plasma Membrane; Murphy, A.S., Peer, W., Schulz, B., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; Volume 19, pp. 3–30. [Google Scholar]
- Li, L.; Han, J.; Wang, Z.; Liu, J.; Wei, J.; Xiong, S.; Zhao, Z. Mass spectrometry methodology in lipid analysis. Int. J. Mol. Sci. 2014, 15, 10492–10507. [Google Scholar] [CrossRef]
- Borrell, J.H.; Domènech, Ò.; Keough, K.M.W. Molecular membrane biochemistry. In Membrane Protein-Lipid Interactions: Physics and Chemistry in the Bilayer; Borrell, J.H., Domènech, Ò., Keough, K.M.W., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 1–29. [Google Scholar]
- Sperling, P.; Franke, S.; Lüthje, S.; Heinz, E. Are glucocerebrosides the predominant sphingolipids in plant plasma membranes? Plant Physiol. Biochem. 2005, 43, 1031–1038. [Google Scholar] [CrossRef]
- Chao, D.-Y.; Gable, K.; Chen, M.; Baxter, I.; Dietrich, C.R.; Cahoon, E.B.; Guerinot, M.L.; Lahner, B.; Lü, S.; Markham, J.E.; et al. Sphingolipids in the root play an important role in regulating the leaf ionome in arabidopsis thaliana. Plant Cell 2011, 23, 1061–1081. [Google Scholar] [CrossRef]
- Pata, M.O.; Hannun, Y.A.; Ng, C.K.Y. Plant sphingolipids: Decoding the enigma of the Sphinx. New Phytol. 2010, 185, 611–630. [Google Scholar] [CrossRef]
- Markham, J.E.; Lynch, D.V.; A Napier, J.; Dunn, T.M.; Cahoon, E.B. Plant sphingolipids: Function follows form. Curr. Opin. Plant Boil. 2013, 16, 350–357. [Google Scholar] [CrossRef]
- Rennie, E.A.; Ebert, B.; Miles, G.P.; Cahoon, R.E.; Christiansen, K.M.; Stonebloom, S.; Khatab, H.; Twell, D.; Petzold, C.J.; Adams, P.D.; et al. Identification of a sphingolipid α-glucuronosyltransferase that Is essential for pollen function in arabidopsis. Plant Cell 2014, 26, 3314–3325. [Google Scholar] [CrossRef]
- Guillas, I.; Guellim, A.; Rézé, N.; Baudouin, E. Long chain base changes triggered by a short exposure of Arabidopsis to low temperature are altered by AHb1 non-symbiotic haemoglobin overexpression. Plant Physiol. Biochem. 2013, 63, 191–195. [Google Scholar] [CrossRef]
- Cantrel, C.; Vazquez, T.; Puyaubert, J.; Rezé, N.; Lesch, M.; Kaiser, W.M.; Dutilleul, C.; Guillas, I.; Zachowski, A.; Baudouin, E. Nitric oxide participates in cold-responsive phosphosphingolipid formation and gene expression in Arabidopsis thaliana. New Phytol. 2011, 189, 415–427. [Google Scholar] [CrossRef]
- Guo, D.-A.; Venkatramesh, M.; Nes, W.D. Developmental regulation of sterol biosynthesis in Zea mays. Lipids 1995, 30, 203–219. [Google Scholar] [CrossRef]
- Schaeffer, A.; Bronner, R.; Benveniste, P.; Schaller, H. The ratio of campesterol to sitosterol that modulates growth in Arabidopsis is controlled by STEROL METHYLTRANSFERASE 2;1. Plant J. 2001, 25, 605–615. [Google Scholar] [CrossRef] [PubMed]
- López-Pérez, L.; Martínez-Ballesta, M.D.C.; Maurel, C.; Carvajal, M. Changes in plasma membrane lipids, aquaporins and proton pump of broccoli roots, as an adaptation mechanism to salinity. Phytochem. 2009, 70, 492–500. [Google Scholar] [CrossRef] [PubMed]
- Norberg, P.; Liljenberg, C. Lipids of plasma membranes prepared from oat root cells: Effects of induced water-deficit tolerance. Plant Physiol. 1991, 96, 1136–1141. [Google Scholar] [CrossRef]
- Valitova, J.N.; Sulkarnayeva, A.G.; Minibayeva, F. Plant sterols: Diversity, biosynthesis, and physiological functions. Biochemistry (Moscow) 2016, 81, 819–834. [Google Scholar] [CrossRef] [PubMed]
- Beck, J.G.; Mathieu, D.; Loudet, C.; Buchoux, S.; Dufourc, E.J. Plant sterols in “rafts”: A better way to regulate membrane thermal shocks. FASEB J. 2007, 21, 1714–1723. [Google Scholar] [CrossRef] [PubMed]
- Kalisch, B.; Dörmann, P.; Hölzl, G. DGDG and Glycolipids in plants and algae. Subcellular Biochemistry 2016, 86, 51–83. [Google Scholar] [PubMed]
- Frolov, V.A.; Shnyrova, A.V.; Zimmerberg, J. Lipid polymorphisms and membrane shape. Cold Spring Harb. Perspect. Boil. 2011, 3, a004747. [Google Scholar] [CrossRef] [PubMed]
- Tsydendambaev, V.D.; Ivanova, T.V.; Khalilova, L.A.; Kurkova, E.B.; Myasoedov, N.A.; Balnokin, Y.V. Fatty acid composition of lipids in vegetative organs of the halophyte Suaeda altissima under different levels of salinity. Russ. J. Plant Physiol. 2013, 60, 661–671. [Google Scholar] [CrossRef]
- Lin, W.; Oliver, D.J. Role of triacylglycerols in leaves. Plant Sci. 2008, 175, 233–237. [Google Scholar] [CrossRef]
- Rochester, C.P.; Kjellbom, P.; Andersson, B.; Larsson, C. Lipid composition of plasma membranes isolated from light-grown barley (Hordeum vulgare) leaves: Identification of cerebroside as a major component. Arch. Biochem. Biophys. 1987, 255, 385–391. [Google Scholar] [CrossRef]
- Brown, D.J.; Dupont, F.M. Lipid composition of plasma membranes and endomembranes prepared from roots of barley (Hordeum vulgare L.). Plant Physiol. 1989, 90, 955–961. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, S.; Uemura, M. Lipid composition of plasma membranes and tonoplasts isolated from etiolated seedling of mung bean (Vigna radiata L.). Plant Physiol. 1986, 82, 807–812. [Google Scholar] [CrossRef] [PubMed]
- Sheffer, M.; Fried, A.; Gottlieb, H.E.; Tietz, A.; Avron, M. Lipid composition of the plasma-membrane of the halotolerant alga, Dunaliella salina. Biochim Biophys. Acta (BBA)—Biomembr. 1986, 857, 165–172. [Google Scholar] [CrossRef]
- Liljenberg, C.; Kates, M. Changes in lipid composition of oat root membranes as a function of water-deficit stress. Can. J. Biochem. Cell Boil. 1985, 63, 77–84. [Google Scholar] [CrossRef]
- Navari-Izzo, F.; Ricci, F.; Vazzana, C.; Quartacci, M.F. Unusual composition of thylakoid membranes of the resurrection plant Boea hygroscopica: Changes in lipids upon dehydration and rehydration. Physiol. Plant. 1995, 94, 135–142. [Google Scholar] [CrossRef]
- Tuna, A.L.; Kaya, C.; Ashraf, M.; Altunlu, H.; Yokas, I.; Yağmur, B. The effects of calcium sulphate on growth, membrane stability and nutrient uptake of tomato plants grown under salt stress. Environ. Exp. Bot. 2007, 59, 173–178. [Google Scholar] [CrossRef]
- Yang, F.; Xiao, X.; Zhang, S.; Korpelainen, H.; Li, C. Salt stress responses in Populus cathayana Rehder. Plant Sci. 2009, 176, 669–677. [Google Scholar] [CrossRef]
- Yasar, F.; Uzal, O.; Ozpay, T. Changes of the lipid perocidation and chlorophyll amount of green bean genotypes under drought stress. Afr. J. Agric. Res. 2010, 5, 2705–2709. [Google Scholar]
- Li, Q.-Y.; Niu, H.-B.; Yin, J.; Wang, M.-B.; Shao, H.-B.; Deng, D.-Z.; Chen, X.-X.; Ren, J.-P.; Li, Y.-C. Protective role of exogenous nitric oxide against oxidative-stress induced by salt stress in barley (Hordeum vulgare). Colloids Surfaces B: Biointerfaces 2008, 65, 220–225. [Google Scholar] [CrossRef]
- Molitor, V.; Trnka, M.; Erber, W.; Steffan, I.; Arrio, B.; Springer-Lederer, H.; Peschek, G.A. Impact of salt adaptation on esterified fatty acids and cytochrome oxidase in plasma and thylakoid membranes from the cyanobacterium Anacystis nidulans. Arch. Microbiol. 1990, 154, 112–119. [Google Scholar] [CrossRef]
- Takagi, M.; Karseno; Yoshida, T. Effect of salt concentration on intracellular accumulation of lipids and triacylglyceride in marine microalgae Dunaliella cells. J. Biosci. Bioeng. 2006, 101, 223–226. [Google Scholar] [CrossRef] [PubMed]
- Lu, N.; Wei, D.; Jiang, X.-L.; Chen, F.; Yang, S.-T. Regulation of lipid metabolism in the snow alga Chlamydomonas nivalis in response to NaCl stress: An integrated analysis by cytomic and lipidomic approaches. Process. Biochem. 2012, 47, 1163–1170. [Google Scholar] [CrossRef]
- Yu, B.J.; Lam, H.M.; Shao, G.H.; Liu, Y.L. Effects of salinity on activities of H+-ATPase, H+-PPase and membrane lipid composition in plasma membrane and tonoplast vesicles isolated from soybean (Glycine max L.) seedlings. J. Environ. Sci. 2005, 17, 259–262. [Google Scholar]
- Xue, H.; Chen, X.; Mei, Y. Function and regulation of phospholipid signalling in plants. Biochem. J. 2009, 421, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Erdei, L.; Stuiver, B.E.P.; Kuiper, P.J.C. The effect of salinity on lipid composition and on activity of Ca2+- and Mg2+-stimulated ATPases in salt-sensitive and salt-tolerant Plantago species. Physiol. Plantarum 1980, 49, 315–319. [Google Scholar] [CrossRef]
- Rodríguez-Rosales, M.P.; Kerkeb, L.; Bueno, P.; Donaire, J.P. Changes induced by NaCl in lipid content and composition, lipoxygenase, plasma membrane H+-ATPase and antioxidant enzyme activities of tomato (Lycopersicon esculentum. Mill) calli. Plant Sci. 1999, 143, 143–150. [Google Scholar] [CrossRef]
- Mansour, M.M.F.; Stadelmann, E.J.; Lee-Stadelmann, O.Y. Salt acclimation of Triticum aestivum by choline chloride: Plant growth, mineral content, and cell permeability. Plant Physiol. Biochem. 1993, 31, 341–348. [Google Scholar]
- Tasseva, G.; Richard, L.; Zachowski, A. Regulation of phosphatidylcholine biosynthesis under salt stress involves choline kinases in Arabidopsis thaliana. FEBS Lett. 2004, 566, 115–120. [Google Scholar] [CrossRef]
- Sathishkumar, R.; Manoharan, K. Lipid changes due to growth-factor supplements in callus and plasma membrane-enriched fraction of rice cultures. Phytochem. 1996, 43, 1171–1174. [Google Scholar] [CrossRef]
- Russell, N.J. Function of lipids: Structural roles and membrane function. In Microbial Lipids; Ratledge, C., Wilkinson, S.C., Eds.; Academic Press: London, UK, 1989; pp. 279–365. [Google Scholar]
- Zhu, J.-K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Boil. 2002, 53, 247–273. [Google Scholar] [CrossRef] [PubMed]
- Awai, K.; Ohta, H.; Sato, N. Oxygenic photosynthesis without galactolipids. Proc. Natl. Acad. Sci. USA 2014, 111, 13571–13575. [Google Scholar] [CrossRef] [PubMed]
- Aoki, M.; Sato, N.; Meguro, A.; Tsuzuki, M. Differing involvement of sulfoquinovosyl diacylglycerol in photosystem II in two species of unicellular cyanobacteria. JBIC J. Boil. Inorg. Chem. 2004, 271, 685–693. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Hagio, M.; Sakurai, I.; Sato, S.; Tabata, S.; Wada, H. Phosphatidylglycerol is essential for the development of thylakoid membranes in Arabidopsis thaliana. Plant Cell Physiol. 2002, 43, 1456–1464. [Google Scholar]
- Dörmann, P.; Benning, C. Galactolipids rule in seed plants. Trends Plant Sci. 2002, 7, 112–118. [Google Scholar] [CrossRef]
- Yamane, K.; Rahman, S.; Kawasaki, M.; Taniguchi, M.; Miyake, H. Pretreatment with a low concentration of methyl viologen decreases the effects of salt stress on chloroplast ultrastructure in rice leaves (Oryza sativa L.). Plant Prod. Sci. 2015, 7, 435–441. [Google Scholar] [CrossRef]
- Shu, S.; Guo, S.-R.; Sun, J.; Yuan, L.-Y. Effects of salt stress on the structure and function of the photosynthetic apparatus in Cucumis sativus and its protection by exogenous putrescine. Physiol. Plant. 2012, 146, 285–296. [Google Scholar] [CrossRef] [PubMed]
- De Paula, F.; Thi, A.; De Silva, J.; Justin, A.; Demandre, C.; Mazliak, P. Effects of water stress on the molecular species composition of polar lipids from Vigna unguiculata L. leaves. Plant Sci. 1990, 66, 185–193. [Google Scholar] [CrossRef]
- Wang, S.; Uddin, M.I.; Tanaka, K.; Yin, L.; Shi, Z.; Qi, Y.; Mano, J.; Matsui, K.; Shimomura, N.; Sakaki, T.; et al. Maintenance of chloroplast structure and function by overexpression of the rice monogalactosyldiacylglycerol synthase gene leads to enhanced salt tolerance in tobacco. Plant Physiol 2014, 165, 1144–1155. [Google Scholar] [CrossRef]
- Guimarães, F.V.A.; De Lacerda, C.F.; Marques, E.C.; De Miranda, M.R.A.; De Abreu, C.E.B.; Prisco, J.T.; Gomes-Filho, E.; Lacerda, C.F.; Miranda, M.R.A.; Abreu, C.E.B. Calcium can moderate changes on membrane structure and lipid composition in cowpea plants under salt stress. Plant Growth Regul. 2011, 65, 55–63. [Google Scholar] [CrossRef]
- Welti, R. Profiling membrane lipids in plant stress responses. Role of phospholipase D alpha in freezing-induced lipid changes in Arabidopsis. J. Boil. Chem. 2002, 277, 31994–32002. [Google Scholar] [CrossRef]
- Gigon, A.; Matos, A.-R.; Laffray, D.; Zuily-Fodil, Y.; Pham-Thi, A.-T. Effect of drought stress on lipid metabolism in the leaves of Arabidopsis thaliana (Ecotype Columbia). Ann. Bot. 2004, 94, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Alam, M.M.; Rahman, A.; Hasanuzzaman, M.; Nahar, K.; Fujita, M. Exogenous proline and glycine betaine mediated upregulation of antioxidant defense and glyoxalase systems provides better protection against salt-induced oxidative stress in two rice (Oryza sativa L.) Varieties. BioMed Res. Int. 2014, 2014, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Skórzyńska, E.; Urbanik-Sypniewska, T.; Russa, R.; Baszyński, T. Galactolipase activity of chloroplasts in cadmium-treated runner bean plants. J. Plant Physiol. 1991, 138, 454–459. [Google Scholar] [CrossRef]
- Lee, A.G. Membrane lipids: It’s only a phase. Curr. Boil. 2000, 10, R377–R380. [Google Scholar] [CrossRef]
- Simidjiev, I.; Stoylova, S.; Amenitsch, H.; Jávorfi, T.; Mustardy, L.; Laggner, P.; Holzenburg, A. Self-assembly of large, ordered lamellae from non-bilayer lipids and integral membrane proteins in vitro. Proc. Natl. Acad. Sci. USA 2000, 97, 1473–1476. [Google Scholar] [CrossRef] [PubMed]
- Garab, G.; Lohner, K.; Laggner, P.; Farkas, T. Self-regulation of the lipid content of membranes by non-bilayer lipids: A hypothesis. Trends Plant Sci. 2000, 5, 489–494. [Google Scholar] [CrossRef]
- Williams, W.P. The physical properties of thylakoid membrane lipids and their relation to photosynthesis. In Lipids in Photosynthesis: Structure, Function and Genetics; Paul-André, S., Norio, M., Eds.; Springer: Dordrecht, The Netherlands, 1998; 6p. [Google Scholar]
- Schuler, I.; Milon, A.; Nakatani, Y.; Ourisson, G.; Albrecht, A.M.; Benveniste, P.; A Hartman, M. Differential effects of plant sterols on water permeability and on acyl chain ordering of soybean phosphatidylcholine bilayers. Proc. Natl. Acad. Sci. USA 1991, 88, 6926–6930. [Google Scholar] [CrossRef]
- Krumova, S.B.; Laptenok, S.P.; Kovács, L.; Tóth, T.; Van Hoek, A.; Garab, G.; Van Amerongen, H. Digalactosyl-diacylglycerol-deficiency lowers the thermal stability of thylakoid membranes. Photosynth. Res. 2010, 105, 229–242. [Google Scholar] [CrossRef]
- Omoto, E.; Nagao, H.; Taniguchi, M.; Miyake, H. Localization of reactive oxygen species and change of antioxidant capacities in mesophyll and bundle sheath chloroplasts of maize under salinity. Physiol. Plantarum 2013, 149, 1–12. [Google Scholar] [CrossRef]
- Block, M.A.; Dorne, A.J.; Joyard, J.; Douce, R. Preparation and characterization of membrane fractions enriched in outer and inner envelope membranes from spinach chloroplasts. II. Biochemical characterization. J. Biol. Chem. 1983, 258, 13281–13286. [Google Scholar] [PubMed]
- Sato, N.; Sonoike, K.; Tsuzuk, M.; Kawaguchi, A. Impaired photosystem ii in a mutant of Chlamydomonas Reinhardtii defective in sulfoquinovosyl diacylglycerol. J. Boil. Inorg. Chem. 1995, 234, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Taran, N.; Okanenko, A.; Musienko, N. Sulpholipid reflects plant resistance to stress-factor action. Biochem. Soc. Trans. 2000, 28, 922–924. [Google Scholar] [CrossRef] [PubMed]
- Minoda, A.; Sonoike, K.; Okada, K.; Sato, N.; Tsuzuki, M. Decrease in the efficiency of the electron donation to tyrosine Z of photosystem II in an SQDG-deficient mutant of Chlamydomonas. FEBS Lett. 2003, 553, 109–112. [Google Scholar] [CrossRef]
- Seigneurin-Berny, D.; Rolland, N.; Dorne, A.-J.; Joyard, J. Sulfolipid Is a potential candidate for annexin binding to the outer surface of chloroplast. Biochem. Biophys. Res. Commun. 2000, 272, 519–524. [Google Scholar] [CrossRef] [PubMed]
- Domon, M.; Nasir, M.N.; Matar, G.; Pikula, S.; Besson, F.; Bandorowicz-Pikula, J. Annexins as organizers of cholesterol- and sphingomyelin-enriched membrane microdomains in Niemann-Pick type C disease. Cell Mol. Life Sci. 2012, 69, 1773–1785. [Google Scholar] [CrossRef] [PubMed]
- Blackwood, R.A.; Ernst, J.D. Characterization of Ca2+-dependent phospholipid binding, vesicle aggregation and membrane fusion by annexins. Biochem. J. 1990, 266, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Gerke, V.; Creutz, C.E.; Moss, S.E. Annexins: Linking Ca2+ signalling to membrane dynamics. Nat. Rev. Nat. Rev. Mol. Cell Biol. 2005, 6, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Kuiper, P.J.C. Lipids in grape roots in relation to chloride transport. Plant Physiol. 1968, 43, 1367–1371. [Google Scholar] [CrossRef][Green Version]
- Kumari, P.; Kumar, M.; Reddy, C.; Jha, B. Algal lipids, fatty acids and sterols. In Functional Ingredients from Algae for Foods and Nutraceuticals; Elsevier: Amsterdam, The Netherlands, 2013; pp. 87–134. [Google Scholar]
- Douglas, T.J.; Csiro, S.R.S. Phospholipid, galactolipid and free sterol composition of fibrous roots from citrus genotypes differing in chloride exclusion ability. Plant Cell Environ. 1985, 8, 693–699. [Google Scholar]
- Salama, K.H.A.; Mansour, M.M.F.; Hassan, N.S. Choline priming improves salt tolerance in wheat (Triticum aestivum L.). Aust. J. Basic Appl. Sci. 2011, 5, 126–132. [Google Scholar] [CrossRef]
- Schuler, I.; Duportail, G.; Glasser, N.; Benveniste, P.; Hartmann, M.-A. Soybean phosphatidylcholine vesicles containing plant sterols: A fluorescence anisotropy study. Biochim. Biophys. Acta (BBA)—Biomembr. 1990, 1028, 82–88. [Google Scholar] [CrossRef]
- Wu, J.; Seliskar, D.M.; Gallagher, J.L. Stress tolerance in the marsh plant Spartina patens: Impact of NaCl on growth and root plasma membrane lipid composition. Physiol. Plant. 1998, 102, 307–317. [Google Scholar] [CrossRef]
- Adler, L.; Liljenberg, C. Sterol content, fatty acid composition of phospholipids, and permeability of labeled ethylene glycols in relation to salt-tolerance of yeasts. Physiol. Plant. 1981, 53, 368–374. [Google Scholar] [CrossRef]
- Grandmougin-Ferjani, A.; Schuler-Muller, I.; Hartmann, M.A. Sterol modulation of the plasma membrane H+-ATPase activity from corn roots reconstituted into soybean lipids. Plant Physiol. 1997, 113, 163–174. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, M. Plant sterols and the membrane environment. Trends Plant Sci. 1998, 3, 170–175. [Google Scholar] [CrossRef]
- Hajlaoui, H.; Denden, M.; El Ayeb, N. Changes in fatty acids composition, hydrogen peroxide generation and lipid peroxidation of salt-stressed corn (Zea mays L.) roots. Acta Physiol. Plant. 2009, 31, 787–796. [Google Scholar] [CrossRef]
- Kuiper, P.J.C. Functioning of plant cell membrane under saline conditions: Membrane lipid composition and ATPase. In Salinity Tolerance in Plants; Staples, R.C., Toenniessen, G.H., Eds.; Wiley: New York, NY, USA, 1984; pp. 71–91. [Google Scholar]
- Xu, X.-Q.; Beardall, J. Effect of salinity on fatty acid composition of a green microalga from an antarctic hypersaline lake. Phytochem. 1997, 45, 655–658. [Google Scholar] [CrossRef]
- Muller, T.; Bleiß, W.; Martin, C.-D.; Rogaschewski, S.; Fuhr, G. Snow algae from northwest Svalbard: Their identification, distribution, pigment and nutrient content. Polar Boil. 1998, 20, 14–32. [Google Scholar] [CrossRef]
- Sudhakar, C.; Lakshmi, A.; Giridarakumar, S. Changes in the antioxidant enzyme efficacy in two high yielding genotypes of mulberry (Morus alba L.) under NaCl salinity. Plant Sci. 2001, 161, 613–619. [Google Scholar] [CrossRef]
- Zhang, M.; Barg, R.; Yin, M.; Salts, Y.; Shabtai, S.; Gueta-Dahan, Y.; Leikin-Frenkel, A.; Ben-Hayyim, G. Modulated fatty acid desaturation via overexpression of two distinct ω-3 desaturases differentially alters tolerance to various abiotic stresses in transgenic tobacco cells and plants. Plant J. 2005, 44, 361–371. [Google Scholar] [CrossRef] [PubMed]
- Allakhverdiev, S.I.; Nishiyama, Y.; Suzuki, I.; Tasaka, Y.; Murata, N. Genetic engineering of the unsaturation of fatty acids in membrane lipids alters the tolerance of Synechocystis to salt stress. Proc. Natl. Acad. Sci. USA 1999, 96, 5862–5867. [Google Scholar] [CrossRef] [PubMed]
- Allakhverdiev, S.; Kinoshita, M.; Inaba, M.; Suzuki, I.; Murata, N. Unsaturated fatty acids in membrane lipids protect the photosynthetic machinery against salt-induced damage in Synechococcus. Plant Physiol. 2001, 125, 1842–1853. [Google Scholar] [CrossRef] [PubMed]
- Légeret, B.; Schulz-Raffelt, M.; Nguyen, H.M.; Auroy, P.; Beisson, F.; Peltier, G.; Blanc, G.; Li-Beisson, Y. Lipidomic and transcriptomic analyses of Chlamydomonas reinhardtii under heat stress unveil a direct route for the conversion of membrane lipids into storage lipids. Plant Cell Environ. 2016, 39, 834–847. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, S.; Tamura, P.J.; Roth, M.R.; Prasad, P.V.V.; Welti, R. Wheat leaf lipids during heat stress: I. High day and night temperatures result in major lipid alterations. Plant Cell Environ. 2016, 39, 787–803. [Google Scholar] [CrossRef] [PubMed]
- Moellering, E.R.; Muthan, B.; Benning, C. Freezing tolerance in plants requires lipid remodeling at the outer chloroplast membrane. Science 2010, 330, 226–228. [Google Scholar] [CrossRef] [PubMed]
- Perlikowski, D.; Kierszniowska, S.; Sawikowska, A.; Krajewski, P.; Rapacz, M.; Eckhardt, Ä; Kosmala, A. Remodeling of leaf cellular glycerolipid composition under drought and re-hydration conditions in grasses from the Lolium-Festuca complex. Front. Plant Sci. 2016, 7, 10960. [Google Scholar] [CrossRef] [PubMed]
- Hung, C.-H.; Kanehara, K.; Nakamura, Y. Isolation and characterization of a mutant defective in triacylglycerol accumulation in nitrogen-starved Chlamydomonas reinhardtii. Biochim Biophys. Acta (BBA)—Mol. Cell Boil. Lipids 2016, 1861, 1282–1293. [Google Scholar] [CrossRef] [PubMed]
- Vu, H.S.; Roston, R.; Shiva, S.; Hur, M.; Wurtele, E.S.; Wang, X.; Shah, J.; Welti, R. Modifications of membrane lipids in response to wounding of Arabidopsis thaliana leaves. Plant Signal. Behav. 2015, 10, e1056422. [Google Scholar] [CrossRef]
- Kim, B.-H.; Ramanan, R.; Kang, Z.; Cho, D.-H.; Oh, H.-M.; Kim, H.-S. Chlorella sorokiniana HS1, a novel freshwater green algal strain, grows and hyperaccumulates lipid droplets in seawater salinity. Biomass- Bioenergy 2016, 85, 300–305. [Google Scholar] [CrossRef]
- Rahman, S.; Matsumuro, T.; Miyake, H.; Takeoka, Y. Salinity-induced ultrastructural alterations in leaf cells of rice (Oryza sativa L.). Plant Prod. Sci. 2000, 3, 422–429. [Google Scholar] [CrossRef]
- Munnik, T.; Meijer, H.J.G.; Ter Riet, B.; Hirt, H.; Frank, W.; Bartels, D.; Musgrave, A. Hyperosmotic stress stimulates phospholipase D activity and elevates the levels of phosphatidic acid and diacylglycerol pyrophosphate. Plant J. 2000, 22, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Ryu, S.B.; Wang, X. Increase in free linolenic and linoleic acids associated with phospholipase D-mediated hydrolysis of phospholipids in wounded castor bean leaves. Biochim Biophys. Acta (BBA)—Lipids Lipid Metab. 1998, 1393, 193–202. [Google Scholar] [CrossRef]
- Hong, Y.; Devaiah, S.P.; Bahn, S.C.; Thamasandra, B.N.; Li, M.; Welti, R.; Wang, X. Phospholipase D epsilon and phosphatidic acid enhance Arabidopsis nitrogen signaling and growth. Plant J. 2009, 58, 376–387. [Google Scholar] [CrossRef] [PubMed]
- Katagiri, T.; Takahashi, S.; Shinozaki, K. Involvement of a novel Arabidopsis phospholipase D, AtPLDδ, in dehydration-inducible accumulation of phosphatidic acid in stress signalling. Plant J. 2001, 26, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Pokotylo, I.; Kolesnikov, Y.; Kravets, V.; Zachowski, A.; Ruelland, E. Plant phosphoinositide-dependent phospholipases C: Variations around a canonical theme. Biochemistry 2014, 96, 144–157. [Google Scholar] [CrossRef]
- Arisz, S.A.; Testerink, C.; Munnik, T. Plant PA signaling via diacylglycerol kinase. Biochim Biophys. Acta (BBA)—Mol. Cell Boil. Lipids 2009, 1791, 869–875. [Google Scholar] [CrossRef]
- Vrije, T.D.; Munnik, T.; Irvine, R.F.; Musgrave, A. Identification of diacylglycerol pyrophosphate as a novel metabolic product of phosphatidic acid during g-protein activation in plants. J. Boil. Chem. 1996, 271, 15708–15715. [Google Scholar]
- Bargmann, B.O.; Laxalt, A.M.; ter Riet, B.; van Schooten, B.; Merquiol, E.; Testerink, C.; Haring, M.A.; Bartels, D.; Munnik, T. Multiple PLDs required for high salinity and water deficit tolerance in plants. Plant Cell Physiol. 2009, 50, 78–89. [Google Scholar] [CrossRef]
- Yu, L.; Nie, J.; Cao, C.; Jin, Y.; Yan, M.; Wang, F.; Liu, J.; Xiao, Y.; Liang, Y.; Zhang, W. Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol. 2010, 188, 762–773. [Google Scholar] [CrossRef]
- Zonia, L.; Munnik, T. Osmotically induced cell swelling versus cell shrinking elicits specific changes in phospholipid signals in tobacco pollen tubes1. Plant Physiol. 2004, 134, 813–823. [Google Scholar] [CrossRef] [PubMed]
- Darwish, E.; Testerink, C.; Khalil, M.; El-Shihy, O.; Munnik, T. Phospholipid signaling responses in salt-stressed rice leaves. Plant Cell Physiol. 2009, 50, 986–997. [Google Scholar] [CrossRef] [PubMed]
- Testerink, C.; Dekker, H.L.; Lim, Z.-Y.; Johns, M.K.; Holmes, A.B.; Koster, C.G.; Ktistakis, N.T.; Munnik, T. Isolation and identification of phosphatidic acid targets from plants. Plant J. 2004, 39, 527–536. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Lin, F.; Mao, T.; Nie, J.; Yan, M.; Yuan, M.; Zhang, W. Phosphatidic acid regulates microtubule organization by interacting with MAP65-1 in response to salt stress in Arabidopsis. Plant Cell 2012, 24, 4555–4576. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, L.; Liu, Y.; Zhang, Q.; Wei, Q.; Zhang, W. Nitric oxide enhances salt tolerance in maize seedlings through increasing activities of proton-pump and Na+/H+ antiport in the tonoplast. Planta 2006, 224, 545–555. [Google Scholar] [CrossRef]
- Pical, C.; Westergren, T.; Dove, S.K.; Larsson, C.; Sommarin, M. Salinity and hyperosmotic stress induce rapid increases in phosphatidylinositol 4,5-bisphosphate, diacylglycerol pyrophosphate, and phosphatidylcholine in Arabidopsis thaliana Cells. J. Boil. Chem. 1999, 274, 38232–38240. [Google Scholar] [CrossRef]
- Van Leeuwen, W.; Vermeer, J.E.; Gadella, T.W.; Munnik, T. Visualization of phosphatidylinositol 4,5-bisphosphate in the plasma membrane of suspension-cultured tobacco BY-2 cells and whole Arabidopsis seedlings. Plant J. 2007, 52, 1014–1026. [Google Scholar] [CrossRef]
- Einspahr, K.J.; Maeda, M.; A Thompson, G. Concurrent changes in Dunaliella salina ultrastructure and membrane phospholipid metabolism after hyperosmotic shock. J. Cell Boil. 1988, 107, 529–538. [Google Scholar] [CrossRef]
- Beilby, M.J.; Al Khazaaly, S. Chara action potential: II. The action potential form under salinity stress. AIMS Biophys. 2017, 4, 298–315. [Google Scholar] [CrossRef]
- Biskup, B.; Gradmann, D.; Thiel, G. Calcium release from InsP3 -sensitive internal stores initiates action potential in Chara. FEBS Lett. 1999, 453, 72–76. [Google Scholar] [CrossRef]
- Dewald, D.B. Rapid Accumulation of Phosphatidylinositol 4,5-bisphosphate and inositol 1,4,5-trisphosphate correlates with calcium mobilization in salt-stressed Arabidopsis. Plant Physiol. 2001, 126, 759–769. [Google Scholar] [CrossRef] [PubMed]
- Drøbak, B.K.; Watkins, P.A. Inositol (1,4,5) trisphosphate production in plant cells: An early response to salinity and hyperosmotic stress. FEBS Lett. 2000, 481, 240–244. [Google Scholar] [CrossRef]
- Jiang, Z.; Zhou, X.; Tao, M.; Yuan, F.; Liu, L.; Wu, F.; Wu, X.; Xiang, Y.; Niu, Y.; Liu, F.; et al. Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx. Nature 2019. [Google Scholar] [CrossRef]
- Carr, K.; McAinsh, M.R.; Powell, B.; Hetherington, A.M.; Ng, C.K. Drought-induced guard cell signal transduction involves sphingosine-1-phosphate. Nature 2001, 410, 596–599. [Google Scholar]
- Harwood, J. Plant Acyl Lipids: Structure, distribution, and analysis. In Lipids: Structure and Function; Elsevier: Amsterdam, The Netherlands, 1980; pp. 1–55. [Google Scholar]
- Li-Beisson, Y.; Shorrosh, B.; Beisson, F.; Andersson, M.X.; Arondel, V.; Bates, P.D.; Baud, S.; Bird, D.; DeBono, A.; Durrett, T.P.; et al. Acyl-Lipid metabolism. Arabidopsis Book 2013. [Google Scholar] [CrossRef] [PubMed]
- Badea, C.; Basu, S.K. The effect of low temperature on metabolism of membrane lipids in plants and associated gene expression. Plant Omics 2009, 2, 78–84. [Google Scholar]
- Gong, Q.; Li, P.; Ma, S.; Rupassara, S.I.; Bohnert, H.J. Salinity stress adaptation competence in the extremophile Thellungiella halophila in comparison with its relative Arabidopsis thaliana. Plant J. 2005, 44, 826–839. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.A.; Alam, I.; Kim, Y.-G.; Ahn, N.-Y.; Heo, S.-H.; Lee, D.-G.; Liu, G.; Lee, B.-H. Screening for salt-responsive proteins in two contrasting alfalfa cultivars using a comparative proteome approach. Plant Physiol. Biochem. 2015, 89, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; He, L.; Guo, S.; Li, J.; Yang, Y.; Yan, B.; Sun, J.; Li, J. Proteomics reveal cucumber Spd-responses under normal condition and salt stress. Plant Physiol. Biochem. 2013, 67, 7–14. [Google Scholar] [CrossRef]
- Bajji, M.; Kinet, J.-M.; Lutts, S. Salt stress effects on roots and leaves of Atriplex halimus L. and their corresponding callus cultures. Plant Sci. 1998, 137, 131–142. [Google Scholar] [CrossRef]
- Dooki, A.D.; Askari, H.; Zaiee, A.-A.; Salekdeh, G.H.; Mayer-Posner, F.J. Proteomic responses of rice young panicles to salinity. Proteomics 2006, 6, 6498–6507. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Lao, Y.M.; Jiang, J.G. Effects of salinities on the gene expression of a (NAD+)-dependent glycerol-3-phosphate dehydrogenase in Dunaliella salina. Sci. Total Environ. 2011, 409, 1291–1297. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-H.; Rao, X.-L.; Shi, H.-T.; Li, R.-J.; Lu, Y.-T. Overexpression of a cytosolic glyceraldehyde-3-phosphate dehydrogenase gene OsGAPC3 confers salt tolerance in rice. Plant Cell Tissue Organ Cult. (PCTOC) 2011, 107, 1–11. [Google Scholar] [CrossRef]
- Jeong, M.J.; Park, S.C.; O Byun, M. Improvement of salt tolerance in transgenic potato plants by glyceraldehyde-3 phosphate dehydrogenase gene transfer. Mol. Cells 2001, 12, 185–189. [Google Scholar] [PubMed]
- Cheng, R.-l.; Feng, J.; Zhang, B.-X.; Huang, Y.; Cheng, J.; Zhang, C.-X. Transcriptome and gene expression analysis of an oleaginous diatom under different salinity conditions. Bioenergy Res. 2014, 7, 192–205. [Google Scholar] [CrossRef]
- Ho, S.-H.; Nakanishi, A.; Kato, Y.; Yamasaki, H.; Chang, J.-S.; Misawa, N.; Hirose, Y.; Minagawa, J.; Hasunuma, T.; Kondo, A. Dynamic metabolic profiling together with transcription analysis reveals salinity-induced starch-to-lipid biosynthesis in alga Chlamydomonas sp. JSC4. Sci. Rep. 2017, 7, 45471. [Google Scholar] [CrossRef] [PubMed]
- Yokthongwattana, C.; Mahong, B.; Roytrakul, S.; Phaonaklop, N.; Narangajavana, J.; Yokthongwattana, K. Proteomic analysis of salinity-stressed Chlamydomonas reinhardtii revealed differential suppression and induction of a large number of important housekeeping proteins. Planta 2012, 235, 649–659. [Google Scholar] [CrossRef] [PubMed]
- Kanesaki, Y.; Suzuki, I.; Allakhverdiev, S.I.; Mikami, K.; Murata, N. Salt Stress and hyperosmotic stress regulate the expression of different sets of genes in Synechocystis sp. PCC 6803. Biochem. Biophys. Res. Commun. 2002, 290, 339–348. [Google Scholar] [CrossRef]
- Jeong, M.-J.; Park, S.-C.; Kwon, H.-B.; Byun, M.-O. Isolation and characterization of the gene encoding glyceraldehyde-3-phosphate dehydrogenase. Biochem. Biophys. Res. Commun. 2000, 278, 192–196. [Google Scholar] [CrossRef]
- Sui, N.; Tian, S.; Wang, W.; Wang, M.; Fan, H. Overexpression of glycerol-3-phosphate acyltransferase from Suaeda salsa improves salt tolerance in Arabidopsis. Front. Plant Sci. 2017, 8, 1337. [Google Scholar] [CrossRef]
- Gupta, S.M.; Pandey, P.; Grover, A.; Patade, V.Y.; Singh, S.; Ahmed, Z. Cloning and characterization of GPAT gene from Lepidium latifolium L.: A step towards translational research in agri-genomics for food and fuel. Mol. Boil. Rep. 2013, 40, 4235–4240. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Sauve, R.; Fish, T.; Thannhauser, T.W. Salt-induced and salt-suppressed proteins in tomato leaves. J. Am. Soc. Hortic. Sci. 2009, 134, 289–294. [Google Scholar] [CrossRef]
- Mostek, A.; Börner, A.; Badowiec, A.; Weidner, S. Alterations in root proteome of salt-sensitive and tolerant barley lines under salt stress conditions. J. Plant Physiol. 2015, 174, 166–176. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Tang, Z.; Su, W.; Sun, W. Proteomic analysis of salt stress-responsive proteins in rice root. Proteomics 2005, 5, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Yu, Z.; Wang, F.; Li, W.; Ye, C.; Li, J.; Tang, J.; Ding, J.; Zhao, J.; Wang, B. Cloning, characterization, and transformation of the phosphoethanolamine N-methyltransferase gene (ZmPEAMT1) in maize (Zea mays L.). Mol. Biotechnol. 2007, 36, 102–112. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Zhang, Y.; Zhu, N.; Koh, J.; Ma, C.; Pan, Y.; Yu, B.; Chen, S.; Li, H. Proteomic analysis of salt tolerance in sugar beet monosomic addition line M14. J. Proteome Res. 2013, 12, 4931–4950. [Google Scholar] [CrossRef] [PubMed]
- Summers, P.S.; Weretilnyk, E.A. Choline synthesis in spinach in relation to salt stress. Plant Physiol. 1993, 103, 1269–1276. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kocourková, D.; Krčková, Z.; Pejchar, P.; Veselková, Š; Valentová, O.; Wimalasekera, R.; Scherer, G.F.E.; Martinec, J. The phosphatidylcholine-hydrolysing phospholipase C NPC4 plays a role in response of Arabidopsis roots to salt stress. J. Exp. Bot. 2011, 62, 3753–3763. [Google Scholar]
- Peters, C.; Kim, S.-C.; Devaiah, S.; Li, M.; Wang, X. Non-specific phospholipase C5 and diacylglycerol promote lateral root development under mild salt stress in Arabidopsis. Plant Cell Environ. 2014, 37, 2002–2013. [Google Scholar] [CrossRef]
- Sun, Y.L.; Li, F.; Su, N.; Sun, X.L.; Zhao, S.J.; Meng, Q.W. The increase in unsaturation of fatty acids of phosphatidylglycerol in thylakoid membrane enhanced salt tolerance in tomato. Photosynth 2010, 48, 400–408. [Google Scholar] [CrossRef]
- Okazaki, Y.; Shimojima, M.; Sawada, Y.; Toyooka, K.; Narisawa, T.; Mochida, K.; Tanaka, H.; Matsuda, F.; Hirai, A.; Hirai, M.Y.; et al. A chloroplastic UDP-glucose pyrophosphorylase from arabidopsis is the committed enzyme for the first step of sulfolipid biosynthesis. Plant Cell 2009, 21, 892–909. [Google Scholar] [CrossRef] [PubMed]
- Meng, M.; Geisler, M.; Johansson, H.; Harholt, J.; Scheller, H.V.; Mellerowicz, E.J.; Kleczkowski, L.A. UDP-Glucose pyrophosphorylase is not rate limiting, but is essential in Arabidopsis. Plant Cell Physiol. 2009, 50, 998–1011. [Google Scholar] [CrossRef] [PubMed]
- Coleman, H.D.; Ellis, D.D.; Gilbert, M.; Mansfield, S.D. Up-regulation of sucrose synthase and UDP-glucose pyrophosphorylase impacts plant growth and metabolism. Plant Biotechnol. J. 2006, 4, 87–101. [Google Scholar] [CrossRef] [PubMed]
- Walia, H.; Wilson, C.; Wahid, A.; Condamine, P.; Cui, X.; Close, T.J. Expression analysis of barley (Hordeum vulgare L.) during salinity stress. Funct. Integr. Genom. 2006, 6, 143–156. [Google Scholar] [CrossRef] [PubMed]
- Ueda, A.; Shi, W.; Nakamura, T.; Takabe, T. Analysis of salt-inducible genes in barley roots by differential display. J. Plant Res. 2002, 115, 119–130. [Google Scholar] [CrossRef]
- Mou, Z.; Wang, X.; Fu, Z.; Dai, Y.; Han, C.; Ouyang, J.; Bao, F.; Hu, Y.; Li, J. Silencing of Phosphoethanolamine N-methyltransferase results in temperature-sensitive male sterility and salt hypersensitivity in Arabidopsis. Plant Cell 2002, 14, 2031–2043. [Google Scholar] [CrossRef] [PubMed]
- Tabuchi, T.; Okada, T.; Takashima, Y.; Azuma, T.; Nanmori, T.; Yasuda, T. Transcriptional response of glycinebetaine-related genes to salt stress and light in leaf beet. Plant Biotechnol. 2006, 23, 317–320. [Google Scholar] [CrossRef]
- Tabuchi, T.; Kawaguchi, Y.; Azuma, T.; Nanmori, T.; Yasuda, T. Similar regulation patterns of choline monooxygenase, phosphoethanolamine N-methyltransferase and S-adenosyl-L-methionine synthetase in leaves of the halophyte Atriplex nummularia L. Plant Cell Physiol. 2005, 46, 505–513. [Google Scholar] [CrossRef]
- Nakamura, Y. Plant phospholipid diversity: Emerging functions in metabolism and protein–lipid interactions. Trends Plant Sci. 2017, 22, 1027–1040. [Google Scholar] [CrossRef]
- Larsson, K.E.; Nyström, B.; Liljenberg, C. A phosphatidylserine decarboxylase activity in root cells of oat (Avena sativa) is involved in altering membrane phospholipid composition during drought stress acclimation. Plant Physiol. Biochem. 2006, 44, 211–219. [Google Scholar] [CrossRef]
- Qiu, Q.S.; Guo, Y.; Quintero, F.J.; Pardo, J.M.; Schumaker, K.S.; Zhu, J.K. Regulation of vacuolar Na+/H+ exchange in Arabidopsis thaliana by the salt-overly-sensitive (SOS) pathway. J. Biol. Chem. 2004, 279, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Barkla, B.J.; Vera-Estrella, R.; Raymond, C. Single-cell-type quantitative proteomic and ionomic analysis of epidermal bladder cells from the halophyte model plant Mesembryanthemum crystallinum to identify salt-responsive proteins. BMC Plant Boil. 2016, 16, 637. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zeng, L.; Chen, S.; Sun, H.; Ma, S. Transcription profile analysis of Lycopersicum esculentum leaves, unravels volatile emissions and gene expression under salinity stress. Plant Physiol. Biochem. 2018, 126, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Meng, Y.; Li, B.; Ma, X.; Lai, Y.; Si, E.; Yang, K.; Xu, X.; Shang, X.; Wang, H.; et al. Physiological and proteomic analyses of salt stress response in the halophyte Halogeton glomeratus. Plant. Cell Environ. 2015, 38, 655–669. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Gollop, N.; Heuer, B. Proteomic analysis of salt-stressed tomato (Solanum lycopersicum) seedlings: Effect of genotype and exogenous application of glycinebetaine. J. Exp. Bot. 2009, 60, 2005–2019. [Google Scholar] [CrossRef]
- Zhang, A.; Han, D.; Wang, Y.; Mu, H.; Zhang, T.; Yan, X.; Pang, Q. Transcriptomic and proteomic feature of salt stress-regulated network in Jerusalem artichoke (Helianthus tuberosus L.) root based on de novo assembly sequencing analysis. Planta 2018, 247, 715–732. [Google Scholar] [CrossRef]
- Heilmann, I. Plasma membrane phosphatidylinositol 4,5-bisphosphate levels decrease with time in culture. Plant Physiol. 2001, 126, 1507–1518. [Google Scholar] [CrossRef][Green Version]
- Zhang, J.; Liu, H.; Sun, J.; Li, B.; Zhu, Q.; Chen, S.; Zhang, H. Arabidopsis fatty acid desaturase FAD2 is required for salt tolerance during seed germination and early seedling growth. PLoS ONE 2012, 7, e30355. [Google Scholar] [CrossRef]
- Wang, H.S.; Yu, C.; Tang, X.F.; Zhu, Z.J.; Ma, N.N.; Meng, Q.W. A tomato endoplasmic reticulum (ER)-type omega-3 fatty acid desaturase (LeFAD3) functions in early seedling tolerance to salinity stress. Plant. Cell Rep. 2014, 33, 131–142. [Google Scholar] [CrossRef]
- Zhang, J.-T.; Zhu, J.-Q.; Zhu, Q.; Liu, H.; Gao, X.-S.; Zhang, H.-X. Fatty acid desaturase-6 (Fad6) is required for salt tolerance in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2009, 390, 469–474. [Google Scholar] [CrossRef]

| Species | Tissue | Salt Tolerance Level a | NaCl Treatment | Treatment Duration | LCsalt-treated/LCcontrol | Ref. |
|---|---|---|---|---|---|---|
| Catharanthus roseus | cell culture | salt-sensitive | 50 mM | 1 week | 1.5 * | [25] |
| 50 mM | 32 weeks | 1.5 * | ||||
| 100 mM | 1 week | 1.2 * | ||||
| Hordeum vulgare | roots | salt-tolerant | 50 mM | 4.3 weeks | 0.7 * | [26] |
| 100 mM | 4.3 weeks | 0.7 * | ||||
| 150 mM | 4.3 weeks | 0.6 * | ||||
| 200 mM | 4.3 weeks | 0.4 * | ||||
| Hordeum maritimum | roots | salt-tolerant | 50 mM | 4.3 weeks | 1.4 * | [26] |
| 100 mM | 4.3 weeks | 1.1 | ||||
| 150 mM | 4.3 weeks | 0.9 | ||||
| 200 mM | 4.3 weeks | 0.7 * | ||||
| Crithmum maritimum | leaves | halophyte | 50 mM | 5 weeks | 1.3 * | [27] |
| 100 mM | 5 weeks | 1.3 * | ||||
| Crithmum maritimum | leaves | halophyte | 200 mM | 5 weeks | 0.9 * | [27] |
| Lipid Class | Species | Tissue | Salt Tolerant Level b | Membrane Class | NaCl Treatment | Treatment Duration | LCsalt-treated/LCcontrol | Ref. |
|---|---|---|---|---|---|---|---|---|
| total PL | Triticum aestivum | roots | salt-sensitive | PM | 100 mM | 1 week | 0.7 * | [28] |
| Buchloe dactyloides | roots | salt-sensitive | PM | 100 mM | 4 days | 0.7 * | [29] | |
| Anacardium occidentale | seedling | salt-sensitive | PM | 8 dS m−1 c | 4 weeks | 1.9 * | [30] | |
| Catharanthus roseus | cell culture | salt-sensitive | total | 50 mM | 1 week | 1.0 * | [25] | |
| Catharanthus roseus | cell culture | salt-sensitive | total | 50 mM | 32 weeks | 1.2 * | [25] | |
| Catharanthus roseus | cell culture | salt-sensitive | total | 100 mM | 1 week | 1.9 * | [25] | |
| Zea mays | roots | salt-sensitive | PM | 150 mM | 2.1 weeks | 2.6 # | [31] | |
| Triticum sativum | roots | salt-sensitive | PM | 150 mM | 3 weeks | 1.7 * | [32] | |
| Anacardium occidentale | seedling | salt-tolerant | PM | 8 dS m−1 c | 4 weeks | 1.1 | [30] | |
| Buchloe dactyloides | roots | salt-tolerant | PM | 100 mM | 4 days | 0.9 * | [29] | |
| Lycopersicon esculentum | calli | salt-tolerant | PM | 50 mM | 24 weeks | 1.1 # | [33] | |
| Lycopersicon esculentum | calli | salt-tolerant | PM | 100 mM | 24 weeks | 1.5 # | [33] | |
| Kosteletzkya virginica | roots | halophyte | PM | 85 mol m−3 | 2 weeks | 2.3 | [34] | |
| Kosteletzkya virginica | cell culture | halophyte | PM | 85 mol m−3 | 2 weeks | 3.2 * | [34] | |
| Mesembryanthemum crystallinum | epidermal bladder cells | halophyte | total | 200 mM | 2 weeks | 1.5 * | [35] | |
| Spartina patens | cell culture | halophyte | PM | 170 mM | 10 weeks | 1.2 | [36] | |
| Spartina patens | cell culture | halophyte | PM | 340 mM | 10 weeks | 1.6 | [36] | |
| Zea mays | roots | salt-sensitive | PM | 150 mM | 2.1 weeks | 0.6 # | [31] | |
| PC | Triticum aestivum | roots | salt-sensitive | PM | 100 mM | 1 week | 0.9 * | [28] |
| Catharanthus roseus | cell culture | salt-sensitive | total | 50 mM | 1 week | 1.0 | [25] | |
| Catharanthus roseus | cell culture | salt-sensitive | total | 50 mM | 32 weeks | 1.1 * | [25] | |
| Catharanthus roseus | cell culture | salt-sensitive | total | 100 mM | 1 week | 1.6 * | [25] | |
| Zea mays | roots | salt-sensitive | PM | 150 mM | 2.1 weeks | 0.8 # | [31] | |
| Triticum sativum | roots | salt-sensitive | PM | 150 mM | 3 weeks | 0.7 * | [32] | |
| Buchloe dactyloides | roots | salt-sensitive | PM | 100 mM | 4 days | 0.4 * | [29] | |
| Lycopersicon esculentum | calli | salt-tolerant | PM | 50 mM | 24 weeks | 1.0 # | [33] | |
| Lycopersicon esculentum | calli | salt-tolerant | PM | 100 mM | 24 weeks | 1.4 # | [33] | |
| Zea mays | roots | salt-tolerant | PM | 150 mM | 2.1 weeks | 0.4 # | [31] | |
| Buchloe dactyloides | roots | salt-tolerant | PM | 100 mM | 4 days | 1.5 * | [29] | |
| Spartina patens | cell culture | halophyte | PM | 170 mM | 10 weeks | ≈1.1 | [36] | |
| Spartina patens | cell culture | halophyte | PM | 340 mM | 10 weeks | ≈1.8 * | [36] | |
| Mesembryanthemum crystallinum | epidermal bladder cells | halophyte | total | 200 mM | 2 weeks | 2.2 * | [35] | |
| PE | Triticum aestivum | roots | salt-sensitive | PM | 100 mM | 1 week | 1.0 | [28] |
| Catharanthus roseus | cell culture | salt-sensitive | total | 50 mM | 1 week | 1.0 | [25] | |
| Catharanthus roseus | cell culture | salt-sensitive | total | 50 mM | 32 weeks | 1.1 | [25] | |
| Buchloe dactyloides | roots | salt-sensitive | PM | 100 mM | 4 days | 1.0 | [29] | |
| Buchloe dactyloides | roots | salt-sensitive | PM | 100 mM | 1 week | 1.9 * | [29] | |
| Zea mays | roots | salt-sensitive | PM | 150 mM | 2.1 weeks | 1.0 # | [31] | |
| Triticum sativum | roots | salt-sensitive | PM | 150 mM | 3 weeks | 1.8 * | [32] | |
| Lycopersicon esculentum | calli | salt-tolerant | PM | 50 mM | 24 weeks | 1.2 # | [33] | |
| Lycopersicon esculentum | calli | salt-tolerant | PM | 100 mM | 24 weeks | 1.4 # | [33] | |
| Zea mays | roots | salt-tolerant | PM | 150 mM | 2.1 weeks | 0.6 # | [31] | |
| Buchloe dactyloides | roots | salt-sensitive | PM | 100 mM | 4 days | 0.9 | [29] | |
| Mesembryanthemum crystallinum | epidermal bladder cells | halophyte | total | 200 mM | 2 weeks | 1.1 | [35] | |
| Spartina patens | cell culture | halophyte | PM | 170 mM | 10 weeks | ≈0.7 | [36] | |
| Spartina patens | cell culture | halophyte | PM | 340 mM | 10 weeks | ≈1.0 | [36] | |
| PC/PE | Catharanthus roseus | cell culture | salt-sensitive | total | 50 mM | 1 week | 1.0 # | [25] |
| Catharanthus roseus | cell culture | salt-sensitive | total | 50 mM | 32 weeks | 1.1 # | [25] | |
| Catharanthus roseus | cell culture | salt-sensitive | total | 100 mM | 1 week | 0.8 # | [25] | |
| Triticum sativum | roots | salt-sensitive | PM | 100 mM | 1 week | 0.8 | [28] | |
| Zea mays | roots | salt-sensitive | PM | 150 mM | 2.1 weeks | 0.8 # | [31] | |
| Triticum sativum | roots | salt-sensitive | PM | 150 mM | 3 weeks | 0.4 * | [32] | |
| Zea mays | roots | salt-tolerant | PM | 150 mM | 2.1 weeks | 0.6 # | [31] | |
| PS | Zea mays | roots | salt-sensitive | PM | 150 mM | 2.1 weeks | 1.4 # | [31] |
| Buchloe dactyloides | roots | salt-sensitive | PM | 100 mM | 4 days | 0.4 * | [29] | |
| Triticum sativum | roots | salt-sensitive | PM | 150 mM | 3 weeks | 2.2 * | [32] | |
| Zea mays | roots | salt-tolerant | PM | 150 mM | 2.1 weeks | 0.7 # | [31] | |
| Buchloe dactyloides | roots | salt-tolerant | PM | 100 mM | 4 days | 1.3 * | [29] | |
| Spartina patens | cell culture | halophyte | PM | 170 mM | 10 weeks | ≈7.2 * | [36] | |
| Spartina patens | cell culture | halophyte | PM | 340 mM | 10 weeks | ≈7.8 * | [36] | |
| PI | Zea mays | roots | salt-sensitive | PM | 150 mM | 2.1 weeks | 1.4 # | [31] |
| Buchloe dactyloides | roots | salt-sensitive | PM | 100 mM | 4 days | 0.5 * | [29] | |
| Triticum aestivum | roots | salt-sensitive | PM | 100 mM | 1 week | 1.1 | [28] | |
| Triticum sativum | roots | salt-sensitive | PM | 150 mM | 3 weeks | 1.5 * | [32] | |
| Zea mays | roots | salt-tolerant | PM | 150 mM | 2.1 weeks | 0.6# | [31] | |
| Buchloe dactyloides | roots | salt-tolerant | PM | 100 mM | 4 days | 1.9 * | [29] | |
| Spartina patens | cell culture | halophyte | PM | 170 mM | 10 weeks | ≈3.5 * | [36] | |
| Spartina patens | cell culture | halophyte | PM | 340 mM | 10 weeks | ≈2.4 * | [36] | |
| PG | Triticum aestivum | roots | salt-sensitive | PM | 100 mM | 1 week | 1.0 | [28] |
| Zea mays | roots | salt-sensitive | PM | 150 mM | 2.1 weeks | 0.9# | [31] | |
| Triticum sativum | roots | salt-sensitive | PM | 150 mM | 3 weeks | 0.8 | [32] | |
| Buchloe dactyloides | roots | salt-sensitive | PM | 100 mM | 4 days | 1.4 | [29] | |
| Arabidopsis thaliana | leaves | salt-sensitive | total | 100 mM | 5 days | 0.7 * | [37] | |
| Arabidopsis thaliana | leaves | salt-sensitive | total | 200 mM | 5 days | 0.5 * | [37] | |
| Zea mays | roots | salt-tolerant | PM | 150 mM | 2.1 weeks | 2.8 # | [31] | |
| Buchloe dactyloides | roots | salt-tolerant | PM | 100 mM | 4 days | 2.2 * | [29] | |
| Thellungiella halophila | leaves | halophyte | total | 100 mM | 5 days | 1.0 | [37] | |
| Thellungiella halophila | leaves | halophyte | total | 200 mM | 5 days | 1.3 * | [37] | |
| Thellungiella halophila | leaves | halophyte | total | 300 mM | 5 days | 1.3 * | [37] | |
| Mesembryanthemum crystallinum | epidermal bladder cells | halophyte | total | 200 mM | 2 weeks | 1.5 * | [35] | |
| PA | Triticum aestivum | roots | salt-sensitive | PM | 100 mM | 1 week | 1.3 | [28] |
| Zea mays | roots | salt-sensitive | PM | 150 mM | 2.1 weeks | 0.7 # | [31] | |
| Triticum sativum | roots | salt-sensitive | PM | 150 mM | 3 weeks | 0.7 | [32] | |
| Lycopersicon esculentum | calli | salt-tolerant | PM | 50 mM | 24 weeks | 1.2 # | [33] | |
| Lycopersicon esculentum | calli | salt-tolerant | PM | 100 mM | 24 weeks | 1.7 # | [33] | |
| Zea mays | roots | salt-tolerant | PM | 150 mM | 2.1 weeks | 0.7 # | [31] |
| Lipid Class. | Species | Tissue | Salt Tolerant Level b | Membrane Class | NaCl Treatment | Treatment Duration | LCsalt-treated/LCcontrol | Ref. |
|---|---|---|---|---|---|---|---|---|
| total GL | Triticum aestivum | roots | salt-sensitive | PM | 100 mM | 1 week | 1.1 | [28] |
| Zea mays | roots | salt-sensitive | PM | 150 mM | 2.1 weeks | 0.8 # | [31] | |
| Brassica Napus | roots | salt-sensitive | PM | 50 mM | 4.3 weeks | 0.8 # c | [38] | |
| Brassica Napus | roots | salt-sensitive | PM | 100 mM | 4.3 weeks | 0.8 # c | [38] | |
| Brassica Napus | roots | salt-sensitive | PM | 150 mM | 4.3 weeks | 0.7 # c | [38] | |
| Brassica Napus | roots | salt-sensitive | PM | 200mM | 4.3 weeks | 0.7 # c | [38] | |
| Zea mays | roots | salt-tolerant | PM | 150 mM | 2.1 weeks | 0.6 # | [31] | |
| Brassica Napus | roots | salt-tolerant | PM | 50 mM | 4.3 weeks | 0.7 # d | [38] | |
| Brassica Napus | roots | salt-tolerant | PM | 100 mM | 4.3 weeks | 0.7 # d | [38] | |
| Brassica Napus | roots | salt-tolerant | PM | 150 mM | 4.3 weeks | 0.7 # d | [38] | |
| Brassica Napus | roots | salt-tolerant | PM | 200mM | 4.3 weeks | 0.6 # d | [38] | |
| Spartina patens | cell culture | halophyte | PM | 170 mM | 10 weeks | 1.2 | [36] | |
| Spartina patens | cell culture | halophyte | PM | 340 mM | 10 weeks | 1.1 | [36] | |
| MGDG | Catharanthus roseus | cell culture | salt-sensitive | total | 50 mM | 1 week | 0.9 | [25] |
| Catharanthus roseus | cell culture | salt-sensitive | total | 50 mM | 32 weeks | 0.8 * | [25] | |
| Catharanthus roseus | cell culture | salt-sensitive | total | 100 mM | 1 week | 0.6 * | [25] | |
| Arabidopsis thaliana | leaves | salt-sensitive | total | 100 mM | 5 days | 1.1 | [37] | |
| Arabidopsis thaliana | leaves | salt-sensitive | total | 200 mM | 5 days | 1.2 * | [37] | |
| Zea mays | mesophyll | salt-sensitive e | chloroplasts | 3% | 5 days | ≈0.6 * | [39] | |
| Sulla carnosa | leaves | salt-sensitive | total | 200 mM | 2.9 weeks | ≈0.3 | [40] | |
| Sulla coronaria | leaves | salt-sensitive | total | 200 mM | 2.9 weeks | ≈0.5 * | [40] | |
| Zea mays | bundle sheath | salt-tolerant e | chloroplasts | 3% | 5 days | ≈1.0 | [39] | |
| Thellungiella halophila | leaves | halophyte | total | 100 mM | 5 days | 1.0 | [37] | |
| Thellungiella halophila | leaves | halophyte | total | 200 mM | 5 days | 0.9 * | [37] | |
| Thellungiella halophila | leaves | halophyte | total | 300 mM | 5 days | 0.9 * | [37] | |
| Mesembryanthemum crystallinum | epidermal bladder cells | halophyte | total | 200 mM | 2 weeks | 1.2 | [35] | |
| Crithmum maritimum | leaves | halophyte | total | 50 mM | 5 weeks | 1.3 * | [27] | |
| Crithmum maritimum | leaves | halophyte | total | 100 mM | 5 weeks | 1.2 | [27] | |
| Crithmum maritimum | leaves | halophyte | total | 200 mM | 5 weeks | 0.8 | [27] | |
| DGDG | Catharanthus roseus | cell culture | salt-sensitive | total | 50 mM | 1 week | 1.3 * | [25] |
| Catharanthus roseus | cell culture | salt-sensitive | total | 50 mM | 32 weeks | 1.3 * | [25] | |
| Catharanthus roseus | cell culture | salt-sensitive | total | 100 mM | 1 week | 0.7 * | [25] | |
| Arabidopsis thaliana | leaves | salt-sensitive | total | 100 mM | 5 days | 0.8 * | [37] | |
| Arabidopsis thaliana | leaves | salt-sensitive | total | 200 mM | 5 days | 0.7 * | [37] | |
| Zea mays | mesophyll | salt-sensitive e | chloroplasts | 3% | 5 days | ≈1.1 | [39] | |
| Sulla carnosa | leaves | salt-sensitive | total | 200 mM | 2.9 weeks | ≈0.3 * | [40] | |
| Sulla coronaria | leaves | salt-sensitive | total | 200 mM | 2.9 weeks | ≈0.5 * | [40] | |
| Zea mays | bundle sheath | salt-tolerant e | chloroplasts | 3% | 5 days | 1.09 | [39] | |
| Thellungiella halophila | leaves | halophyte | total | 100 mM | 5 days | 0.8 * | [37] | |
| Thellungiella halophila | leaves | halophyte | total | 200 mM | 5 days | 0.6 * | [37] | |
| Thellungiella halophila | leaves | halophyte | total | 300 mM | 5 days | 0.6 * | [37] | |
| Mesembryanthemum crystallinum | epidermal bladder cells | halophyte | total | 200 mM | 2 weeks | 1.2 | [35] | |
| Crithmum maritimum | leaves | halophyte | total | 50 mM | 5 weeks | 1.4 | [27] | |
| Crithmum maritimum | leaves | halophyte | total | 100 mM | 5 weeks | 1.3 | [27] | |
| Crithmum maritimum | leaves | halophyte | total | 200 mM | 5 weeks | 0.7 * | [27] | |
| SQDG | Catharanthus roseus | cell culture | salt-sensitive | total | 50 mM | 1 week | 1.3 | [25] |
| Catharanthus roseus | cell culture | salt-sensitive | total | 50 mM | 32 weeks | 2.9 * | [25] | |
| Catharanthus roseus | cell culture | salt-sensitive | total | 100 mM | 1 week | 2.3 * | [25] | |
| Arabidopsis thaliana | leaves | salt-sensitive | total | 100 mM | 5 days | 1.2 * | [37] | |
| Arabidopsis thaliana | leaves | salt-sensitive | total | 200 mM | 5 days | 1.2 * | [37] | |
| Zea mays | mesophyll | salt-sensitive e | chloroplasts | 3% | 5 days | 0.6 * | [39] | |
| Sulla carnosa | leaves | salt-sensitive | total | 200 mM | 2.9 weeks | ≈0.4 * | [40] | |
| Sulla coronaria | leaves | salt-sensitive | total | 200 mM | 2.9 weeks | ≈0.6 * | [40] | |
| Zea mays | bundle sheath | salt-tolerant e | chloroplasts | 3% | 5 days | 0.7 * | [39] | |
| Crithmum maritimum | leaves | halophyte | total | 50 mM | 5 weeks | 3.3 * | [27] | |
| Crithmum maritimum | leaves | halophyte | total | 100 mM | 5 weeks | 2.8 * | [27] | |
| Crithmum maritimum | leaves | halophyte | total | 200 mM | 5 weeks | 2.6 | [27] | |
| Aster tripolium | leaves | halophyte | total | 258 mM | 1.4 weeks | ≈1.3 # | [41] | |
| Aster tripolium | leaves | halophyte | total | 517 mM | 1.4 weeks | ≈1.5 # | [41] | |
| Sesuvium portulacastrum | leaves | halophyte | total | 428 mM | 1.4 weeks | ≈1.6 # | [41] | |
| Sesuvium portulacastrum | leaves | halophyte | total | 856 mM | 1.4 weeks | ≈2.1 # | [41] | |
| Thellungiella halophila | leaves | halophyte | total | 100 mM | 5 days | 1.5 * | [37] | |
| Thellungiella halophila | leaves | halophyte | total | 200 mM | 5 days | 1.7 * | [37] | |
| Thellungiella halophila | leaves | halophyte | total | 300 mM | 5 days | 1.8 * | [37] | |
| MGDG/DGDG | Arabidopsis thaliana | leaves | salt-sensitive | total | 100 mM | 5 days | 1.2 # | [37] |
| Arabidopsis thaliana | leaves | salt-sensitive | total | 200 mM | 5 days | 1.7 # | [37] | |
| Sulla coronaria | leaves | salt-sensitive | total | 200 mM | 2.9 weeks | ≈0.9 | [40] | |
| Sulla carnosa | leaves | salt-sensitive | total | 200 mM | 2.9 weeks | ≈3.1 * | [40] | |
| Thellungiella halophila | leaves | halophyte | total | 100 mM | 5 days | 1.3 # | [37] | |
| Thellungiella halophila | leaves | halophyte | total | 200 mM | 5 days | 1.4 # | [37] | |
| Thellungiella halophila | leaves | halophyte | total | 300 mM | 5 days | 1.4 # | [37] |
| Species | Tissue | Salt Tolerant Level b | Membrane Preparation Method | Membrane Class | PA | PS | PI | PC | PE | PG | MGDG | DGDG | SQDG | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Spinacia oleracea | leaves | salt-sensitive | Two-phase partitioning | PM | 6.1 | - | - | 25.4 | 24.4 | 3.1 | nd | 2.7 c | - | [64] |
| Brassica oleracea | buds | salt-sensitive | Two-phase partitioning | PM | 13.1 | - | - | 17.6 | 13.7 | 3.4 | 0.3 c | 1.0 c | - | [64] |
| Hordeum vulgare | leaves | salt-sensitive | Two-phase partitioning | PM | 9.2 | - | - | 17.5 | 10.8 | 2.1 | 3.7 c | 1.4 c | - | [64] |
| Hordeum vulgare | roots | salt-sensitive | Two-phase partitioning | PM | 1.7 | - | - | 8.2 | 6.8 | 0.8 | 1.0 c | 0.3 c | - | [64] |
| Hordeum vulgare | roots | salt-sensitive | density gradient centrifugation | PM | 5.6 | 6.6 | 5.1 d | 12.7 | 33.8 | 3.6 | - | - | - | [65] |
| Hordeum vulgare | roots | salt-sensitive | density gradient centrifugation | ER | 1.0 | 1.5 | 9.8 d | 59.5 | 20.6 | 6.9 | - | - | - | [65] |
| Hordeum vulgare | roots | salt-sensitive | density gradient centrifugation | Tonoplast + Golgi | 1.8 | 2.5 | 7.4 d | 57.6 | 23.2 | 6.3 | - | - | - | [65] |
| Zea mays | roots | salt-sensitive | Two-phase partitioning | PM | 6.8 | 13.6 | 12.3 | 20.4 | 13.6 | 21.1 | - | - | - | [31] |
| Vigna radiata | hypocotyl | salt-sensitive | density gradient centrifugation | PM | 8.0 | 1.5 | 2.6 | 16 | 18.6 | 2.2 | 0.2 c | 0.8 c | - | [66] |
| Vigna radiata | hypocotyl | salt-sensitive | density gradient centrifugation | Tonoplast | 1.1 | 2.2 | 5.7 | 23.7 | 16.0 | 2.3 | 1.0 c | 3.4 c | - | [66] |
| Triticum aestivum | roots | salt-sensitive | Two-phase partitioning | PM | 13.1 | 9.2 | 5.9 | 18 | 10 | 28.3 | - | - | - | [32] |
| Zea mays | roots | salt-tolerant | Two-phase partitioning | PM | 7.5 | 19.4 | 14.9 | 22.4 | 19.4 | 16.4 | - | - | - | [31] |
| Dunalieila salina (alga) | cell culture | halotolerant | density gradient centrifugation | PM | - | - | - | 13.2 | 10.7 | 5.3 | 3.1 c | 2.9 c | 1.7 c | [67] |
| Species | Tissue | Salt Tolerant Level a | Membrane Type | 14:0 | 16:0 | 16:1 | 17:0 | 18:0 | 18:1 | 18:2 | 18:3 | 20:0 | 20:1 | 20:2 | 20:3 | 22:0 | 22:1 | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Arabidopsis thaliana | leaves | salt-sensitive | total | - | 22.3 | 2.5 | - | 22.8 | 22.7 | 7.8 | 21.9 | - | - | - | - | - | - | [37] |
| Spinacia oleracea | leaves | salt-sensitive | plasma membrane | - | 29.7 | 1.4 | - | 3.2 | 10.4 | 27.1 | 27.3 | nd | 1.0 | - | - | nd | - | [64] |
| Brassica oleracea | inflorescences | salt-sensitive | plasma membrane | - | 20.8 | 1.6 | - | 2.4 | 13.4 | 17.4 | 42.3 | 1.4 | 0.7 | - | - | tr | - | [64] |
| Hordeum vulgare | leaves | salt-sensitive | plasma membrane | - | 31.5 | 0.9 | - | 4.1 | 3.1 | 32.9 | 25.8 | tr | tr | - | - | 1.8 | - | [64] |
| Hordeum vulgare | roots | salt-sensitive | plasma membrane | - | 43.9 | 1.5 | - | 2.7 | 2.3 | 33.5 | 12.8 | 0.4 | 2.1 | - | - | 0.9 | - | [64] |
| Zea mays | roots | salt-sensitive | plasma membrane | - | 19.2 | - | 5.5 | 21.7 | 27.6 | 7.1 | - | 18.5 | - | - | - | - | - | [31] |
| Vigna radiata | hypocotyl | salt-sensitive | plasma membrane | - | 35.0 | - | - | 5.9 | 9.2 | 21.4 | 19.0 | - | 1.6 | 1.6 | 2.2 | - | 1.1 | [66] |
| Vigna radiata | hypocotyl | salt-sensitive | Tonoplast | - | 39.4 | - | - | 6.1 | 9.1 | 22.2 | 19.8 | - | 1.5 | 1.2 | 0.8 | - | 2.1 | [66] |
| Avena sativa | roots | salt-sensitive | plasma membrane | nd | 22.5 | tr | - | tr | 3.0 | 48.0 | 25.0 | - | - | - | - | - | - | [68] |
| Triticum aestivum | roots | salt-sensitive | plasma membrane | - | 17.4 | 9.6 | 13.2 | 13.6 | 12.3 | 14.6 | - | 19.3 | - | - | - | - | - | [32] |
| Zea mays | roots | salt-tolerant | plasma membrane | - | 6.1 | - | 26.3 | 4.1 | 23.4 | 14.6 | - | 25.3 | - | - | - | - | - | [31] |
| Boea hygroscopica | leaves | not sure | thylakoid membranes | 2.0 | 28.0 | 2.0 | 7.0 | 8.0 | 36.0 | 17.0 | - | - | - | - | - | - | [69] | |
| Thellungiella halophila | leaves | halophyte | total | - | 41.0 | 6.0 | - | 23 | 7.7 | 0.9 | 21.3 | - | - | - | - | - | - | [37] |
| Spartina patens | Callus tissue | halophyte | plasma membrane | - | 12.9 | - | 0.3 | 0.8 | - | 15.8 | - | 0.5 | - | - | - | 1.0 | - | [36] |
| Protein | Organism | Protein Abundance | Transcript Regulation | Ref. |
|---|---|---|---|---|
| acyl carrier proteins 1 | Thellungiella halophila; Arabidopsis thaliana | down | [165] | |
| acyl carrier proteins 4 | Thellungiella halophila; Arabidopsis thaliana | down | [165] | |
| acetyl-CoA carboxylase carboxyl transferase subunit beta | Oleaginous Diatom | up | [173] | |
| carboxyltransferase β -subunit | Chlamydomonas sp. JSC4 | up | [174] | |
| dihydrolipoyllysine-residue acetyltransferase 2 | Thellungiella halophila; Arabidopsis thaliana | down | [165] | |
| biotin carboxylase | Chlamydomonas reinhardtii | decreased | [175] | |
| Cucumis sativus | increased | [167] | ||
| Medicago truncatula | decreased | [166] | ||
| Thellungiella halophila | up | [165] | ||
| Chlamydomonas sp. JSC4 | up | [174] | ||
| long-chain acyl-CoA synthetase 2 | Thellungiella halophila | up | [165] | |
| ketoacyl-ACP reductase | Synechocystis sp. PCC | up | [176] | |
| enoyl-ACP reductase | Thellungiella halophila; Arabidopsis thaliana | down | [165] | |
| Oryza sativa | increased | [169] | ||
| glyceraldehyde-3-phosphate dehydrogenase | Oryza sativa | up | [171] | |
| Pleurotus sajor-caju | up | [177] | ||
| glycerol-3-phosphate dehydrogenase | Dunaliella salina | down | [170] | |
| Suaeda salsa | up | [178] | ||
| glycerol-3-phosphate acyltransferase | Lepidium latifolium | down | [179] | |
| UDP-glucose pyrophosphorylase | Solanum lycopersicum | up | [180] | |
| Hordeum vulgare | up | [181] | ||
| Oryza sativa | down | [182] | ||
| phosphoethanolamine N-methyltransferase | Zea mays | up | [183] | |
| Sugar beet monosomic addition line | increased | [184] | ||
| choline kinase | Spinacia oleracea | no-change | [185] | |
| non-specific phospholipase C4 | Arabidopsis thaliana | up | [186] | |
| non-specific phospholipase C5 | Arabidopsis thaliana | up | [187] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, Q.; Liu, L.; Barkla, B.J. Membrane Lipid Remodeling in Response to Salinity. Int. J. Mol. Sci. 2019, 20, 4264. https://doi.org/10.3390/ijms20174264
Guo Q, Liu L, Barkla BJ. Membrane Lipid Remodeling in Response to Salinity. International Journal of Molecular Sciences. 2019; 20(17):4264. https://doi.org/10.3390/ijms20174264
Chicago/Turabian StyleGuo, Qi, Lei Liu, and Bronwyn J. Barkla. 2019. "Membrane Lipid Remodeling in Response to Salinity" International Journal of Molecular Sciences 20, no. 17: 4264. https://doi.org/10.3390/ijms20174264
APA StyleGuo, Q., Liu, L., & Barkla, B. J. (2019). Membrane Lipid Remodeling in Response to Salinity. International Journal of Molecular Sciences, 20(17), 4264. https://doi.org/10.3390/ijms20174264