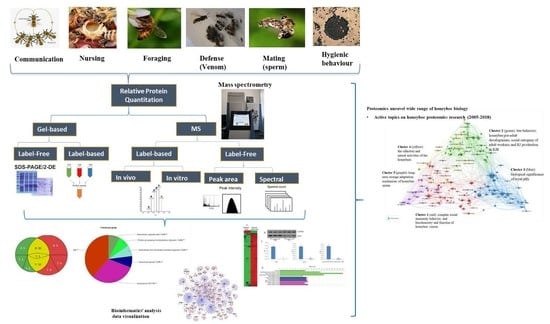
The Emerging Proteomic Research Facilitates in-Depth Understanding of the Biology of Honeybees
Abstract
1. Introduction
2. Proteomics Unravels the Molecular Basis for a Wide Range of Honeybee Biology
2.1. The Molecular Basis Behind a Complex Social Immunity Behavior
2.2. Understanding of Biochemistry and Function of Honeybee Venom
2.3. The Molecular Basis of Neurology Implicated in the Behavior of Honeybees
2.4. The Molecular Biology Behind the Honeybee Pre-Adult Developments
2.5. Social Ontogeny of Adult Workers
2.6. The Molecular Basis for Enhanced RJ Production in RJB
2.7. Proteome, Phosphoproteome, and Glycoproteome Analyses of RJ from Different Bee Species or Strains Reveal the Biological Significance to Honeybees
2.8. Antennal Proteomics Reveal the Olfactory and Neural Activity of Honeybees
2.9. Molecular Basis for Long-Term Storage Adaptation of Honeybee Sperm
3. Future Research Directions
Acknowledgments
Conflicts of Interest
References
- Michener, C.D. The Bees of the World, 2nd ed.; The Johns Hopkins University Press: Baltimore, MD, USA, 2007; ISBN 9780801861338. [Google Scholar]
- Klein, A.-M.; Vaissiere, B.E.; Cane, J.H.; Steffan-Dewenter, I.; Cunningham, S.A.; Kremen, C.; Tscharntke, T. Importance of pollinators in changing landscapes for world crops. Proc. R. Soc. B Biol. Sci. 2007, 274, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Abrol, D.P. Asiatic Honeybee Apis cerana: Biodiversity Conservation and Agricultural Production; Springer Science: Dordrecht, The Netherlands, 2013. [Google Scholar]
- Chen, C.; Wang, H.; Liu, Z.; Chen, X.; Tang, J.; Meng, F.; Shi, W. Population genomics provide insights into the evolution and adaptation of the eastern honey bee (Apis cerana). Mol. Biol. Evol. 2018, 35, 2260–2271. [Google Scholar] [CrossRef] [PubMed]
- Hora, Z.A.; Altaye, S.Z.; Wubie, A.J.; Li, J. Proteomics Improves the New Understanding of Honeybee Biology. J. Agric. Food Chem. 2018, 66, 3605–3615. [Google Scholar] [CrossRef] [PubMed]
- The Honeybee Genome Sequencing Consortium. Insights into social insects from the genome of the honeybee Apis mellifera. Nature 2006, 443, 931–949. [Google Scholar] [CrossRef] [PubMed]
- Peiren, N.; Vanrobaeys, F.; De Graaf, D.C.; Devreese, B.; Van Beeumen, J.; Jacobs, F.J. The protein composition of honeybee venom reconsidered by a proteomic approach. Biochim. Biophys. Acta 2005, 1752, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Chan, Q.W.T.; Howes, C.G.; Foster, L.J. Quantitative Comparison of Caste Differences in Honeybee Hemolymph. Mol. Cell. Proteom. 2006, 5, 2252–2262. [Google Scholar] [CrossRef]
- Bogaerts, A.; Baggerman, G.; Vierstraete, E.; Schoofs, L.; Verleyen, P. The hemolymph proteome of the honeybee: Gel-based or gel-free? Proteomics 2009, 9, 3201–3208. [Google Scholar] [CrossRef]
- Li, J.; Huawei, L.; Zhaohui, Z.; Yinghong, P. Identification of the proteome complement of high royal jelly producing bees (Apis mellifera) during worker larval development. Apidologie 2007, 38, 545–557. [Google Scholar] [CrossRef]
- Zhang, L.; Li, J.K.; Wu, L.M. Profile Analysis of the Proteome of the Egg of the High Royal Jelly Producing Bees (Apis mellifera L.). Agric. Sci. China 2007, 6, 1138–1148. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Mann, M.; Kelleher, N.L. Precision proteomics: The case for high resolution and high mass accuracy. Proc. Natl. Acad. Sci. USA 2008, 105, 18132–18138. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Q.; Li, R.; Dai, J.; Li, Q.; Su, Z.; Guo, Y.; Li, C.; Shyr, Y.; Zeng, R. Preprocessing Significantly Improves the Peptide/Protein Identification Sensitivity of High-resolution Isobarically Labeled Tandem Mass Spectrometry Data. Mol. Cell. Proteom. 2015, 14, 405–417. [Google Scholar] [CrossRef] [PubMed]
- Chan, Q.W.T.; Yi Chan, M.; Logan, M.; Fang, Y.; Higo, H.; Foster, L.J. Honey bee protein atlas at organ-level resolution. Genome Res. 2013, 23, 1951–1960. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Bienefeld, K.; Wegener, J.; Zautke, F.; Hao, Y.; Feng, M.; Han, B.; Fang, Y.; Wubie, A.J.; Li, J. Proteome analysis of the hemolymph, mushroom body, and antenna provides novel insight into honeybee resistance against varroa infestation. J. Proteome Res. 2016, 15, 2841–2854. [Google Scholar] [CrossRef] [PubMed]
- Altaye, S.Z.; Meng, L.; Li, J. Molecular insights into the enhanced performance of royal jelly secretion by a stock of honeybee (Apis mellifera ligustica) selected for increasing royal jelly production. Apidologie 2019, 1–18. [Google Scholar] [CrossRef]
- Han, B.; Fang, Y.; Feng, M.; Hu, H.; Qi, Y.; Huo, X.; Meng, L.; Wu, B.; Li, J. Quantitative Neuropeptidome Analysis Reveals Neuropeptides Are Correlated with Social Behavior Regulation of the Honeybee Workers. J. Proteome Res. 2015, 14, 4382–4393. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Fang, Y.; Feng, M.; Hu, H.; Hao, Y.; Ma, C.; Huo, X.; Meng, L.; Zhang, X.; Wu, F.; et al. Brain Membrane Proteome and Phosphoproteome Reveal Molecular Basis Associating with Nursing and Foraging Behaviors of Honeybee Workers. J. Proteome Res. 2017, 16, 3646–3663. [Google Scholar] [CrossRef]
- Meng, L.; Huo, X.; Feng, M.; Fang, Y.; Han, B.; Hu, H.; Wu, F.; Li, J. Proteomics Reveals the Molecular Underpinnings of Stronger Learning and Memory in Eastern Compared to Western Bees. Mol. Cell. Proteom. 2017, 17, 255–269. [Google Scholar] [CrossRef]
- Hernández, L.G.; Lu, B.; Cruz, G.C.N.; Calábria, L.K.; Martins, F.; Togawa, R.; Espindola, F.S.; Iii, J.R.Y.; Ricardo, B.; Sousa, M.V. De The worker honeybee brain proteome. J. Proteome Res. 2013, 11, 1485–1493. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Feng, M.; Begna, D.; Yu, F.; Aijuan, Z. Proteome comparison of hypopharyngeal gland development between Italian and royal jelly producing worker honeybees (Apis mellifera L.). J. Proteome Res. 2010, 9, 6578–6594. [Google Scholar]
- Hu, H.; Bezabih, G.; Feng, M.; Wei, Q.; Zhang, X.; Wu, F.; Meng, L.; Fang, Y.; Han, B.; Ma, C.; et al. In-depth Proteome of the Hypopharyngeal Glands of Honeybee Workers Reveals Highly Activated Protein and Energy Metabolism in Priming the Secretion of Royal Jelly. Mol. Cell. Proteom. 2019, 18, 606–621. [Google Scholar] [CrossRef] [PubMed]
- Huo, X.; Wu, B.; Feng, M.; Han, B.; Fang, Y.; Hao, Y.; Meng, L.; Wubie, A.J.; Fan, P.; Hu, H.; et al. Proteomic Analysis Reveals the Molecular Underpinnings of Mandibular Gland Development and Lipid Metabolism in Two Lines of Honeybees (Apis mellifera ligustica). J. Proteome Res. 2016, 15, 3342–3357. [Google Scholar] [CrossRef] [PubMed]
- Feng, M.; Ramadan, H.; Han, B.; Fang, Y.; Li, J. Hemolymph proteome changes during worker brood development match the biological divergences between western honey bees (Apis mellifera) and eastern honey bees (Apis cerana). BMC Genom. 2014, 15, 563. [Google Scholar] [CrossRef] [PubMed]
- Ararso, Z.; Ma, C.; Qi, Y.; Feng, M.; Han, B.; Hu, H.; Meng, L.; Li, J. Proteome Comparisons between Hemolymph of Two Honeybee Strains (Apis mellifera ligustica) Reveal Divergent Molecular Basis in Driving Hemolymph Function and High Royal Jelly Secretion. J. Proteome Res. 2018, 17, 402–419. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Feng, M.; Han, B.; Qi, Y.; Hu, H.; Fan, P.; Huo, X.; Meng, L.; Li, J. Proteome analysis unravels mechanism underling the embryogenesis of the honeybee drone and its divergence with the worker (Apis mellifera lingustica). J. Proteome Res. 2015, 14, 4059–4071. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Feng, M.; Han, B.; Lu, X.; Ramadan, H.; Li, J. In-depth Proteomics Characterization of Embryogenesis of the Honey Bee Worker (Apis mellifera ligustica). Mol. Cell. Proteom. 2014, 13, 2306–2320. [Google Scholar] [CrossRef] [PubMed]
- Matysiak, J.; Hajduk, J.; Pietrzak, Ł.; Schmelzer, C.E.H.; Kokot, Z.J. Shotgun proteome analysis of honeybee venom using targeted enrichment strategies. Toxicon 2014, 90, 255–264. [Google Scholar] [CrossRef]
- Matysiak, J.; Hajduk, J.; Mayer, F.; Hebeler, R.; Kokot, Z.J. Hyphenated LC—MALDI—ToF/ToF and LC—ESI—QToF approach in proteomic characterization of honeybee venom. J. Pharm. Biomed. Anal. 2016, 121, 69–76. [Google Scholar] [CrossRef]
- Matysiak, J.; Hajduk, J.; Agata, Y.; T, N.N.; Kokot, Z.J. Proteomic analysis of Apis mellifera venom determined by liquid chromatography (LC) coupled with nano-LC-MALDI-TOF/TOF MS. Acta Pol. Pharm. Res. 2017, 74, 53–65. [Google Scholar]
- Parker, R.; Guarna, M.M.; Melathopoulos, A.P.; Moon, K.; White, R.; Huxter, E. Correlation of proteome-wide changes with social immunity behaviors provides insight into resistance to the parasitic mite, Varroa destructor, in the honey bee (Apis mellifera). Genome Biol. 2012, 13. [Google Scholar] [CrossRef]
- den Boer, S.P.A.; Boomsma, J.J.; Baer, B. Honey bee males and queens use glandular secretions to enhance sperm viability before and after storage. J. Insect Physiol. 2009, 55, 538–543. [Google Scholar] [CrossRef] [PubMed]
- Baer, B.; Zareie, R.; Paynter, E.; Poland, V.; Millar, A.H. Seminal fluid proteins differ in abundance between genetic lineages of honeybees. J. Proteom. 2012, 75, 5646–5653. [Google Scholar] [CrossRef] [PubMed]
- Baer, B.; Eubel, H.; Taylor, N.L.; O’Toole, N.; Millar, A.H. Insights into female sperm storage from the spermathecal fluid proteome of the honeybee Apis mellifera. Genome Biol. 2009, 10, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Zareie, R.; Eubel, H.; Millar, A.H.; Baer, B. Long-term survival of high quality sperm: Insights into the sperm proteome of the honeybee Apis mellifera. J. Proteome Res. 2013, 12, 5180–5188. [Google Scholar] [CrossRef] [PubMed]
- Baer, B.; Heazlewood, J.L.; Taylor, N.L.; Eubel, H.; Millar, A.H. The seminal fluid proteome of the honeybee Apis mellifera. Proteomics 2009, 9, 2085–2097. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.-F.; Zheng, H.-Q.; Pirk, C.W.W.; Hu, F.-L.; Xu, Z.-W. High Royal Jelly-Producing Honeybees (Apis mellifera ligustica) (Hymenoptera: Apidae) in Review. Apidologie 2016, 109, 510–514. [Google Scholar] [CrossRef] [PubMed]
- Chan, Q.W.T.; Foster, L.J. Changes in protein expression during honey bee larval development. Genome Biol. 2008, 9, R156. [Google Scholar] [CrossRef]
- Cardoen, D.; Ernst, U.R.; van Vaerenbergh, M.; Boerjan, B.; de Graaf, D.C.; Wenseleers, T.; Schoofs, L.; Verleyen, P. Differential proteomics in dequeened honeybee colonies reveals lower viral load in hemolymph of fertile worker bees. PLoS ONE 2011, 6, e20043. [Google Scholar] [CrossRef]
- Cardoen, D.; Ernst, U.R.; Boerjan, B.; Bogaerts, A.; Formesyn, E.; De Graaf, D.C.; Wenseleers, T.; Schoofs, L.; Verleyen, P. Worker honeybee sterility: A proteomic analysis of suppressed ovary activation. J. Proteome Res. 2012, 11, 2838–2850. [Google Scholar] [CrossRef][Green Version]
- Erban, T.; Jedelsky, P.L.; Titera, D. Two-dimensional proteomic analysis of honeybee, Apis mellifera, winter worker hemolymph. Apidologie 2013, 44, 404–418. [Google Scholar] [CrossRef][Green Version]
- Woltedji, D.; Song, F.; Zhang, L.; Gala, A.; Han, B.; Feng, M.; Fang, Y.; Li, J. Western honeybee drones and workers (Apis mellifera ligustica) have different olfactory mechanisms than eastern honeybees (Apis cerana cerana). J. Proteome Res. 2012, 11, 4526–4540. [Google Scholar] [CrossRef] [PubMed]
- Harbo, J.R.; Harris, J.W. Suppressed mite reproduction explained by the behaviour of adult bees. J. Apic. Res. 2005, 44, 21–23. [Google Scholar] [CrossRef]
- Harbo, J.R.; Harris, J.W. Responses to Varroa by honey bees with different levels of Varroa Sensitive Hygiene. J. Apic. Res. 2009, 48, 156–161. [Google Scholar] [CrossRef]
- De Graaf, D.C.; Aerts, M.; Danneels, E.; Devreese, B. Bee, wasp and ant venomics pave the way for a component-resolved diagnosis of sting allergy. J. Proteom. 2009, 72, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Calvete, J.J.; Ju, P.; Sanz, L. Snake venomics. Strategy and applications. J. Mass Spectrom. 2007, 42, 1405–1414. [Google Scholar] [CrossRef]
- Li, R.; Zhang, L.; Fang, Y.; Han, B.; Lu, X.; Zhou, T.; Feng, M.; Li, J. Proteome and phosphoproteome analysis of honeybee (Apis mellifera) venom collected from electrical stimulation and manual extraction of the venom gland. BMC Genom. 2013, 14, 766. [Google Scholar] [CrossRef]
- Resende, V.M.F.; Vasilj, A.; Santos, K.S.; Palma, M.S.; Shevchenko, A. Proteome and phosphoproteome of Africanized and European honeybee venoms. Proteomics 2013, 13, 2638–2648. [Google Scholar] [CrossRef]
- Van Vaerenbergh, M.; Debyser, G.; Devreese, B.; de Graaf, D.C. Exploring the hidden honeybee (Apis mellifera) venom proteome by integrating a combinatorial peptide ligand library approach with FTMS. J. Proteom. 2013, 99, 169–178. [Google Scholar] [CrossRef]
- Danneels, E.L.; Van Vaerenbergh, M.; Debyser, G.; Devreese, B.; de Graaf, D.C. Honeybee Venom Proteome Profile of Queens and Winter Bees as Determined by a Mass Spectrometric Approach. Toxins 2015, 7, 4468–4483. [Google Scholar] [CrossRef]
- Calábria, L.K.; Cunha, R.B. Worker Honeybee Brain Proteome. J. Proteome Res 2011, 11, 1485–1493. [Google Scholar]
- Page, R.E.; Amdam, G.V. The making of a social insect: Developmental architectures of social design. BioEssays 2007, 29, 334–343. [Google Scholar] [CrossRef] [PubMed]
- Robinson, G.E. Regulation of honey bee age polyethism by juvenile hormone. Behav. Ecol. Sociobiol. 1987, 20, 329–338. [Google Scholar] [CrossRef]
- Duarte, A.; Weissing, F.J.; Pen, I.; Keller, L. An Evolutionary Perspective on Self-Organized Division of Labor in Social Insects. Annu. Rev. Ecol. Evol. Syst. 2011, 42, 91–110. [Google Scholar] [CrossRef]
- Amdam, G.V.; Norberg, K.; Hagen, A.; Omholt, S.W. Social exploitation of vitellogenin. Proc. Natl. Acad. Sci. USA 2003, 100, 1799–1802. [Google Scholar] [CrossRef] [PubMed]
- Whitfield, C.W.; Cziko, A.M.; Robinson, G.E. Gene expression profiles in the brain predict behavior in individual honey bees. Science 2003, 302, 296–299. [Google Scholar] [CrossRef] [PubMed]
- Whitfield, C.W.; Ben-Shahar, Y.; Brillet, C.; Leoncini, I.; Crauser, D.; Leconte, Y.; Rodriguez-Zas, S.; Robinson, G.E. Genomic dissection of behavioral maturation in the honey bee. Proc. Natl. Acad. Sci. USA 2006, 103, 16068–16075. [Google Scholar] [CrossRef] [PubMed]
- Garcia, L.; Garcia, C.H.S.; Karen, L.; Costa, G.; Cruz, N.; Sa, A.; Ba, S.N.; Fontes, W.; Ricart, C.A.O.; Espindola, F.S.; et al. Proteomic Analysis of Honey Bee Brain upon Ontogenetic and Behavioral Development Proteomic Analysis of Honey Bee Brain upon Ontogenetic and Behavioral Development. J. Proteome Res. 2009, 8, 1464–1473. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, L.G.; Lu, B.; Da Cruz, G.C.N.; Calabria, L.K.; Martins, N.F.; Togawa, R.; Espindola, F.S.; Yates, J.R.; Cunha, R.B.; De Sousa, M.V. Worker honeybee brain proteome. J. Proteome Res. 2012, 11, 1485–1493. [Google Scholar] [CrossRef]
- Uno, Y.; Fujiyuki, T.; Morioka, M.; Takeuchi, H.; Kubo, T. Identification of proteins whose expression is up- or down-regulated in the mushroom bodies in the honeybee brain using proteomics. FEBS Lett. 2007, 581, 97–101. [Google Scholar] [CrossRef]
- Uno, Y.; Fujiyuki, T.; Morioka, M.; Kubo, T. Mushroom body-preferential expression of proteins/genes involved in endoplasmic reticulum Ca2+-transport in the worker honeybee (Apis mellifera L.) brain. Insect Mol. Biol. 2013, 22, 52–61. [Google Scholar] [CrossRef]
- Wolschin, F.; Münch, D.; Amdam, G. V Structural and proteomic analyses reveal regional brain differences during honeybee aging. J. Exp. Biol. 2009, 212, 4027–4032. [Google Scholar] [CrossRef] [PubMed]
- Baker, N.; Wolschin, F.; Amdam, G.V. Age-related learning deficits can be reversible in honeybees Apis mellifera. Exp. Gerontol. 2012, 47, 764–772. [Google Scholar] [CrossRef] [PubMed]
- da Silva Menegasso, A.R.; Pratavieira, M.; da Gama Fischer, J.D.S.; Carvalho, P.C.; Roat, T.C.; Malaspina, O.; Palma, M.S. Profiling the proteomics in honeybee worker brains submitted to the proboscis extension reflex. J. Proteom. 2016, 151, 131–144. [Google Scholar]
- Roat, T.C.; Santos, K.S.; Malaspina, O.; Palma, M.S. Modification of the brain proteome of Africanized honeybees (Apis mellifera) exposed to a sub-lethal doses of the insecticide fipronil. Ecotoxicology 2014, 23, 1659–1670. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, L.; Feng, M.; Zhang, Z.; Pan, Y. Identification of the proteome composition occurring during the course of embryonic development of bees (Apis mellifera). Insect Mol. Biol. 2009, 18, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Gala, A.; Fang, Y.; Woltedji, D.; Zhang, L.; Han, B.; Feng, M.; Li, J. Changes of proteome and phosphoproteome trigger embryo-larva transition of honeybee worker (Apis mellifera ligustica). J. Proteom. 2013, 78, 428–446. [Google Scholar] [CrossRef]
- Li, J.; Fang, Y.; Zhang, L.; Begna, D. Honeybee (Apis mellifera ligustica) drone embryo proteomes. J. Insect Physiol. 2011, 57, 372–384. [Google Scholar] [CrossRef]
- Chan, M.M.Y.; Choi, S.Y.C.; Chan, Q.W.T.; Li, P.; Guarna, M.M.; Foster, L.J. Proteome profile and lentiviral transduction of cultured honey bee (Apis mellifera L.) cells. Insect Mol. Biol. 2010, 19, 653–658. [Google Scholar] [CrossRef]
- Li, J.; Wu, J.; Rundassa, D.B.; Song, F.; Zheng, A.; Fang, Y. Differential protein expression in honeybee (Apis mellifera L.) larvae: Underlying caste differentiation. PLoS ONE 2010, 5, e13455. [Google Scholar] [CrossRef]
- Begna, D.; Fang, Y.; Feng, M.; Li, J. Mitochondrial proteins differential expression during honeybee (Apis mellifera L.) queen and worker larvae caste determination. J. Proteome Res. 2011, 10, 4263–4280. [Google Scholar] [CrossRef]
- Begna, D.; Han, B.; Feng, M.; Fang, Y.; Li, J. Differential expressions of nuclear proteomes between honeybee (Apis mellifera L.) queen and worker larvae: A deep insight into caste pathway decisions. J. Proteome Res. 2012, 11, 1317–1329. [Google Scholar] [CrossRef] [PubMed]
- Kamakura, M. Royalactin induces queen differentiation in honeybees. Seikagaku 2012, 84, 994–1003. [Google Scholar] [CrossRef] [PubMed]
- Michelette, E. de F.; Soares, A. Characterization of preimaginal developmental stages in Africanized honey bee workers (Apis mellifera L). Apidologie 1993, 24, 431–440. [Google Scholar] [CrossRef]
- Zheng, A.; Li, J.; Begna, D.; Fang, Y.; Feng, M.; Song, F. Proteomic analysis of honeybee (Apis mellifera L.) pupae head development. PLoS ONE 2011, 6, e20428. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Erban, T.; Petrova, D.; Harant, K.; Jedelsky, P.L.; Titera, D. Two-dimensional gel proteome analysis of honeybee, Apis mellifera, worker red-eye pupa hemolymph. Apidologie 2014, 45, 53–72. [Google Scholar] [CrossRef]
- Erban, T.; Harant, K.; Kamler, M.; Markovic, M.; Titera, D. Detailed proteome mapping of newly emerged honeybee worker hemolymph and comparison with the red-eye pupal stage. Apidologie 2016, 47, 805–817. [Google Scholar] [CrossRef]
- Wolschin, F.; Amdam, G.V. Comparative proteomics reveal characteristics of life-history transitions in a social insect. Proteome Sci. 2007, 5, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wolschin, F.; Amdam, G.V. Plasticity and robustness of protein patterns during reversible development in the honey bee (Apis mellifera). Anal. Bioanal. Chem. 2007, 389, 1095–1100. [Google Scholar] [CrossRef]
- Chan, Q.W.T.; Mutti, N.S.; Foster, L.J.; Kocher, S.D.; Amdam, G.V.; Wolschin, F. The worker honeybee fat body proteome is extensively remodeled preceding a major life-history transition. PLoS ONE 2011, 6, e24794. [Google Scholar] [CrossRef]
- Garcia, L.; Garcia, C.H.S.; Calabria, L.K.; Da Cruz, G.C.N.; Puentes, A.S.; Bao, S.N.; Fontes, W.; Ricart, C.O.; Espindola, F.S.; De Sousa, M.V. Proteomic analysis of honey bee brain upon ontogenetic and behavioral development. J. Proteome Res. 2009, 8, 1464–1473. [Google Scholar] [CrossRef]
- Qi, Y.; Fan, P.; Hao, Y.; Han, B.; Fang, Y.; Feng, M.; Cui, Z.; Li, J. Phosphoproteomic analysis of protein phosphorylation networks in the hypopharyngeal gland of honeybee workers (Apis mellifera ligustica). J. Proteome Res. 2015, 14, 4647–4661. [Google Scholar] [CrossRef] [PubMed]
- Randolt, K.; Gimple, O.; Geissendörfer, J.; Reinders, J.; Prusko, C.; Mueller, M.J.; Albert, S.; Tautz, J.; Beier, H. Immune-related proteins induced in the hemolymph after aseptic and septic injury differ in honey bee worker larvae and adults. Arch. Insect Biochem. Physiol. 2008, 69, 155–167. [Google Scholar] [CrossRef]
- Peixoto, L.G.; Calábria, L.K.; Garcia, L.; Capparelli, F.E.; Goulart, L.R.; de Sousa, M.V.; Espindola, F.S. Identification of major royal jelly proteins in the brain of the honeybee Apis mellifera. J. Insect Physiol. 2009, 55, 671–677. [Google Scholar] [CrossRef] [PubMed]
- Fujita, T.; Kozuka-Hata, H.; Ao-Kondo, H.; Kunieda, T.; Oyama, M.; Kubo, T. Proteomic analysis of the royal jelly and characterization of the functions of its derivation glands in the honeybee. J. Proteome Res. 2013, 12, 404–411. [Google Scholar] [CrossRef] [PubMed]
- Patel, N.G.; Haydak, M.H.; Gochnauer, T.A. Electrophoretic Components of the Proteins in Honeybee Larval Food. Nature 1960, 186, 633. [Google Scholar] [CrossRef] [PubMed]
- Callow, R.K.; Johnston, N.C.; Simpson, J. 10-Hydroxy-Δ2-decenoic acid in the honeybee (Apis mellifera). Experientia 1959, 15, 421–422. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, Z.L.; Tian, L.Q.; Qin, Q.H.; Wu, X.B.; Yan, W.Y.; Zeng, Z.J. Transcriptome differences in the hypopharyngeal gland between Western Honeybees (Apis mellifera) and Eastern Honeybees (Apis cerana). BMC Genom. 2014, 15, 744. [Google Scholar] [CrossRef] [PubMed]
- Santos, K.S.; Delazari Dos Santos, L.; Anita Mendes, M.; Monson De Souza, B.; Malaspina, O.; Palma, M.S. Profiling the proteome complement of the secretion from hypopharyngeal gland of Africanized nurse-honeybees (Apis mellifera L.). Insect Biochem. Mol. Biol. 2005, 35, 85–91. [Google Scholar] [CrossRef]
- Tamura, S.; Amano, S.; Kono, T.; Kondoh, J.; Yamaguchi, K.; Kobayashi, S.; Ayabe, T.; Moriyama, T. Molecular characteristics and physiological functions of major royal jelly protein 1 oligomer. Proteomics 2009, 9, 5534–5543. [Google Scholar] [CrossRef]
- Albert, S.; Klaudiny, J. The MRJP/YELLOW protein family of Apis mellifera: Identification of new members in the EST library. J. Insect Physiol. 2004, 50, 51–59. [Google Scholar] [CrossRef]
- Schmitzová, J.; Klaudiny, J.; Albert, S.; Schröder, W.; Schreckengost, W.; Hanes, J.; Júdová, J.; Šimúth, J. A family of major royal jelly proteins of the honeybee Apis mellifera L. Cell. Mol. Life Sci. 1998, 54, 1020–1030. [Google Scholar] [CrossRef] [PubMed]
- Simuth, J. Some properties of the main protein of honeybee (Apis mellifera) royal jelly. Apidologie 2001, 32, 69–80. [Google Scholar] [CrossRef]
- Li, J.; Wang, T.; Zhang, Z.; Pan, Y. Proteomic analysis of royal jelly from three strains of western honeybees (Apis mellifera). J. Agric. Food Chem. 2007, 55, 8411–8422. [Google Scholar] [CrossRef] [PubMed]
- Isidorov, V.; Czyżewska, U.; Isidorova, A.; Bakier, S. Gas chromatographic and mass spectrometric characterization of the organic acids extracted from some preparations containing lyophilized royal jelly. J. Chromatogr. B 2009, 877, 3776–3780. [Google Scholar] [CrossRef]
- Ramadan, M.F.; Al-Ghamdi, A. Bioactive compounds and health-promoting properties of royal jelly: A review. J. Funct. Foods 2012, 4, 39–52. [Google Scholar] [CrossRef]
- Yu, F.; Mao, F.; Li, J. Royal Jelly Proteome Comparison between A. mellifera ligustica and A. cerana cerana. J. Proteome Res. 2010, 2207–2215. [Google Scholar] [CrossRef]
- Han, B.; Fang, Y.; Feng, M.; Lu, X.; Huo, X.; Meng, L.; Wu, B.; Li, J. In-depth phosphoproteomic analysis of royal jelly derived from western and eastern honeybee species. J. Proteome Res. 2014, 13, 5928–5943. [Google Scholar] [CrossRef]
- Chen, S.-L.; Li, J.-K.; Zhong, B.-X.; Su, S. Microsatellite analysis of royal jelly producing traits of Italian honeybee (Apis mellifera Liguatica). Acta Genet. Sin. 2005, 32, 1037–1044. [Google Scholar] [PubMed]
- Han, B.; Li, C.; Zhang, L.; Fang, Y.; Feng, M.; Li, J. Novel royal jelly proteins identified by gel-based and gel-free proteomics. J. Agric. Food Chem. 2011, 59, 10346–10355. [Google Scholar] [CrossRef]
- Zhang, L.; Han, B.; Li, R.; Lu, X.; Nie, A.; Guo, L.; Fang, Y.; Feng, M.; Li, J. Comprehensive identification of novel proteins and N-glycosylation sites in royal jelly. BMC Genom. 2014, 15, 135. [Google Scholar] [CrossRef]
- Feng, M.; Fang, Y.; Han, B.; Xu, X.; Fan, P.; Hao, Y.; Qi, Y.; Hu, H.; Huo, X.; Meng, L.; et al. In-Depth N-Glycosylation Reveals Species-Specific Modifications and Functions of the Royal Jelly Protein from Western (Apis mellifera) and Eastern Honeybees (Apis cerana). J. Proteome Res. 2015, 14, 5327–5340. [Google Scholar] [CrossRef] [PubMed]
- Trhlin, M.; Rajchard, J. Chemical communication in the honeybee (Apis mellifera L.): A review. Vet. Med. 2011, 2011, 265–273. [Google Scholar] [CrossRef]
- Dani, F.R.; Iovinella, I.; Felicioli, A.; Niccolini, A.; Calvello, M.A.; Carucci, M.G.; Qiao, H.; Pieraccini, G.; Turillazzi, S.; Moneti, G.; et al. Mapping the expression of soluble olfactory proteins in the honeybee. J. Proteome Res. 2010, 9, 1822–1833. [Google Scholar] [CrossRef] [PubMed]
- Feng, M.; Song, F.; Aleku, D.W.; Han, B.; Fang, Y.; Li, J. Antennal proteome comparison of sexually mature drone and forager honeybees. J. Proteome Res. 2011, 10, 3246–3260. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Song, F.; Zhang, L.; Aleku, D.W.; Han, B.; Feng, M.; Li, J. Differential antennal proteome comparison of adult honeybee drone, worker and queen (Apis mellifera L.). J. Proteom. 2012, 75, 756–773. [Google Scholar] [CrossRef] [PubMed]
- Guarna, M.M.; Melathopoulos, A.P.; Huxter, E.; Iovinella, I.; Parker, R.; Stoynov, N.; Tam, A.; Moon, K.M.; Chan, Q.W.T.; Pelosi, P.; et al. A search for protein biomarkers links olfactory signal transduction to social immunity. BMC Genom. 2015, 16, 63. [Google Scholar] [CrossRef] [PubMed]
- Collins, A.M.; Caperna, T.J.; Williams, V.; Garrett, W.M.; Evans, J.D. Proteomic analyses of male contributions to honey bee Abstract. Insect Mol. Biol. 2006, 15, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Poland, V.; Eubel, H.; King, M.; Solheim, C.; Harvey Millar, A.; Baer, B. Stored sperm differs from ejaculated sperm by proteome alterations associated with energy metabolism in the honeybee Apis mellifera. Mol. Ecol. 2011, 20, 2643–2654. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Altaye, S.Z.; Meng, L.; Lu, Y.; Li, J. The Emerging Proteomic Research Facilitates in-Depth Understanding of the Biology of Honeybees. Int. J. Mol. Sci. 2019, 20, 4252. https://doi.org/10.3390/ijms20174252
Altaye SZ, Meng L, Lu Y, Li J. The Emerging Proteomic Research Facilitates in-Depth Understanding of the Biology of Honeybees. International Journal of Molecular Sciences. 2019; 20(17):4252. https://doi.org/10.3390/ijms20174252
Chicago/Turabian StyleAltaye, Solomon Zewdu, Lifeng Meng, Yao Lu, and Jianke Li. 2019. "The Emerging Proteomic Research Facilitates in-Depth Understanding of the Biology of Honeybees" International Journal of Molecular Sciences 20, no. 17: 4252. https://doi.org/10.3390/ijms20174252
APA StyleAltaye, S. Z., Meng, L., Lu, Y., & Li, J. (2019). The Emerging Proteomic Research Facilitates in-Depth Understanding of the Biology of Honeybees. International Journal of Molecular Sciences, 20(17), 4252. https://doi.org/10.3390/ijms20174252