Mitochondrial Genomes of Two Thaparocleidus Species (Platyhelminthes: Monogenea) Reveal the First rRNA Gene Rearrangement among the Neodermata
Abstract
1. Background
2. Results
2.1. Genome Organization and Base Composition
2.2. Non-Coding Regions
2.3. Phylogeny
2.4. Gene Orders
3. Discussion
4. Materials and methods
4.1. Specimen Collection and Identification
4.2. DNA Extraction, Amplification and Sequencing
4.3. Sequence Annotation and Analyses
4.4. Phylogenetic Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bychowsky, B.E. Monogenetic Trematodes, Their Systematics and Phylogeny; Academy of Sciences: Moscow, Leningrad, USSR, Russia, 1957; p. 509. [Google Scholar]
- Price, C. Two New Subfamilies of Monogenenetic Trematodes. Q. J. Fla. Acad. Sci. 1966, 29, 199–201. [Google Scholar]
- Gusev, A.V. The new subfamily of monogenean parasites Monogenoidea. Dokl. Akad. Nauk. 1961, 139, 1480–1482. [Google Scholar]
- Bychowsky, B.; Nagibina, L. Revision of Ancyrocephalinae Bychowsky, 1937. Parazitol. Sb. 1978, 28, 5–15. [Google Scholar]
- Kritsky, D.C.; Boeger, W.A. The phylogenetic status of the Ancyrocephalidae Bychowsky, 1937 (Monogenea: Dactylogyroidea). J. Parasitol. 1989, 75, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Lim, L.H.S.; Timofeeva, T.A.; Gibson, D.I. Dactylogyridean monogeneans of the siluriform fishes of the Old World. Syst. Parasitol. 2001, 50, 159–197. [Google Scholar] [CrossRef] [PubMed]
- Simkova, A.; Plaisance, L.; Matejusova, I.; Morand, S.; Verneau, O. Phylogenetic relationships of the Dactylogyridae Bychowsky, 1933 (Monogenea: Dactylogyridea): The need for the systematic revision of the Ancyrocephalinae Bychowsky, 1937. Syst. Parasitol. 2003, 54, 1–11. [Google Scholar] [CrossRef]
- Mendoza-Palmero, C.A.; Blasco-Costa, I.; Scholz, T. Molecular phylogeny of Neotropical monogeneans (Platyhelminthes: Monogenea) from catfishes (Siluriformes). Parasit. Vectors 2015, 8, 164. [Google Scholar] [CrossRef]
- Plaisance, L.; Littlewood, D.T.J.; Olson, P.D.; Morand, S. Molecular phylogeny of gill monogeneans (Platyhelminthes, Monogenea, Dactylogyridae) and colonization of Indo-West Pacific butterflyfish hosts (Perciformes, Chaetodontidae). Zool. Scr. 2005, 34, 425–436. [Google Scholar] [CrossRef]
- Blasco-Costa, I.; Miguez-Lozano, R.; Sarabeev, V.; Balbuena, J.A. Molecular phylogeny of species of Ligophorus (Monogenea: Dactylogyridae) and their affinities within the Dactylogyridae. Parasitol. Int. 2012, 61, 619–627. [Google Scholar] [CrossRef]
- Zhang, D.; Zou, H.; Wu, S.G.; Li, M.; Jakovlić, I.; Zhang, J.; Chen, R.; Wang, G.T.; Li, W.X. Sequencing of the complete mitochondrial genome of a fish-parasitic flatworm Paratetraonchoides inermis (Platyhelminthes: Monogenea): tRNA gene arrangement reshuffling and implications for phylogeny. Parasit. Vectors 2017, 10, 462. [Google Scholar] [CrossRef]
- Perkins, E.M.; Donnellan, S.C.; Bertozzi, T.; Chisholm, L.A.; Whittington, I.D. Looks can deceive: Molecular phylogeny of a family of flatworm ectoparasites (Monogenea: Capsalidae) does not reflect current morphological classification. Mol. Phylogenet. Evol. 2009, 52, 705–714. [Google Scholar] [CrossRef] [PubMed]
- Poulin, R.; Morand, S. The diversity of parasites. Q. Rev. Biol. 2000, 75, 277–293. [Google Scholar] [CrossRef]
- Huyse, T.; Plaisance, L.; Webster, B.L.; Mo, T.A.; Bakke, T.A.; Bachmann, L.; Littlewood, D.T. The mitochondrial genome of Gyrodactylus salaris (Platyhelminthes: Monogenea), a pathogen of Atlantic salmon (Salmo salar). Parasitology 2007, 134, 739–747. [Google Scholar] [CrossRef] [PubMed]
- Philippe, H.; Brinkmann, H.; Lavrov, D.V.; Littlewood, D.T.J.; Manuel, M.; Wörheide, G.; Baurain, D. Resolving difficult phylogenetic questions: Why more sequences are not enough. Plos Biol. 2011, 9, e1000602. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zou, H.; Hua, C.-J.; Li, W.-X.; Mahboob, S.; Al-Ghanim, K.A.; Al-Misned, F.; Jakovlić, I.; Wang, G.-T. Mitochondrial architecture rearrangements produce asymmetrical nonadaptive mutational pressures that subvert the phylogenetic reconstruction in Isopoda. Genome Biol. Evol. 2019, 11, 1797–1812. [Google Scholar] [CrossRef]
- Doolittle, W.F. The practice of classification and the theory of evolution, and what the demise of Charles Darwin’s tree of life hypothesis means for both of them. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2009, 364, 2221–2228. [Google Scholar] [CrossRef]
- Shao, R.; Barker, S. Mitochondrial genomes of parasitic arthropods: Implications for studies of population genetics and evolution. Parasitology 2007, 134, 153–167. [Google Scholar] [CrossRef]
- Perkins, E.M.; Donnellan, S.C.; Bertozzi, T.; Whittington, I.D. Closing the mitochondrial circle on paraphyly of the Monogenea (Platyhelminthes) infers evolution in the diet of parasitic flatworms. Int. J. Parasitol. 2010, 40, 1237–1245. [Google Scholar] [CrossRef]
- Park, J.K.; Kim, K.H.; Kang, S.; Kim, W.; Eom, K.S.; Littlewood, D.T. A common origin of complex life cycles in parasitic flatworms: Evidence from the complete mitochondrial genome of Microcotyle sebastis (Monogenea: Platyhelminthes). Bmc Evol. Biol. 2007, 7, 11. [Google Scholar] [CrossRef]
- Huyse, T.; Buchmann, K.; Littlewood, D.T. The mitochondrial genome of Gyrodactylus derjavinoides (Platyhelminthes: Monogenea) – a mitogenomic approach for Gyrodactylus species and strain identification. Gene 2008, 417, 27–34. [Google Scholar] [CrossRef]
- Wey-Fabrizius, A.R.; Podsiadlowski, L.; Herlyn, H.; Hankeln, T. Platyzoan mitochondrial genomes. Mol. Phylogenet. Evol. 2013, 69, 365–375. [Google Scholar] [CrossRef] [PubMed]
- Ross, E.; Blair, D.; Guerrero-Hernandez, C.; Sanchez Alvarado, A. Comparative and transcriptome analyses uncover key aspects of coding-and long noncoding RNAs in flatworm mitochondrial genomes. G3 (Bethesda) 2016, 6, 1191–1200. [Google Scholar] [CrossRef] [PubMed]
- Naylor, G.J.; Collins, T.M.; Brown, W.M. Hydrophobicity and phylogeny. Nature 1995, 373, 565–566. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, D.; Zou, H.; Wu, S.G.; Li, M.; Jakovlic, I.; Zhang, J.; Chen, R.; Wang, G.T.; Li, W.X. Sequencing, characterization and phylogenomics of the complete mitochondrial genome of Dactylogyrus lamellatus (Monogenea: Dactylogyridae). J. Helminthol. 2018, 92, 455–466. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Li, W.X.; Zou, H.; Wu, S.G.; Li, M.; Jakovlic, I.; Zhang, J.; Chen, R.; Wang, G.T. Mitochondrial genomes of two diplectanids (Platyhelminthes: Monogenea) expose paraphyly of the order Dactylogyridea and extensive tRNA gene rearrangements. Parasit. Vectors 2018, 11, 601. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Li, W.X.; Zou, H.; Wu, S.G.; Li, M.; Jakovlic, I.; Zhang, J.; Chen, R.; Wang, G.T. Mitochondrial genomes and 28S rDNA contradict the proposed obsoletion of the order Tetraonchidea (Platyhelminthes: Monogenea). Int. J. Biol. Macromol. 2019. under review. [Google Scholar]
- Erpenbeck, D.; Voigt, O.; Worheide, G.; Lavrov, D.V. The mitochondrial genomes of sponges provide evidence for multiple invasions by Repetitive Hairpin-forming Elements (RHE). BMC Genom. 2009, 10, 591. [Google Scholar] [CrossRef]
- Zhang, J.; Wu, X.; Xie, M.; Li, A. The complete mitochondrial genome of Pseudochauhanea macrorchis (Monogenea: Chauhaneidae) revealed a highly repetitive region and a gene rearrangement hot spot in Polyopisthocotylea. Mol. Biol. Rep. 2012, 39, 8115–8125. [Google Scholar] [CrossRef]
- Le, T.H.; Blair, D.; McManus, D.P. Mitochondrial genomes of parasitic flatworms. Trends Parasitol. 2002, 18, 206–213. [Google Scholar] [CrossRef]
- Fumagalli, L.; Taberlet, P.; Favre, L.; Hausser, J. Origin and evolution of homologous repeated sequences in the mitochondrial DNA control region of shrews. Mol. Biol. Evol. 1996, 13, 31–46. [Google Scholar] [CrossRef]
- McMurray, A.A.; Sulston, J.E.; Quail, M.A. Short-insert libraries as a method of problem solving in genome sequencing. Genome Res. 1998, 8, 562–566. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hu, M.; Jex, A.R.; Campbell, B.E.; Gasser, R.B. Long PCR amplification of the entire mitochondrial genome from individual helminths for direct sequencing. Nat. Protoc. 2007, 2, 2339–2344. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Li, W.X.; Zou, H.; Wu, S.G.; Li, M.; Jakovlić, I.; Zhang, J.; Chen, R.; Wang, G.T. Homoplasy or plesiomorphy? Reconstruction of the evolutionary history of mitochondrial gene order rearrangements in the subphylum Neodermata. Int. J. Parasitol. 2019, 49, 819–829. [Google Scholar] [CrossRef] [PubMed]
- Li, W.X.; Zhang, D.; Boyce, K.; Xi, B.W.; Zou, H.; Wu, S.G.; Li, M.; Wang, G.T. The complete mitochondrial DNA of three monozoic tapeworms in the Caryophyllidea: A mitogenomic perspective on the phylogeny of eucestodes. Parasit. Vectors 2017, 10, 314. [Google Scholar] [CrossRef] [PubMed]
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef] [PubMed]
- Boore, J. The use of genome-level characters for phylogenetic reconstruction. Trends Ecol. Evol. 2006, 21, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, L.; Fromm, B.; de Azambuja, L.P.; Boeger, W.A. The mitochondrial genome of the egg-laying flatworm Aglaiogyrodactylus forficulatus (Platyhelminthes: Monogenoidea). Parasit. Vectors 2016, 9, 285. [Google Scholar] [CrossRef]
- Wu, B.H.; Lang, S.; Wang, W.J. Fauna Sinica: Platyhelminthes: Monogenea; Science Press: Beijing, China, 2000. [Google Scholar]
- Hassouna, N.; Mithot, B.; Bachellerie, J.-P. The complete nucleotide sequence of mouse 28S rRNA gene. Implications for the process of size increase of the large subunit rRNA in higher eukaryotes. Nucleic Acids Res. 1984, 12, 3563–3583. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2002, Chapter 2. Unit 2.3. [Google Scholar] [CrossRef]
- Lalitha, S. Primer Premier 5. Biot. Soft. Int. Rep. 2000, 1, 270–272. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Zou, H.; Jakovlic, I.; Chen, R.; Zhang, D.; Zhang, J.; Li, W.X.; Wang, G.T. The complete mitochondrial genome of parasitic nematode Camallanus cotti: Extreme discontinuity in the rate of mitogenomic architecture evolution within the Chromadorea class. BMC Genom. 2017, 18, 840. [Google Scholar] [CrossRef] [PubMed]
- Burland, T.G. DNASTAR’s Lasergene sequence analysis software. Methods Mol. Biol. 2000, 132, 71–91. [Google Scholar] [PubMed]
- Bernt, M.; Donath, A.; Juhling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Putz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Laslett, D.; Canback, B. ARWEN: A program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 2008, 24, 172–175. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Li, W.X.; Jakovlić, I.; Zou, H.; Zhang, J.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. bioRxiv 2018. [Google Scholar] [CrossRef]
- Hadley, W. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009. [Google Scholar]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [PubMed]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Cock, P.J.; Antao, T.; Chang, J.T.; Chapman, B.A.; Cox, C.J.; Dalke, A.; Friedberg, I.; Hamelryck, T.; Kauff, F.; Wilczynski, B.; et al. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 2009, 25, 1422–1423. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; Mark, P.V.D.; Ayres, D.L.; Darling, A.; Höhna, S. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Lartillot, N.; Rodrigue, N.; Stubbs, D.; Richer, J. PhyloBayes MPI: Phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst. Biol. 2013, 62, 611–615. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef] [PubMed]
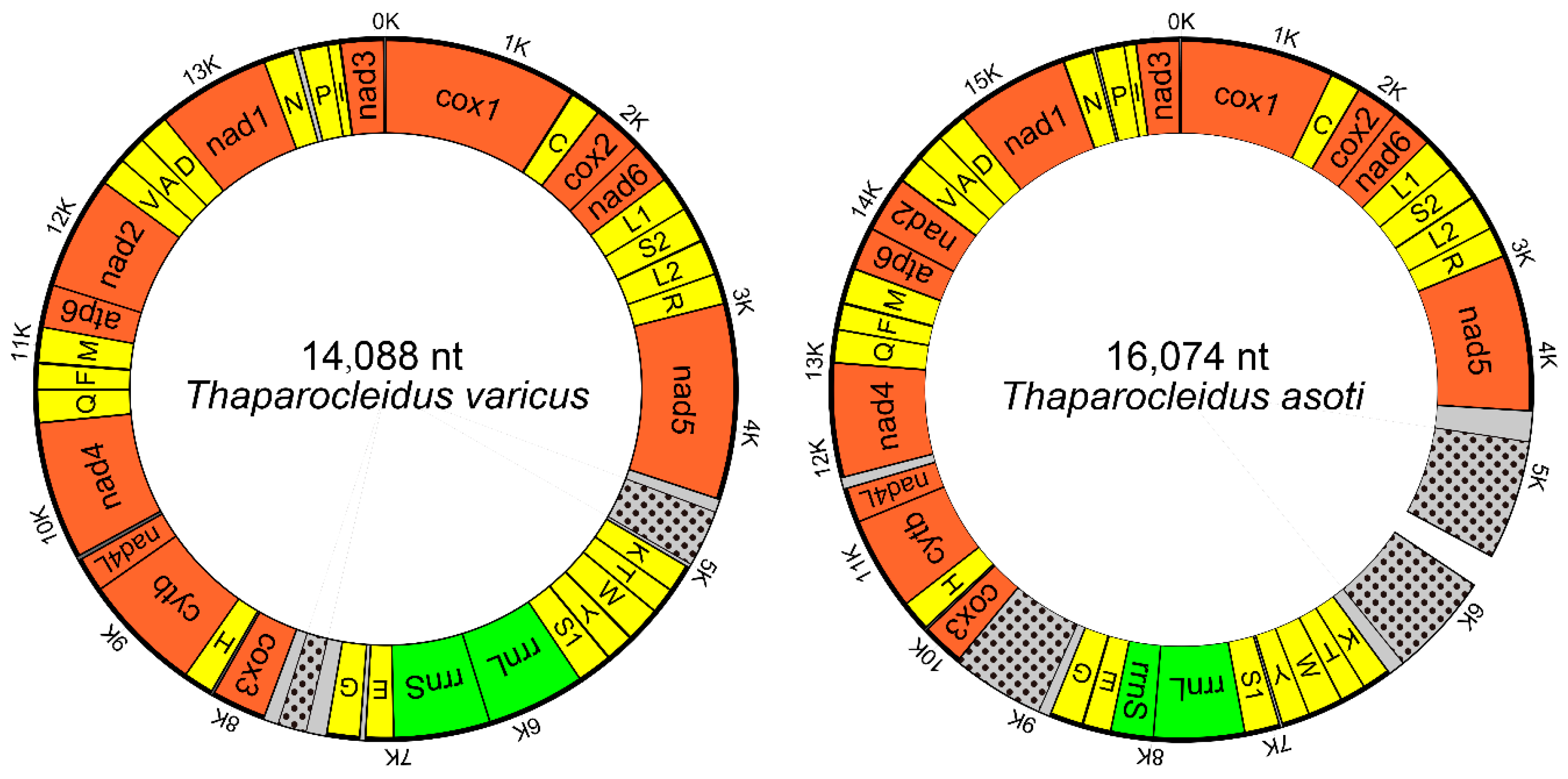
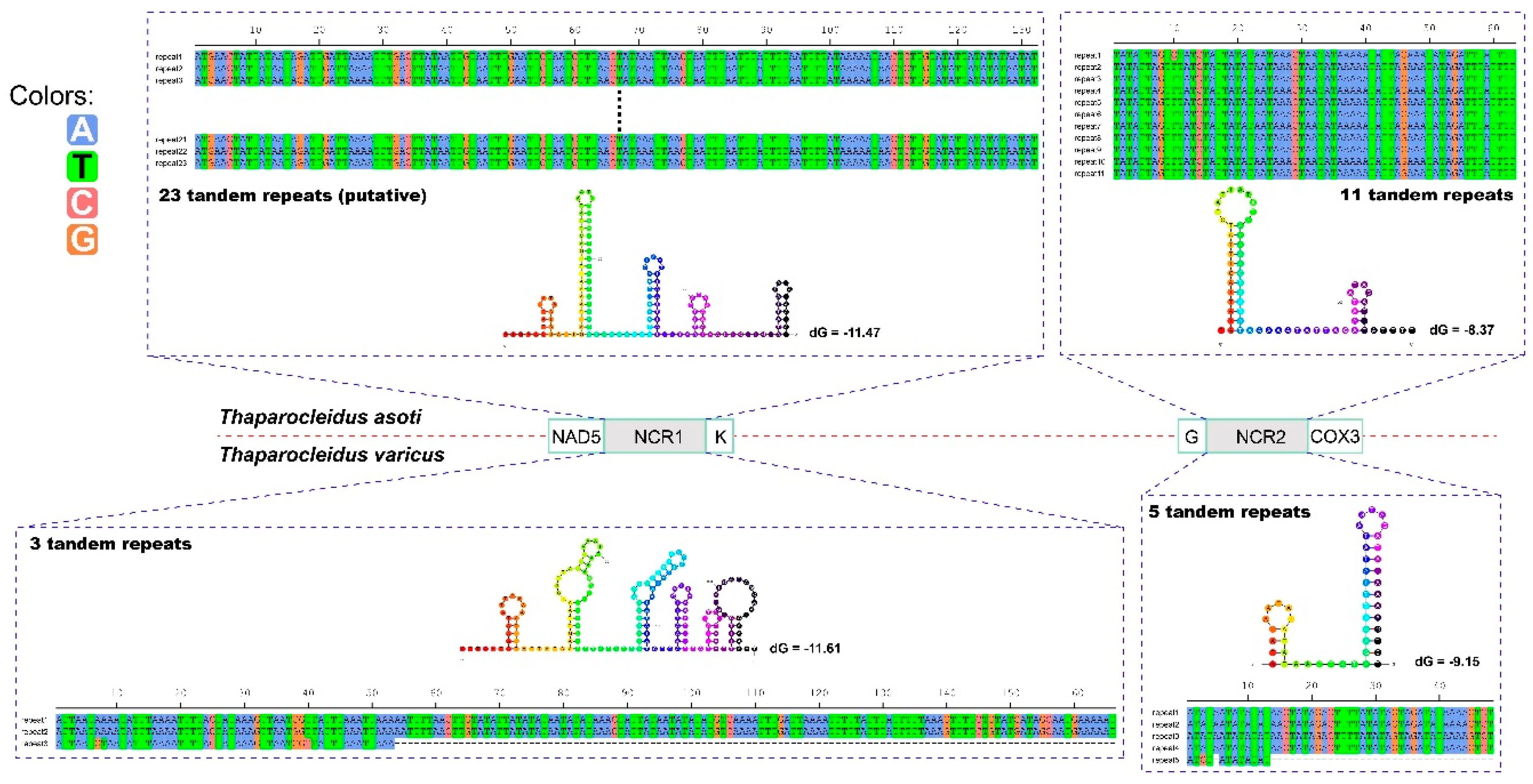

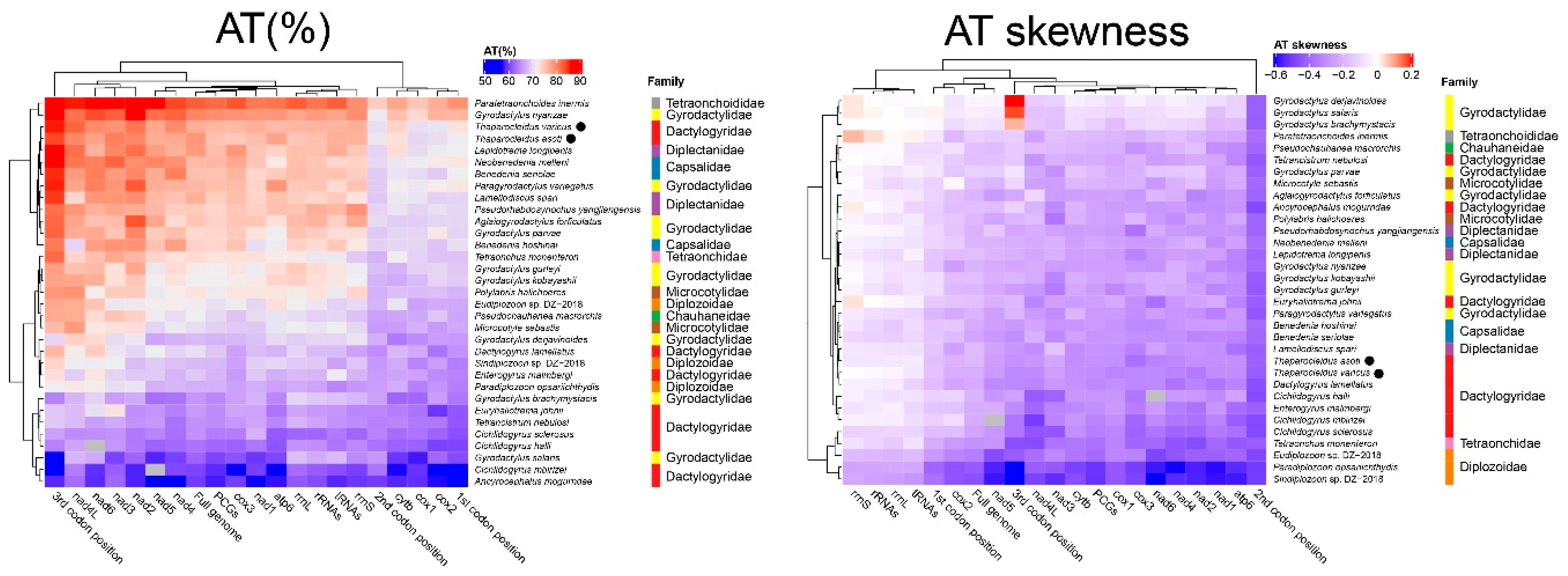
| Gene | Position | Size | Intergenic Nucleotides | Codon | Anti-codon | Identity | ||
|---|---|---|---|---|---|---|---|---|
| From | To | Start | Stop | |||||
| Thaparocleidus asoti/Thaparocleidus varicus | ||||||||
| cox1 | 1/1 | 1554/1557 | 1554/1557 | ATG/ATT | TAG/TAG | 82.59 | ||
| trnC | 1554/1566 | 1617/1630 | 64/65 | −1/8 | GCA/GCA | 87.69 | ||
| cox2 | 1621/1634 | 2245/2260 | 625/627 | 3/3 | ATG/GTG | T/TAA | 77.35 | |
| nad6 | 2247/2261 | 2693/2707 | 447/447 | 1/0 | GTG/ATG | TAA/TAG | 72.48 | |
| trnL1 | 2694/2708 | 2758/2776 | 65/69 | TAG/TAG | 78.26 | |||
| trnS2 | 2759/2777 | 2825/2843 | 67/67 | TGA/TGA | 77.61 | |||
| trnL2 | 2831/2850 | 2895/2914 | 65/65 | 5/6 | TAA/TAA | 87.69 | ||
| trnR | 2896/2916 | 2963/2981 | 68/66 | 0/1 | TCG/TCG | 72.46 | ||
| nad5 | 2966/2983 | 4537/4548 | 1572/1566 | 2/1 | ATG/ATG | TAA/TAA | 68.89 | |
| trnK | 6558/5027 | 6622/5091 | 65/65 | 2020/478 | CTT/CTT | 86.36 | ||
| trnT | 6623/5094 | 6686/5159 | 64/66 | 0/2 | TGT/TGT | 86.36 | ||
| trnW | 6689/5162 | 6751/5225 | 63/64 | 2/2 | TCA/TCA | 93.75 | ||
| trnY | 6752/5232 | 6814/5295 | 63/64 | 0/6 | GTA/GTA | 93.85 | ||
| trnS1 | 6838/5301 | 6894/5357 | 57/57 | 23/5 | GCT/GCT | 82.46 | ||
| rrnL | 6895/5358 | 7828/6297 | 934/940 | 84.93 | ||||
| rrnS | 7829/6298 | 8550/7030 | 722/733 | 85.56 | ||||
| trnE | 8551/7031 | 8611/7093 | 61/63 | TTC/TTC | 77.78 | |||
| trnG | 8620/7131 | 8686/7196 | 67/66 | 8/37 | TCC/TCC | 80.6 | ||
| cox3 | 9479/7613 | 10150/8284 | 672/672 | 792/416 | ATG/ATG | TAA/TAA | 76.64 | |
| trnH | 10131/8265 | 10192/8328 | 62/64 | −20/−20 | GTG/GTG | 89.06 | ||
| cytb | 10193/8329 | 11269/9405 | 1077/1077 | ATG/ATG | TAA/TAA | 84.22 | ||
| nad4L | 11269/9405 | 11520/9656 | 252/252 | −1/−1 | ATG/ATG | TAG/TAG | 76.59 | |
| nad4 | 11608/9629 | 12852/10846 | 1245/1218 | 87/−28 | ATG/TTG | TAG/TAA | 71.73 | |
| trnQ | 12856/10849 | 12916/10911 | 61/63 | 3/2 | TTG/TTG | 85.71 | ||
| trnF | 12915/10910 | 12979/10974 | 65/65 | −2/−2 | GAA/GAA | 98.46 | ||
| trnM | 12971/10967 | 13035/11030 | 65/64 | −9/−8 | CAT/CAT | 92.31 | ||
| atp6 | 13039/11031 | 13548/11543 | 510/513 | 3/0 | ATG/ATG | TAG/TAA | 76.02 | |
| nad2 | 13552/11544 | 14373/12371 | 822/828 | 3/0 | ATG/ATG | TAA/TAA | 69.2 | |
| trnV | 14378/12372 | 14442/12435 | 65/64 | 4/0 | TAC/TAC | 81.54 | ||
| trnA | 14443/12436 | 14506/12503 | 64/68 | TGC/TGC | 82.35 | |||
| trnD | 14506/12504 | 14568/12566 | 63/63 | −1/0 | GTC/GTC | 81.25 | ||
| nad1 | 14569/12567 | 15468/13466 | 900/900 | ATG/GTG | TAA/TAA | 80.56 | ||
| trnN | 15475/13468 | 15538/13530 | 64/63 | 6/1 | GTT/GTT | 84.38 | ||
| trnP | 15562/13573 | 15626/13639 | 65/67 | 23/42 | TGG/TGG | 83.82 | ||
| trnI | 15626/13639 | 15692/13704 | 67/66 | −1/−1 | GAT/GAT | 92.54 | ||
| nad3 | 15696/13711 | 16058/14073 | 363/363 | 3/6 | ATG/ATG | TAA/TAA | 75.21 | |
| N | B | B | T | L | C | D | G | G | A | T | P | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Neobenedenia melleni | 1254 | 546 | 1120 | 294 | 292 | 1186 | 1056 | 622 | 1120 | 302 | 344 | 148 |
| Benedenia seriolae | 546 | 1254 | 610 | 162 | 162 | 580 | 514 | 342 | 552 | 184 | 230 | 84 |
| Benedenia hoshinai | 1120 | 610 | 1254 | 294 | 292 | 1186 | 1056 | 660 | 1120 | 302 | 356 | 146 |
| Thaparocleidus varicus | 294 | 162 | 294 | 1254 | 222 | 326 | 316 | 114 | 294 | 128 | 144 | 102 |
| Lepidotrema longipenis | 292 | 162 | 292 | 222 | 1254 | 322 | 306 | 110 | 292 | 146 | 232 | 182 |
| Cichlidogyrus sclerosus | 1186 | 580 | 1186 | 326 | 322 | 1254 | 1120 | 638 | 1186 | 322 | 370 | 162 |
| Dactylogyrus lamellatus | 1056 | 514 | 1056 | 316 | 306 | 1120 | 1254 | 608 | 1056 | 322 | 336 | 162 |
| Gyrodactylus gurleyi | 622 | 342 | 660 | 114 | 110 | 638 | 608 | 1254 | 688 | 252 | 214 | 94 |
| Gyrodactylus nyanzae | 1120 | 552 | 1120 | 294 | 292 | 1186 | 1056 | 688 | 1254 | 344 | 356 | 146 |
| Aglaiogyrodactylus forficulatus | 302 | 184 | 302 | 128 | 146 | 322 | 322 | 252 | 344 | 1254 | 150 | 108 |
| Tetraonchus monenteron | 344 | 230 | 356 | 144 | 232 | 370 | 336 | 214 | 356 | 150 | 1254 | 430 |
| Paratetraonchoides inermis | 148 | 84 | 146 | 102 | 182 | 162 | 162 | 94 | 146 | 108 | 430 | 1254 |
| Regions | Size (bp) | T(U) | C | A | G | AT(%) | GC(%) | GT(%) | AT Skew | GC Skew |
|---|---|---|---|---|---|---|---|---|---|---|
| Thaparocleidus asoti/Thaparocleidus varicus | ||||||||||
| PCGs | 10038/10020 | 48.6/49.4 | 7.5/7.6 | 26.8/26.8 | 17.1/16.2 | 75.4/76.2 | 24.6/23.8 | 65.7/65.6 | −0.288/−0.297 | 0.393/0.364 |
| 1st codon position | 3346/3340 | 41.7/42.3 | 7.8/7.2 | 29.7/30.5 | 20.8/20.0 | 71.4/72.8 | 28.6/27.2 | 62.5/62.3 | −0.168/−0.162 | 0.455/0.474 |
| 2nd codon position | 3346/3340 | 50.1/50.2 | 11.6/12.0 | 21.0/20.4 | 17.3/17.5 | 71.1/70.6 | 28.9/29.5 | 67.4/67.7 | −0.410/−0.423 | 0.196/0.187 |
| 3rd codon position | 3346/3340 | 53.9/55.8 | 3.0/3.5 | 29.8/29.6 | 13.4/11.1 | 83.7/85.4 | 16.4/14.6 | 67.3/66.9 | −0.288/−0.308 | 0.631/0.516 |
| atp6 | 510/513 | 50.2/49.3 | 6.7/8.2 | 27.6/26.7 | 15.5/15.8 | 77.8/76.0 | 22.2/24.0 | 65.7/65.1 | −0.290/−0.297 | 0.398/0.317 |
| cox1 | 1554/1557 | 45.3/46.6 | 11.0/11.0 | 24.8/24.0 | 18.9/18.4 | 70.1/70.6 | 29.9/29.4 | 64.2/65.0 | −0.293/−0.320 | 0.265/0.252 |
| cox2 | 625/627 | 42.4/43.1 | 9.3/9.7 | 28.2/27.8 | 20.2/19.5 | 70.6/70.9 | 29.5/29.2 | 62.6/62.6 | −0.202/−0.216 | 0.370/0.333 |
| cox3 | 672/672 | 51.3/49.9 | 6.5/6.8 | 23.5/26.2 | 18.6/17.1 | 74.8/76.1 | 25.1/23.9 | 69.9/67.0 | −0.372/−0.311 | 0.479/0.429 |
| cytb | 1077/1077 | 47.3/47.8 | 8.9/8.9 | 25.6/26.2 | 18.2/17.1 | 72.9/74.0 | 27.1/26.0 | 65.5/64.9 | −0.297/−0.292 | 0.342/0.314 |
| nad1 | 900/900 | 48.9/49.9 | 8.1/7.1 | 26.9/27.0 | 16.1/16.0 | 75.8/76.9 | 24.2/23.1 | 65.0/65.9 | −0.290/−0.298 | 0.330/0.385 |
| nad2 | 822/828 | 51.6/54.7 | 5.4/5.1 | 28.2/26.4 | 14.8/13.8 | 79.8/81.1 | 20.2/18.9 | 66.4/68.5 | −0.293/−0.348 | 0.470/0.462 |
| nad3 | 363/363 | 49.6/49.3 | 3.0/5.0 | 30.9/29.8 | 16.5/16.0 | 80.5/79.1 | 19.5/21.0 | 66.1/65.3 | −0.233/−0.247 | 0.690/0.526 |
| nad4 | 1245/1218 | 50.8/52.6 | 7.5/6.7 | 26.5/27.0 | 15.3/13.6 | 77.3/79.6 | 22.8/20.3 | 66.1/66.2 | −0.314/−0.322 | 0.343/0.339 |
| nad4L | 252/252 | 50.8/53.6 | 5.2/5.2 | 29.0/28.6 | 15.1/12.7 | 79.8/82.2 | 20.3/17.9 | 65.9/66.3 | −0.274/−0.304 | 0.490/0.422 |
| nad5 | 1572/1566 | 48.4/48.9 | 5.5/6.3 | 28.7/28.6 | 17.4/16.2 | 77.1/77.5 | 22.9/22.5 | 65.8/65.1 | −0.256/−0.262 | 0.517/0.443 |
| nad6 | 447/447 | 51.7/51.9 | 5.8/5.4 | 26.2/27.7 | 16.3/15.0 | 77.9/79.6 | 22.1/20.4 | 68.0/66.9 | −0.328/−0.303 | 0.475/0.473 |
| rrnL | 934/940 | 39.8/39.3 | 8.5/8.4 | 35.1/37.0 | 16.6/15.3 | 74.9/76.3 | 25.1/23.7 | 56.4/54.6 | −0.063/−0.029 | 0.325/0.291 |
| rrnS | 722/733 | 41.0/38.6 | 8.4/8.3 | 35.6/38.2 | 15.0/14.9 | 76.6/76.8 | 23.4/23.2 | 56.0/53.5 | −0.071/−0.005 | 0.278/0.282 |
| rRNAs | 1656/1673 | 40.3/39.0 | 8.5/8.4 | 35.3/37.5 | 15.9/15.1 | 75.6/76.5 | 24.4/23.5 | 56.2/54.1 | −0.066/−0.019 | 0.305/0.288 |
| tRNAs | 1410/1424 | 40.6/40.8 | 7.8/8.1 | 36.2/35.7 | 15.3/15.4 | 76.8/76.5 | 23.1/23.5 | 55.9/56.2 | −0.057/−0.067 | 0.325/0.313 |
| Full genome | 16074/14088 | 46.5/46.8 | 7.4/7.6 | 31.2/30.1 | 14.8/15.5 | 77.7/76.9 | 22.2/23.1 | 61.3/62.3 | −0.197/−0.217 | 0.334/0.341 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, D.; Zou, H.; Jakovlić, I.; Wu, S.G.; Li, M.; Zhang, J.; Chen, R.; Li, W.X.; Wang, G.T. Mitochondrial Genomes of Two Thaparocleidus Species (Platyhelminthes: Monogenea) Reveal the First rRNA Gene Rearrangement among the Neodermata. Int. J. Mol. Sci. 2019, 20, 4214. https://doi.org/10.3390/ijms20174214
Zhang D, Zou H, Jakovlić I, Wu SG, Li M, Zhang J, Chen R, Li WX, Wang GT. Mitochondrial Genomes of Two Thaparocleidus Species (Platyhelminthes: Monogenea) Reveal the First rRNA Gene Rearrangement among the Neodermata. International Journal of Molecular Sciences. 2019; 20(17):4214. https://doi.org/10.3390/ijms20174214
Chicago/Turabian StyleZhang, Dong, Hong Zou, Ivan Jakovlić, Shan G. Wu, Ming Li, Jin Zhang, Rong Chen, Wen X. Li, and Gui T. Wang. 2019. "Mitochondrial Genomes of Two Thaparocleidus Species (Platyhelminthes: Monogenea) Reveal the First rRNA Gene Rearrangement among the Neodermata" International Journal of Molecular Sciences 20, no. 17: 4214. https://doi.org/10.3390/ijms20174214
APA StyleZhang, D., Zou, H., Jakovlić, I., Wu, S. G., Li, M., Zhang, J., Chen, R., Li, W. X., & Wang, G. T. (2019). Mitochondrial Genomes of Two Thaparocleidus Species (Platyhelminthes: Monogenea) Reveal the First rRNA Gene Rearrangement among the Neodermata. International Journal of Molecular Sciences, 20(17), 4214. https://doi.org/10.3390/ijms20174214





