Abstract
In this narrative review, we summarize recent pieces of evidence of the role of microbiota alterations in Rett syndrome (RTT). Neurological problems are prominent features of the syndrome, but the pathogenic mechanisms modulating its severity are still poorly understood. Gut microbiota was recently demonstrated to be altered both in animal models and humans with different neurodevelopmental disorders and/or epilepsy. By investigating gut microbiota in RTT cohorts, a less rich microbial community was identified which was associated with alterations of fecal microbial short-chain fatty acids. These changes were positively correlated with severe clinical outcomes. Indeed, microbial metabolites can play a crucial role both locally and systemically, having dynamic effects on host metabolism and gene expression in many organs. Similar alterations were found in patients with autism and down syndrome as well, suggesting a potential common pathway of gut microbiota involvement in neurodevelopmental disorders.
1. Introduction
The commensal microbial community inhabiting the mammalian gastrointestinal tract, microbiota, has been demonstrated to exert important functions both prenatally and postnatally [1]. Several studies on germ-free mice established that, at the local level, the presence of pioneering microorganisms triggers the correct development and maturation of the gut-associated lymphoid tissue. Indeed, in the absence of microbiota, animals fail to complete Peyer patches and mesenteric lymph nodes maturation and display longer and thinner villi with a reduced vascular network [2]. This intuitive and intimate relationship between the gut and its microbiota during development also applies to distant districts, with the brain as the most counterintuitive and intriguing. Microbial-derived signals are crucial for the physiological brain development, promoting neurotrophins secretion [3], microglia maturation [4], blood-brain barrier functionality (by enhancing tight-junction expression) [5], and correct myelination [6]. Brain development is tremendously vulnerable to environmental factors, and events occurring in early phases might have long-term effects later in life [7]. Beside the described role in early infancy, the microbiota-gut-brain axis was recently recognized to be a highly complex and tightly regulated bidirectional network. From top to bottom, it encompasses the central nervous system (CNS), the sympathetic and parasympathetic branches of the autonomic nervous system, the enteric nervous system, and the neuroendocrine and neuroimmune systems [8]. This complex network is enriched by the gut microbial community exerting a bottom-up modulation by interacting mainly with the vagus nerve and the neuroendocrine and neuroimmune systems [9].
In this review, we will focus on gut microbiota changes in Rett syndrome (RTT), the most prevalent neurodevelopmental disorder in females, whose pathogenic mechanisms are still poorly understood and whose genetic background cannot fully explain the phenotypic heterogeneity.
2. Gut Microbiome Alterations in Neurodevelopmental Disorders
Neurodevelopmental disorders (NDs) are chronic disorders that affect CNS function during the developmental period in the domains of motor skills, cognition, communication, and/or behavior. The diagnosis of NDs is primarily clinical, based on a constellation of behaviors/symptoms that are specified in the diagnostic and statistical manual of mental disorders (DSM) [10]. The most common ND worldwide is autism spectrum disorder (ASD), for which several studies, conducted since 2000 in different geographical regions and by different teams, have estimated a median prevalence of 62/10,000 [11].
ASD is an extremely heterogeneous neurodevelopmental disorder and its onset can vary. Some children show signs from early infancy, while others exhibit symptoms of regression around 18–24 months. Other specific features may be recognizable only at an older age. Clinically, ASD is characterized by stereotypical behaviors and a deficit in social interaction and communication [12]. In addition to the neuropsychiatric features, children with ASD often experience gastrointestinal (GI) problems that are more frequent and more severe compared with children from the general population. Functional constipation is the most common issue [13], followed by diarrhea/chronic diarrhea, abdominal pain or discomfort, bloating, gas or flatulence, soiling, incontinence or bedwetting, reflux or heartburn, nausea or vomiting, and pain or difficulties in bowel movement [14]. Gastrointestinal symptoms strongly correlate with the severity of autism and with increased irritability, anxiety, and social withdrawal [15,16]. This evidence elicits researches focusing on the connection between ASD and the gut microbiota. Intriguingly, an altered composition of gut microbiome has been demonstrated in subjects with ASD [17,18,19,20], suggesting a possible link between dysbiosis (an aberrant shift in microbial ecology) and neurodevelopmental diseases. Despite differences in cohorts, study designs, and methodologies, a common microbial signature has been identified (reviewed by Ding et al. [21] and by the recent systematic review by Liu et al. [22]). ASD-related major changes involve an increase in the relative abundance of Clostridium spp. and Sutterella spp., Bacteroides spp. and Desulfovibrio spp. and a depletion in Bifidobacterium spp., Blautia spp., Prevotella spp., and Veillonella spp. [19,23,24]. The Clostridium hypothesis, suggesting the involvement of toxigenic bacteria in the onset of the ASD regressive form, was first suggested by Ellen Bolte [25] and later supported by other authors [26,27]. Indeed, clostridia are spore-forming and can exert a toxic activity by producing exotoxins, phenol, and p-cresol [27,28]. P-cresol has been postulated as a possible urinary biomarker in ASD and its concentration correlates with constipation (slow fecal transit) [29]. Other microbial-derived metabolites have been linked to ASD in recent years, including propionate. Propionate is a short-chain fatty acid (SFCA) and one of the main products of microbial fermentation, together with acetate and butyrate. SCFAs are utilized by the host as an energy source and exert other physiological functions. SCFAs participate in glucose homeostasis, affect lipid metabolism, regulate the immune system and inflammatory response, and serve as signaling molecules [30].
Intracerebroventricular injection of propionate in rodents was found to cause social impairment [31], as well as behavioral, neuropathological, and biochemical changes associated with ASD [32]. However, the contribution of microbial metabolites to ASD still must be elucidated, taking into account that fecal SCFAs were reported to be either lower or higher in ASD children [15,33,34].
Among genetic NDs, down syndrome (OMIM #190685, trisomy 21–DS) is the most frequent cause of intellectual disability (ID). ASD features and the high prevalence of gastrointestinal system alterations (about 50%) often characterize DS individuals [35]. Despite DS incidence and the suggestive features of a possible gut microbiota modulation, only one study has investigated the DS gut microbial community thus far. Biagi and coworkers [36] compared 17 DS adults with a matched group of healthy controls. The authors reported specific changes in the microbial community of individuals affected by DS, with some of them being associated with behavioral traits. Although DS gut microbiota does not significantly differ in the relative abundance of the dominant microbial families (i.e., Ruminococcaceae, Lachnospiraceae, Clostridiales, Bifidobacteriaceae, and Bacteroidaceae), some subdominant taxa are differently represented in DS compared with healthy controls. In particular, the relative abundance of Parasporobacterium spp. and Sutterella spp. is increased, whereas Veillonellaceae is reduced. Besides, when considering the severity of neurobehavioral manifestations in DS patients, Sutterella was positively correlated with the aberrant behavior checklist (ABC) total score, while no correlation was found between ABC and Parasporobacterium or Veillonellaceae.
3. Gut Microbiome Alterations in Rett Syndrome
Although Rett syndrome (RTT) is no longer categorized as a pervasive developmental disorder, patients share certain features with ASD. RTT (OMIM #312750) is an X-linked dominant neurodevelopmental disorder and one of the most common causes of ID in females. Of all cases, 90–95% are associated with pathogenic variants in the MECP2 (Methyl CpG Binding Protein 2) gene, encoding a chromatin-associated protein that can both activate and repress transcription [37,38]. The RTT core phenotype mainly consists of neurological problems (partial/complete loss of acquired purposeful hand skills; partial/complete loss of acquired spoken language; gait abnormalities; stereotypic hand movements) [39]. Most girls affected by RTT experience epileptic seizures, with epilepsy representing a major concern of RTT caregivers and having a crucial impact on children and their family’s quality of life [40]. In addition to epilepsy, parental stress is affected by their daughters’ GI pain experience [41]. Indeed, one of the major issues impacting patients affected by RTT on a daily basis is represented by gastrointestinal and nutritional problems [42]. Families reported a series of different symptoms, such as straining with bowel movements, the passage of hard stools, constipation, and prolonged feeding time or chewing difficulty in more than 50% of RTT patients [42]. More recently, cholelithiasis, or gallbladder disease, has been reported to be relatively frequent in RTT, and should be considered one of the causes of abdominal pain [43]. Due to the high frequency of gastrointestinal comorbidities, recommendations for managing GI symptoms have been recently developed [44].
Despite the similar features with ASD and the observation of the frequent gastrointestinal discomfort reported by RTT caregivers, the study of possible microbial alterations characterizing RTT gut microbiota is still relatively new. Only two Italian groups have investigated this aspect in RTT girls thus far. The characteristics and main results of these studies are outlined in Table 1.

Table 1.
Characteristics of gut microbiota studies in Rett syndrome (RTT).
Because of the higher vulnerability to respiratory infections related both to RTT itself and to the different associated comorbidities (e.g. scoliosis, epilepsy, ID) [45], subjects with RTT may be exposed to several antibiotics to prevent/cure recurrent respiratory infections. Antibiotics are responsible for altering the gut microbiome. For this reason, in the studies conducted on RTT girls, a common exclusion criterion was an antibiotic assumption in the three months preceding the collection of the sample.
RTT gut microbiota is characterized by a reduction in α-diversity [46,47]. α-diversity metrics allow studying the richness, i.e., the number of unique microbial taxa within a given sample, as well as the evenness, how uniformly the unique taxa are distributed, and the phylogenetic relatedness between them. The loss of microbial diversity is considered a hallmark of dysbiosis, paving the way to a reduced gut microbiota resilience [48]. Thus, the gut microbial ecosystem of RTT girls can be considered intrinsically more susceptible to disease status. The decrease in diversity observed in RTT patients is more pronounced in severe phenotypes, as patients with a lower diversity have higher clinical severity scale (CSS) scores [47].
β-diversity (Unifrac distances) analyses, which measure dissimilarities in microbial community composition between samples, showed that RTT gut microbial community clustered according to the disease status. On the one hand, Strati and coworkers grouped RTT patients according to GI dysfunction and observed that health status, not GI symptoms, distinguish the patients from healthy controls, whereas constipated RTT and non-constipated RTT were not statistically different. On the other hand, the severity of the disease further stratified RTT patients [47]. In particular, unweighted Unifrac metric, which considers both rare and common bacterial taxa, highlighted a significant dissimilarity between the RTT and healthy subjects, suggesting that, as described for DS [36], subdominant taxa are differently represented. The analyses on the relative microbial abundance at different taxonomic levels showed an enrichment in Erysipelotrichaceae in RTT patients and, at the genus level, of Clostridium spp., Sutterella spp., and Escherichia spp. [46,47]. Other taxa were found to be discordant between the two studies, i.e., Bifidobacterium and Bacteroides. Bifidobacteria data could suffer from an age-related abundance. Indeed, Strati et al. enrolled younger girls affected by RTT, with a consistent number of children younger than 10 years old. This taxon is well-known to inversely correlate with age, with the highest abundance during lactation [49]. Like the microbial alterations reported in ASD patients, Borghi et al. observed a decrease in Bacteroides spp. [47]. This genus, as well as Bacteroidetes, are reported to be negatively correlated with body-mass index (BMI) [50,51,52]. BMI values were lower in the RTT group, justifying this observation. Anthropometric measurements were not specified in the second cohort. Despite differences in the observed taxa relative abundances in the two studies, a common increase in the fecal concentration of branched-chain fatty acids (iso-butyrate and iso-valerate-BCFAs) and, to a lesser extent, of propionate and butyrate, has been observed. BCFAs are byproducts of protein degradation, especially of animal proteins that are rich in branched-chain amino acids, by proteolytic bacteria such as Bacteroidetes. Dietary survey [47] showed a higher protein consumption, mainly due to higher animal protein intake, and a lower fiber intake in RTT compared with healthy controls. No specific dietary recommendations for RTT patients have been reported so far, but proteins might be preferred for the increased ratio nutrients/volume, considering eating difficulties, palatability, and texture. Beside BCFAs, proteolysis results in the production of phenolic and indolic compounds. As already mentioned for ASD, p-cresol, the main phenolic product, is a toxic compound and has been demonstrated to be able to alter the intestinal barrier permeability [53]. Although no study has evaluated its fecal or urinary concentrations in RTT subjects to date, the enrichment in gut microbiota in the main producing species, i.e., Clostridium spp. [46,47] Bacteroides spp. [47], suggests a possible increase of p-cresol also in RTT.
Mouse models of RTT are available and they recapitulate several neurological features observed in patients [54], including altered cortical rhythms and enhanced susceptibility to epileptic seizures [55,56]. Moreover, severe modifications in the colon organization are present in MeCP2 mutant mice [57], suggesting that alterations occurring at the gut level, including gut microbiome, could be associated with the classical neurological impairments observed in RTT mice. Despite this evidence, the gut microbiota of RTT mouse models has not been explored to date, and the aspects related to the gut microbiome-brain axis in this murine model have not been examined.
4. Gut Microbiome Alterations in Epilepsy
Both RTT and ASD are often comorbid with seizure disorders [58]. Although it is not mentioned in the diagnostic criteria [39], up to 70% of girls affected by RTT experience frequent seizures, and 30% of them are resistant to the available antiepileptic drugs (AEDs) [59]. In classic RTT, the mean age of epilepsy onset is 4 years. Seizure frequency and severity usually have an age-dependent course, as girls aged 10–14 years are the most difficult to treat and often require AED polytherapy [60]. Recent findings from the Rett Natural History Study pointed out that seizures in RTT may show a pattern of remission and relapse within the lifetime. Specific MECP2 mutations are not significantly associated with either seizure prevalence or seizure severity, although seizure prevalence is associated with disease severity [61].
The potential role of gut microbiota in epilepsy is currently emerging. He et al. [62] studied a patient with a 17-year history of epilepsy, which improved after fecal microbiota transplantation treatment for Crohn’s disease. More recently, the gut microbial community in patients with drug-resistant epilepsy was found to be significantly altered with an abnormally increased abundance of rare bacteria mainly belonging from the phylum Firmicutes. On the contrary, the gut microbiome of patients with drug-sensitive epilepsy was more similar to that of healthy controls. Therefore, it can be speculated that gut dysbiosis may be involved in the pathogenesis of drug-resistant epilepsy [63]. In particular, patients with seizures responsive to AEDs have a greater amount of bifidobacteria and lactobacilli, which could be interpreted as protective factors for epilepsy [63]. Indeed, a recent study on drug-resistant epilepsy showed that probiotic supplementation with a mixture of bifidobacteria and lactobacilli reduces both seizures and sCD14 serum concentration, a recognized marker for bacterial translocation [64].
Many drugs have been demonstrated to modulate or alter the gut microbial community. In particular, compounds targeting the nervous system exhibited a significant anticommensal activity on a broad range of microorganisms [65]. Although the direct role of AEDs has not been investigated to date, it is plausible, although not yet elucidated [47].
Other pieces of evidence based on diet modification support the possible role of gut microbiota in modulating epilepsy. The ketogenic diet (KD) is a high-fat, adequate-protein, low-carbohydrate diet used as a treatment for neurometabolic disorders, such as glucose transporter 1 (Glut1)-deficiency syndrome, and for drug-resistant epilepsy. Considering the unbalanced macronutrient composition of the KD, it is expected to induce some changes in gut microbiota. Tagliabue et al. [66] evaluated the gut microbiota composition in six children with Glut1-deficiency syndrome after three months of KD treatment and did not find statistically significant differences in Firmicutes and Bacteroidetes. However, they found a statistically significant increase in Desulfovibrio spp., a subdominant taxon reported in inflammatory bowel disease and other inflammatory conditions [67].
Xie and coworkers [68] found that the gut microbiota of Chinese epileptic children differed from age-matched healthy infants and reported that the KD strongly improved gut microbiota alterations, promoting seizure reductions. Another study, which enrolled 20 children with refractory epilepsy, observed distinctive microbial changes in KD-responder (seizure-free or ≥ 50% of seizure reduction) and non-responder patients (<50% of seizure reduction). The KD-responders were characterized by an increase in the relative abundance of Bacteroidetes, whereas non-responders showed a significant increase in the relative abundance of the Firmicutes (Clostridiales, Clostridia, Ruminococcaceae, Lachnospiraceae, Alistipes, and Rikenellaceae) [69]. A further study on KD treatment for severe epilepsy in Swedish children, which applied a shotgun metagenomic DNA sequencing approach, found that alpha diversity was not significantly changed by diet. The study highlighted a decrease in the relative abundance of Actinobacteria and Firmicutes, and an increase of Bacteroidetes and Proteobacteria [70]. Intriguingly, microbiota changes were linked to functional changes, particularly in relation to carbohydrates metabolism pathways.
The KD has been investigated as a possible intervention in RTT [71], especially in the CDKL5-related variant, which is characterized by multiple seizure-type epilepsy and a poor response to AEDs [72]. Major concerns to a broad application of the KD are the reported poor long-term efficacy and lack of adherence.
Despite the growing number of studies on the KD and its impact on microbiota in epilepsy, the findings should be interpreted with caution because of the small number of patients involved, the different epilepsy etiology, and the specific diet composition. Nevertheless, the accumulating evidence suggests the need for a better understanding of the state of dysbiosis to establish strategies to possibly counterbalance it.
5. Discussion
Intriguingly, neurodevelopmental disorders sharing clinical features display a microbial signature or are at least enriched/depleted in the same taxa. The major overlaps, both clinical and microbial, are highlighted in the Venn diagram (Figure 1).
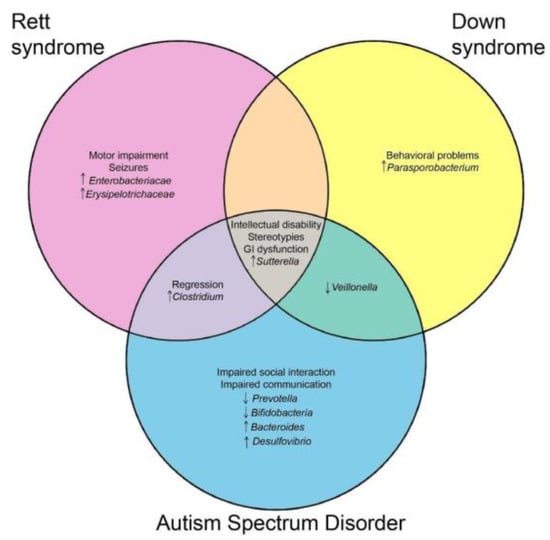
Figure 1.
Venn diagram presenting the shared clinical and microbial features between Rett Syndrome, autism spectrum disorder, and down syndrome.
The diagram was generated (http://bioinfogp.cnb.csic.es/tools/venny/index.html) considering only microbial changes reported in most studies (i.e., discarding discordant findings).
In particular, an enrichment in Sutterella spp. has been reported in all the reviewed NDs. A similar trend was also observed in attention-deficit/hyperactivity disorder (ADHD) [73] and in rodent models with Alzheimer’s disease [74] and ASD [75]. Sutterella belongs to Beta-proteobacteria, gram-negative bacterium. Due to the reported higher relative abundance in several human diseases, Hiippala and coworkers [76] assessed its in vitro pro-inflammatory activity and compared it with the well-known Escherichia coli (gamma-Proteobacteria). Sutterella demonstrated only a mild pro-inflammatory activity on intestinal epithelial cells, which was not sufficient to induce GI homeostasis alterations. On the other hand, this taxon was demonstrated to efficiently adhere to enterocytes [77], and the effect of a direct cross-talk with these cells in the frame of the gut-brain-axis warrants further elucidation.
Similarly, RTT and ASD subjects share an increase in the relative abundance of Clostridium spp. This taxon, together with Bacteroides spp., is well-known for its proteolytic ability, which results in potentially toxic compounds that can impact on gut homeostasis and permeability [53] and can affect the survival of other beneficial microorganisms, such as lactic bacteria [77]. Bifidobacteria and lactobacilli are known to be able to secrete neurotransmitters, particularly the gamma-aminobutyric acid (GABA) [78]. A decrease in GABA and its signaling is suggested to be involved in many ND-associated clinical features, i.e., stereotypies, hypersensitivities, and seizures [79,80,81]. Moreover, the same taxa have been shown to promote intestinal mucosa integrity [82], decreasing local and systemic inflammatory status. CNS is highly susceptible to inflammation, contributing to the pathogenesis of RTT [83] and to associated comorbidities. In particular, neuroinflammation has been demonstrated to trigger seizures in patients with epilepsy [84]. A direct role of gut microbiota in eliciting inflammation and, in turn, seizure occurrence, has been postulated and investigated in animal models [85].
Although speculative conclusions about microbiota-dependent modifications in brain function and behaviors have been raised over recent years (summarized in the context of NDs in Figure 2), no studies have directly assessed the molecular or neurophysiological mechanisms responsible for neurodevelopmental alterations characteristic of NDs [86].
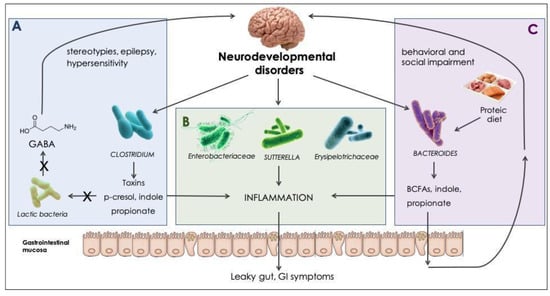
Figure 2.
Microbial modulation in neurodevelopmental disorders. Effects driven directly or indirectly by gut microbes, found to be increased in NDs and their products. (A) Clostridia are spore-forming, toxigenic bacteria, with proteolytic ability [53]; digestion of aromatic amino acids results in phenol, p-cresol, and indole production. High indole concentrations can suppress bifidobacteria and lactobacilli growth [77]. Lactic bacteria are known to endogenously produce gamma-aminobutyric acid (GABA) [78]. GABA decrease has been reported as an underlying mechanism for stereotypies occurrence [79], ASD hypersensitivity [80], and epilepsy [81]. (B) Enterobacteriaceae [87], Sutturella spp. [76], and Erysipelotrichaceae [88] have been reported to exert a pro-inflammatory effect, which can alter gastrointestinal homeostasis and alter the gastrointestinal (GI) barrier permeability. (C) Bacteroides are nutritionally versatile bacteria, with both saccharolytic and proteolytic activity [53]; a rich-protein diet promotes the production of branched-chain fatty acids (BCFAs) and propionate [89]. High propionate concentrations have demonstrated to exert behavioral and social impairment in animal models [31].
Most of the literature has focused either on the clinical aspect or on the microbiome alterations. Nevertheless, NDs are characterized by multifaceted phenotypes that might contribute to the observed differences in the microbiota among various cohorts. A comprehensive approach, combining detailed clinical description with microbiological data, is needed to better understand underlying relationships between symptoms and specific microbial alterations.
6. Conclusions
Although microbiome studies in RTT patients are in their early stages, several authors have investigated this aspect in ASD, which shares several clinical similarities with RTT. Main changes in the gut microbial community suggest enrichment in pro-inflammatory species that could promote alterations in the gut permeability and its barrier function. Within the context of the gut-brain axis, these microbiota-dependent modifications could trigger common behavioral and neurodevelopmental features.
The observation of the presence of a dysbiosis in patients with NDs, especially ASD, triggers the development of innovative therapeutic strategies throughout microbiome-based treatment. Nevertheless, clinical trials with probiotic supplementation are currently limited and lack standardized probiotic regimen, i.e., different administered strains or concentrations and duration of treatment. No clinical trials have been carried out on RTT cohorts thus far.
In addition to probiotic intervention, a careful diet survey on large cohorts of RTT patients would allow the development of diet recommendations that might mitigate microbiome alterations per se.
Author Contributions
E.B. and A.V. conceptualized, wrote, and reviewed the article.
Funding
The FFABR 2017 grant by the Italian Ministry of Education, Universities and Research to A.V. covered the costs to publish in open access.
Acknowledgments
The authors thank Angela Peron for reviewing the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| AED | Antiepileptic drug |
| ASD | Autism spectrum disorder |
| BCFA | Branched-chain fatty acids |
| CDKL5 | Cyclin-dependent kinase-like 5 |
| CNS | Central nervous system |
| CSS | Clinical severity score |
| DS | Down syndrome |
| GABA | Gamma aminobutyric acid |
| GI | Gastrointestinal tract |
| ID | Intellectual disability |
| KD | Ketogenic diet |
| MeCP2 | Methyl CpG binding protein 2 |
| ND | Neurodevelopmental disorder |
| RTT | Rett syndrome |
| SCFA | Short-chain fatty acids |
References
- Round, J.L.; Mazmanian, S.K. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009, 9, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Sharon, G.; Sampson, T.R.; Geschwind, D.H.; Mazmanian, S.K. Central Nervous System and the Gut Microbiome. Cell 2016, 167, 915–932. [Google Scholar] [CrossRef] [PubMed]
- Sommer, F.; Bäckhed, F. The gut microbiota--masters of host development and physiology. Nat. Rev. Microbiol. 2013, 11, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Bercik, P.; Denou, E.; Collins, J.; Jackson, W.; Lu, J.; Jury, J.; Deng, Y.; Blennerhassett, P.; Macri, J.; McCoy, K.D.; et al. The intestinal microbiota affects central levels of brain-derived neurotropic factor and behavior in mice. Gastroenterology 2011, 141, 599–609. [Google Scholar] [CrossRef] [PubMed]
- Erny, D.; Hrabe de Angelis, A.L.; Jaitin, D.; Wieghofer, P.; Staszewski, O.; David, E.; Keren-Shaul, H.; Mahlakoiv, T.; Jakobshagen, K.; Buch, T.; et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat. Neurosci. 2015, 18, 965–977. [Google Scholar] [CrossRef] [PubMed]
- Braniste, V.; Al-Asmakh, M.; Kowal, C.; Anuar, F.; Abbaspour, A.; Tòth, M.; Korecka, A.; Bakocevic, N.; Ng, L.G.; Kundu, P.; et al. The gut microbiota influences blood-brain barrier permeability in mice. Sci. Transl. Med. 2014, 6, 263ra158. [Google Scholar] [CrossRef] [PubMed]
- Hoban, A.E.; Stilling, R.M.; Ryan, F.J.; Shanahan, F.; Dinan, T.G.; Claesson, M.J.; Clarke, G.; Cryan, J.F. Regulation of prefrontal cortex myelination by the microbiota. Transl. Psychiatry 2016, 6, e774. [Google Scholar] [CrossRef] [PubMed]
- Diaz Heijtz, R.; Wang, S.; Anuar, F.; Qian, Y.; Björkholm, B.; Samuelsson, A.; Hibberd, M.L.; Forssberg, H.; Pettersson, S. Normal gut microbiota modulates brain development and behavior. Proc. Natl. Acad. Sci. USA. 2011, 108, 3047–3052. [Google Scholar] [CrossRef] [PubMed]
- Mayer, E.A.; Savidge, T.; Shulman, R.J. Brain-gut microbiome interactions and functional bowel disorders. Gastroenterology 2014, 146, 1500–1512. [Google Scholar] [CrossRef]
- Ismail, F.Y.; Shapiro, B.K. What are neurodevelopmental disorders? Curr. Opin. Neurol. 2019, 32, 611–616. [Google Scholar] [CrossRef]
- Elsabbagh, M.; Divan, G.; Koh, Y.J.; Kim, Y.S.; Kauchali, S.; Marcín, C.; Montiel-Nava, C.; Patel, V.; Paula, C.S.; Wang, C.; et al. Global prevalence of autism and other pervasive developmental disorders. Autism. Res. 2012, 5, 160–179. [Google Scholar] [CrossRef] [PubMed]
- American Psychiatric Association. American Psychiatric Association Diagnostic and Statistical Manual of Mental Disorders, 5th ed.; American Psychiatric Association: Arlington County, VA, USA, 2013. [Google Scholar]
- Marler, S.; Ferguson, B.J.; Lee, E.B.; Peters, B.; Williams, K.C.; McDonnell, E.; Macklin, E.A.; Levitt, P.; Margolis, K.G.; Beversdorf, D.Q.; et al. Association of Rigid-Compulsive Behavior with Functional Constipation in Autism Spectrum Disorder. J. Autism. Dev. Disord. 2017, 47, 1673–1681. [Google Scholar] [CrossRef] [PubMed]
- Holingue, C.; Newill, C.; Lee, L.C.; Pasricha, P.J.; Daniele Fallin, M. Gastrointestinal symptoms in autism spectrum disorder: A review of the literature on ascertainment and prevalence. Autism. Res. 2018, 11, 24–36. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.B.; Johansen, L.J.; Powell, L.D.; Quig, D.; Rubin, R.A. Gastrointestinal flora and gastrointestinal status in children with autism–comparisons to typical children and correlation with autism severity. BMC Gastroenterol. 2011, 16, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Nikolov, R.N.; Bearss, K.E.; Lettinga, J.; Erickson, C.; Rodowski, M.; Aman, M.G.; McCracken, J.T.; McDougle, C.J.; Tierney, E.; Vitiello, B.; et al. Gastrointestinal symptoms in a sample of children with pervasive developmental disorders. J. Autism. Develop. Disord. 2009, 39, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Kohane, I.S.; McMurry, A.; Weber, G.; MacFadden, D.; Rappaport, L.; Kunkel, L.; Bickel, J.; Wattanasin, N.; Spence, S.; Murphy, S.; et al. The co-morbidity burden of children and young adults with autism spectrum disorders. PLoS ONE 2012, 7, e33224. [Google Scholar] [CrossRef]
- Kang, D.W.; Park, J.G.; Ilhan, Z.E.; Wallstrom, G.; Labaer, J.; Adams, J.B.; Krajmalnik-Brown, R. Reduced incidence of Prevotella and other fermenters in intestinal microflora of autistic children. PLoS ONE 2013, 8, e68322. [Google Scholar] [CrossRef]
- Wang, L.; Christophersen, C.T.; Sorich, M.J.; Gerber, J.P.; Angley, M.T.; Conlon, M.A. Increased abundance of Sutterella spp. and Ruminococcus torques in feces of children with autism spectrum disorder. Mol. Autism. 2013, 4, 42. [Google Scholar] [CrossRef] [PubMed]
- Argou-Cardozo, I.; Zeidán-Chuliá, F. Clostridium Bacteria and Autism Spectrum Conditions: A Systematic Review and Hypothetical Contribution of Environmental Glyphosate Levels. Med. Sci. (Basel) 2018, 6, E29. [Google Scholar] [CrossRef]
- Ding, H.T.; Taur, Y.; Walkup, J.T. Gut Microbiota and Autism: Key Concepts and Findings. J. Autism Dev. Disord. 2017, 47, 480–489. [Google Scholar] [CrossRef]
- Liu, F.; Li, J.; Wu, F.; Zheng, H.; Peng, Q.; Zhou, H. Altered composition and function of intestinal microbiota in autism spectrum disorders: A systematic review. Transl. Psychiatry 2019, 9, 43. [Google Scholar] [CrossRef] [PubMed]
- De Angelis, M.; Piccolo, M.; Vannini, L.; Siragusa, S.; De Giacomo, A.; Serrazzanetti, D.I.; Cristofori, F.; Guerzoni, M.E.; Gobbetti, M.; Francavilla, R. Fecal microbiota and metabolome of children with autism and pervasive developmental disorder not otherwise specified. PLoS ONE 2013, 8, e76993. [Google Scholar] [CrossRef] [PubMed]
- Finegold, S.M.; Dowd, S.E.; Gontcharova, V.; Liu, C.; Henley, K.E.; Youn, E.; Summanen, P.H.; Granpeesheh, D.; Dixon, D.; Liu, M.; et al. Pyrosequencing study of fecal microflora of autistic and control children. Anaerobe 2010, 16, 444–453. [Google Scholar] [CrossRef] [PubMed]
- Bolte, E.R. Autism and Clostridium tetani. Med. Hypotheses 1998, 51, 133–144. [Google Scholar] [CrossRef]
- Song, Y.; Liu, C.; Finegold, S.M. Real-time PCR quantitation of clostridia in feces of autistic children. Appl. Environ. Microbiol. 2004, 70, 6459–6465. [Google Scholar] [CrossRef]
- Finegold, S.M. Therapy and epidemiology of autism-clostridial spores as key elements. Med. Hypotheses 2008, 70, 508–511. [Google Scholar] [CrossRef]
- Stiles, B.G.; Pradhan, K.; Fleming, J.M.; Samy, R.P.; Barth, H.; Popoff, M.R. Clostridium and bacillus binary enterotoxins: Bad for the bowels, and eukaryotic being. Toxins (Basel) 2014, 6, 2626–2656. [Google Scholar] [CrossRef]
- Gabriele, S.; Sacco, R.; Altieri, L.; Neri, C.; Urbani, A.; Bravaccio, C.; Riccio, M.P.; Iovene, M.R.; Bombace, F.; De Magistris, L.; et al. Slow intestinal transit contributes to elevate urinary p-cresol level in Italian autistic children. Autism. Res. 2016, 9, 752–759. [Google Scholar] [CrossRef]
- Dalile, B.; Van Oudenhove, L.; Vervliet, B.; Verbeke, K. The role of short-chain fatty acids in microbiota-gut-brain communication. Nat. Rev. Gastroenterol. Hepatol. 2019. [Epub ahead of print]. [Google Scholar] [CrossRef]
- Shultz, S.R.; Aziz, N.A.; Yang, L.; Sun, M.; MacFabe, D.F.; O’Brien, T.J. Intracerebroventricular injection of propionic acid, an enteric metabolite implicated in autism, induces social abnormalities that do not differ between seizure-prone (FAST) and seizure-resistant (SLOW) rats. Behav. Brain Res. 2015, 278, 542–548. [Google Scholar] [CrossRef]
- Macfabe, D.F. Short-chain fatty acid fermentation products of the gut microbiome: Implications in autism spectrum disorders. Microb. Ecol. Health Dis. 2012, 23. [Google Scholar] [CrossRef]
- Wang, L.; Christophersen, C.T.; Sorich, M.J.; Gerber, J.P.; Angley, M.T.; Conlon, M.A. Elevated fecal short chain fatty acid and ammonia concentrations in children with autism spectrum disorder. Dig. Dis. Sci. 2012, 57, 2096–2102. [Google Scholar] [CrossRef]
- Kang, D.W.; Ilhan, Z.E.; Isern, N.G.; Hoyt, D.W.; Howsmon, D.P.; Shaffer, M.; Lozupone, C.A.; Hahn, J.; Adams, J.B.; Krajmalnik-Brown, R. Differences in fecal microbial metabolites and microbiota of children with autism spectrum disorders. Anaerobe 2018, 49, 121–131. [Google Scholar] [CrossRef]
- Vicari, S.; Pontillo, M.; Armando, M. Neurodevelopmental and psychiatric issues in Down’s syndrome: Assessment and intervention. Psychiatr. Genet. 2013, 23, 95–107. [Google Scholar] [CrossRef]
- Biagi, E.; Candela, M.; Centanni, M.; Consolandi, C.; Rampelli, S.; Turroni, S.; Severgnini, M.; Peano, C.; Ghezzo, A.; Scurti, M.; et al. Gut microbiome in Down syndrome. PLoS ONE 2014, 9, e112023. [Google Scholar] [CrossRef]
- Amir, R.E.; Van den Veyver, I.B.; Wan, M.; Tran, C.Q.; Francke, U.; Zoghbi, H.Y. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 1999, 23, 185–188. [Google Scholar] [CrossRef]
- Marschik, P.B.; Kaufmann, W.E.; Sigafoos, J.; Wolin, T.; Zhang, D.; Bartl-Pokorny, K.D.; Pini, G.; Zappella, M.; Tager-Flusberg, H.; Einspieler, C.; et al. Changing the perspective on early development of Rett syndrome. Res. Dev. Disabil. 2013, 34, 1236–1239. [Google Scholar] [CrossRef]
- Neul, J.L.; Kaufmann, W.E.; Glaze, D.G.; Christodoulou, J.; Clarke, A.J.; Bahi-Buisson, N.; Leonard, H.; Bailey, M.E.S.; Schanen, N.C.; Zappella, M.; et al. Rett syndrome: Revised diagnostic criteria and nomenclature. Ann. Neurol. 2010, 68, 944–950. [Google Scholar] [CrossRef]
- Bahi-Buisson, N.; Guellec, I.; Nabbout, R.; Guet, A.; Nguyen, G.; Dulac, O.; Chiron, C. Parental view of epilepsy in Rett Syndrome. Brain Dev. 2008, 30, 126–130. [Google Scholar]
- Byiers, B.J.; Tervo, R.C.; Feyma, T.J.; Symons, F.J. Seizures and pain uncertainty associated with parenting stress and Rett syndrome. J. Child. Neurol. 2014, 29, 526–529. [Google Scholar] [CrossRef]
- Motil, K.J.; Caeg, E.; Barrish, J.O.; Geerts, S.; Lane, J.B.; Percy, A.K.; Annese, F.; McNair, L.; Skinner, S.A.; Lee, H.S.; et al. Gastrointestinal and nutritional problems occur frequently throughout life in girls and women with Rett syndrome. J. Pediatr. Gastroenterol. Nutr. 2012, 55, 292–298. [Google Scholar] [CrossRef]
- Freilinger, M.; Böhm, M.; Lanator, I.; Vergesslich-Rothschild, K.; Huber, W.D.; Anderson, A.; Wong, K.; Baikie, G.; Ravikumara, M.; Downs, J.; et al. Prevalence, clinical investigation, and management of gallbladder disease in Rett syndrome. Dev. Med. Child. Neurol. 2014, 56, 756–762. [Google Scholar] [CrossRef]
- Baikie, G.; Ravikumara, M.; Downs, J.; Naseem, N.; Wong, K.; Percy, A.; Lane, J.; Weiss, B.; Ellaway, C.; Bathgate, K.; et al. Gastrointestinal dysmotility in Rett syndrome. J. Pediatr. Gastroenterol. Nutr. 2014, 58, 237–244. [Google Scholar] [CrossRef]
- MacKay, J.; Leonard, H.; Wong, K.; Wilson, A.; Downs, J. Respiratory morbidity in Rett syndrome: An observational study. Dev. Med. Child Neurol. 2018, 60, 951–957. [Google Scholar] [CrossRef]
- Strati, F.; Cavalieri, D.; Albanese, D.; de Felice, C.; Donati, C.; Hayek, J.; Jousson, O.; Leoncini, S.; Pindo, M.; Renzi, D.; et al. Altered gut microbiota in Rett syndrome. Microbiome 2016, 4, 41. [Google Scholar] [CrossRef]
- Borghi, E.; Borgo, F.; Severgnini, M.; Savini, M.N.; Casiraghi, M.C.; Vignoli, A. Rett syndrome: A focus on gut microbiota. Int J. Mol. Sci. 2017, 18, 344. [Google Scholar] [CrossRef]
- Mosca, A.; Leclerc, M.; Hugot, J.P. Gut microbiota diversity and human diseases: Should we reintroduce key predators in our ecosystem? Front. Microbiol. 2016, 7, 455. [Google Scholar] [CrossRef]
- Turroni, F.; Ribbera, A.; Foroni, E.; van Sinderen, D.; Ventura, M. Human gut microbiota and bifidobacteria: From composition to functionality. Antonie. Van Leeuwenhoek 2008, 94, 35–50. [Google Scholar] [CrossRef]
- Riva, A.; Borgo, F.; Lassandro, C.; Verduci, E.; Morace, G.; Borghi, E.; Berry, D. Pediatric obesity is associated with an altered gut microbiota and discordant shifts in Firmicutes populations. Env. Microbiol. 2017, 9, 95–105. [Google Scholar] [CrossRef]
- Fernandes, J.; Su, W.; Rahat-Rozenbloom, S.; Wolever, T.M.; Comelli, E.M. Adiposity, gut microbiota and faecal short chain fatty acids are linked in adult humans. Nutr. Diabetes 2014, 4, e121. [Google Scholar] [CrossRef]
- Indiani, C.M.D.S.P.; Rizzardi, K.F.; Castelo, P.M.; Ferraz, L.F.C.; Darrieux, M.; Parisotto, T.M. Childhood Obesity and Firmicutes/Bacteroidetes Ratio in the Gut Microbiota: A Systematic Review. Child. Obes. 2018, 14, 501–509. [Google Scholar] [CrossRef]
- Nyangale, E.P.; Mottram, D.S.; Gibson, G.R. Gut microbial activity, implications for health and disease: The potential role of metabolite analysis. J. Proteome Res. 2012, 11, 5573–5585. [Google Scholar] [CrossRef]
- Tan, Q.; Zoghbi, H.Y. Mouse models as a tool for discovering new neurological diseases. Neurobiol. Learn. Mem. 2018. [Google Scholar] [CrossRef]
- Wither, R.G.; Colic, S.; Bardakjian, B.L.; Snead, O.C.; Zhang, L.; Eubanks, J.H. Electrographic and Pharmacological Characterization of a Progressive Epilepsy Phenotype in Female MeCP2-Deficient Mice. Epilepsy Res. 2018, 140, 177–183. [Google Scholar] [CrossRef]
- McLeod, F.; Ganley, R.; Williams, L.; Selfridge, J.; Bird, A.; Cobb, S.R. Reduced Seizure Threshold and Altered Network Oscillatory Properties in a Mouse Model of Rett Syndrome. Neuroscience 2013, 231, 195–205. [Google Scholar] [CrossRef]
- Millar-Büchner, P.; Philp, A.R.; Gutierrez, N.; Villanueva, S.; Kerr, B.; Flores, C.A. Severe changes in colon epithelium in the Mecp2-null mouse model of Rett syndrome. Mol. Cell Pediatr. 2016, 3, 37. [Google Scholar] [CrossRef]
- Tuchman, R.; Rapin, I. Epilepsy in autism. Lancet Neurol. 2002, 1, 352–358. [Google Scholar] [CrossRef]
- Nissenkorn, A.; Levy-Drummer, R.S.; Bondi, O.; Renieri, A.; Villard, L.; Mari, F.; Mencarelli, M.A.; Lo Rizzo, C.; Meloni, I.; Pineda, M.; et al. Epilepsy in Rett syndrome--lessons from the Rett networked database. Epilepsia 2015, 56, 569–576. [Google Scholar] [CrossRef]
- Vignoli, A.; Savini, M.N.; Nowbut, M.S.; Peron, A.; Turner, K.; La Briola, F.; Canevini, M.P. Effectiveness and tolerability of antiepileptic drugs in 104 girls with Rett syndrome. Epilepsy Behav. 2017, 66, 27–33. [Google Scholar] [CrossRef]
- Tarquinio, D.C.; Hou, W.; Berg, A.; Kaufmann, W.E.; Lane, J.B.; Skinner, S.A.; Motil, K.J.; Neul, J.L.; Percy, A.K.; Glaze, D.G. Longitudinal course of epilepsy in Rett syndrome and related disorders. Brain 2017, 140, 306–318. [Google Scholar] [CrossRef]
- He, Z.; Cui, B.T.; Zhang, T.; Li, P.; Long, C.Y.; Ji, G.Z.; Zhang, F.M. Fecal microbiota transplantation cured epilepsy in a case with Crohn’s disease: The first report. World J. Gastroenterol. 2017, 23, 3565–3568. [Google Scholar] [CrossRef]
- Peng, A.; Qiu, X.; Lai, W.; Li, W.; Zhang, L.; Zhu, X.; He, S.; Duan, J.; Chen, L. Altered composition of the gut microbiome in patients with drug-resistant epilepsy. Epilepsy Res. 2018, 147, 102–107. [Google Scholar] [CrossRef]
- Gómez-Eguílaz, M.; Ramón-Trapero, J.L.; Pérez-Martínez, L.; Blanco, J.R. The beneficial effect of probiotics as a supplementary treatment in drug-resistant epilepsy: A pilot study. Benef. Microbes. 2018, 9, 875–881. [Google Scholar] [CrossRef]
- Maier, L.; Pruteanu, M.; Kuhn, M.; Zeller, G.; Telzerow, A.; Anderson, E.E.; Brochado, A.R.; Fernandez, K.C.; Dose, H.; Mori, H.; et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 2018, 555, 623–628. [Google Scholar] [CrossRef]
- Tagliabue, A.; Ferraris, C.; Uggeri, F.; Trentani, C.; Bertoli, S.; de Giorgis, V.; Veggiotti, P.; Elli, M. Short-term impact of a classical ketogenic diet on gut microbiota in GLUT1 Deficiency Syndrome: A 3-month prospective observational study. Clin. Nutr. ESPEN 2017, 17, 33–37. [Google Scholar] [CrossRef]
- Chen, Y.R.; Zhou, L.Z.; Fang, S.T.; Long, H.Y.; Chen, J.Y.; Zhang, G.X. Isolation of Desulfovibrio spp. from human gut microbiota using a next-generation sequencing directed culture method. Lett. Appl. Microbiol. 2019, 68, 553–561. [Google Scholar] [CrossRef]
- Xie, G.; Zhou, Q.; Qiu, C.; Dai, W.; Wang, H.; Li, Y.; Liao, J.; Lu, X.; Lin, S.; Ye, J.; et al. Ketogenic diet poses a significant effect on imbalanced gut microbiota in infants with refractory epilepsy. World J. Gastroenterol. 2017, 23, 6164–6171. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, S.; Zhou, Y.; Yu, L.; Zhang, L.; Wang, Y. Altered gut microbiome composition in children with refractory epilepsy after ketogenic diet. Epilepsy Res. 2018, 145, 163–168. [Google Scholar] [CrossRef]
- Lindefeldt, M.; Eng, A.; Darban, H.; Bjerkner, A.; Zetterström, C.K.; Allander, T.; Andersson, B.; Borenstein, E.; Dahlin, M.; Prast-Nielsen, S. The ketogenic diet influences taxonomic and functional composition of the gut microbiota in children with severe epilepsy. NPJ Biofilms Microbiomes 2019, 5, 5. [Google Scholar] [CrossRef]
- Dolce, A.; Ben-Zeev, B.; Naidu, S.; Kossoff, E.H. Rett syndrome and epilepsy: An update for child neurologists. Pediatr. Neurol. 2013, 48, 337–435. [Google Scholar] [CrossRef]
- Lim, Z.; Wong, K.; Olson, H.E.; Bergin, A.M.; Downs, J.; Leonard, H. Use of the ketogenic diet to manage refractory epilepsy in CDKL5 disorder: Experience of >100 patients. Epilepsia 2017, 58, 1415–1422. [Google Scholar] [CrossRef]
- Wang, L.J.; Yang, C.Y.; Chou, W.J.; Lee, M.J.; Chou, M.C.; Kuo, H.C.; Yeh, Y.M.; Lee, S.Y.; Huang, L.H.; Li, S.C. Gut microbiota and dietary patterns in children with attention-deficit/hyperactivity disorder. Eur. Child Adolesc. Psychiatry 2019. [Epub ahead of print]. [Google Scholar] [CrossRef]
- Bäuerl, C.; Collado, M.C.; Diaz Cuevas, A.; Viña, J.; Pérez Martínez, G. Shifts in gut microbiota composition in an APP/PSS1 transgenic mouse model of Alzheimer’s disease during lifespan. Lett. Appl. Microbiol. 2018, 66, 464–471. [Google Scholar] [CrossRef]
- Coretti, L.; Cristiano, C.; Florio, E.; Scala, G.; Lama, A.; Keller, S.; Cuomo, M.; Russo, R.; Pero, R.; Paciello, O.; et al. Sex-related alterations of gut microbiota composition in the BTBR mouse model of autism spectrum disorder. Sci. Rep. 2017, 7, 45356. [Google Scholar] [CrossRef]
- Hiippala, K.; Kainulainen, V.; Kalliomäki, M.; Arkkila, P.; Satokari, R. Mucosal prevalence and interactions with the epithelium indicate commensalism of Sutterella spp. Front. Microbiol. 2016, 7, 1706. [Google Scholar] [CrossRef]
- Nowak, A.; Libudzisz, Z. Influence of phenol, p-cresol and indole on growth and survival of intestinal lactic acid bacteria. Anaerobe 2006, 12, 80–84. [Google Scholar] [CrossRef]
- Barrett, E.; Ross, R.P.; O’Toole, P.W.; Fitzgerald, G.F.; Stanton, C. γ-Aminobutyric acid production by culturable bacteria from the human intestine. J. Appl. Microbiol. 2012, 113, 411–417. [Google Scholar] [CrossRef]
- Pizzarelli, R.; Cherubini, E. Alterations of GABAergic signaling in autism spectrum disorders. Neural. Plast. 2011, 2011, 297153. [Google Scholar] [CrossRef]
- Sapey-Triomphe, L.A.; Lamberton, F.; Sonié, S.; Mattout, J.; Schmitz, C. Tactile hypersensitivity and GABA concentration in the sensorimotor cortex of adults with autism. Autism. Res. 2019, 12, 562–575. [Google Scholar] [CrossRef]
- Treiman, D.M. GABAergic mechanisms in epilepsy. Epilepsia 2001, 42, 8–12. [Google Scholar] [CrossRef]
- Wei, M.; Wang, Z.; Liu, H.; Jiang, H.; Wang, M.; Liang, S.; Shi, K.; Feng, J. Probiotic Bifidobacterium animalis subsp. lactis Bi-07 alleviates bacterial translocation and ameliorates microinflammation in experimental uraemia. Nephrology 2014, 19, 500–506. [Google Scholar] [CrossRef]
- Pintaudi, M.; Veneselli, E.; Voci, A.; Vignoli, A.; Castiglione, D.; Calevo, M.G.; Grasselli, E.; Ragazzoni, M.; Cogliati, F.; Calzari, L.; et al. Blood oxidative stress and metallothionein expression in Rett syndrome: Probing for markers. World J. Biol. Psychiatry 2016, 17, 198–209. [Google Scholar] [CrossRef]
- Vezzani, A.; Balosso, S.; Ravizza, T. Neuroinflammatory pathways as treatment targets and biomarkers in epilepsy. Nat. Rev. Neurol. 2019, 15, 459–472. [Google Scholar] [CrossRef]
- Medel-Matus, J.S.; Shin, D.; Dorfman, E.; Sankar, R.; Mazarati, A. Facilitation of kindling epileptogenesis by chronic stress may be mediated by intestinal microbiome. Epilepsia Open. 2018, 3, 290–294. [Google Scholar] [CrossRef]
- Tognini, P. Gut Microbiota: A Potential Regulator of Neurodevelopment. Front. Cell Neurosci 2017, 11, 25. [Google Scholar] [CrossRef]
- Hotamisligil, G.S. Inflammation and metabolic disorders. Nature 2006, 444, 860–867. [Google Scholar] [CrossRef]
- Kaakoush, N.O. Insights into the Role of Erysipelotrichaceae in the Human Host. Front. Cell Infect. Microbiol. 2015, 5, 84. [Google Scholar] [CrossRef]
- Rios-Covian, D.; Salazar, N.; Gueimonde, M.; de Los Reyes-Gavilan, C.G. Shaping the Metabolism of Intestinal Bacteroides Population through Diet to Improve Human Health. Front. Microbiol. 2017, 8, 376. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).