Transcriptome Profiling of Placenta through Pregnancy Reveals Dysregulation of Bile Acids Transport and Detoxification Function
Abstract
1. Introduction
2. Results
2.1. RNA Sequencing and Identification of mRNAs in All Porcine Placentas
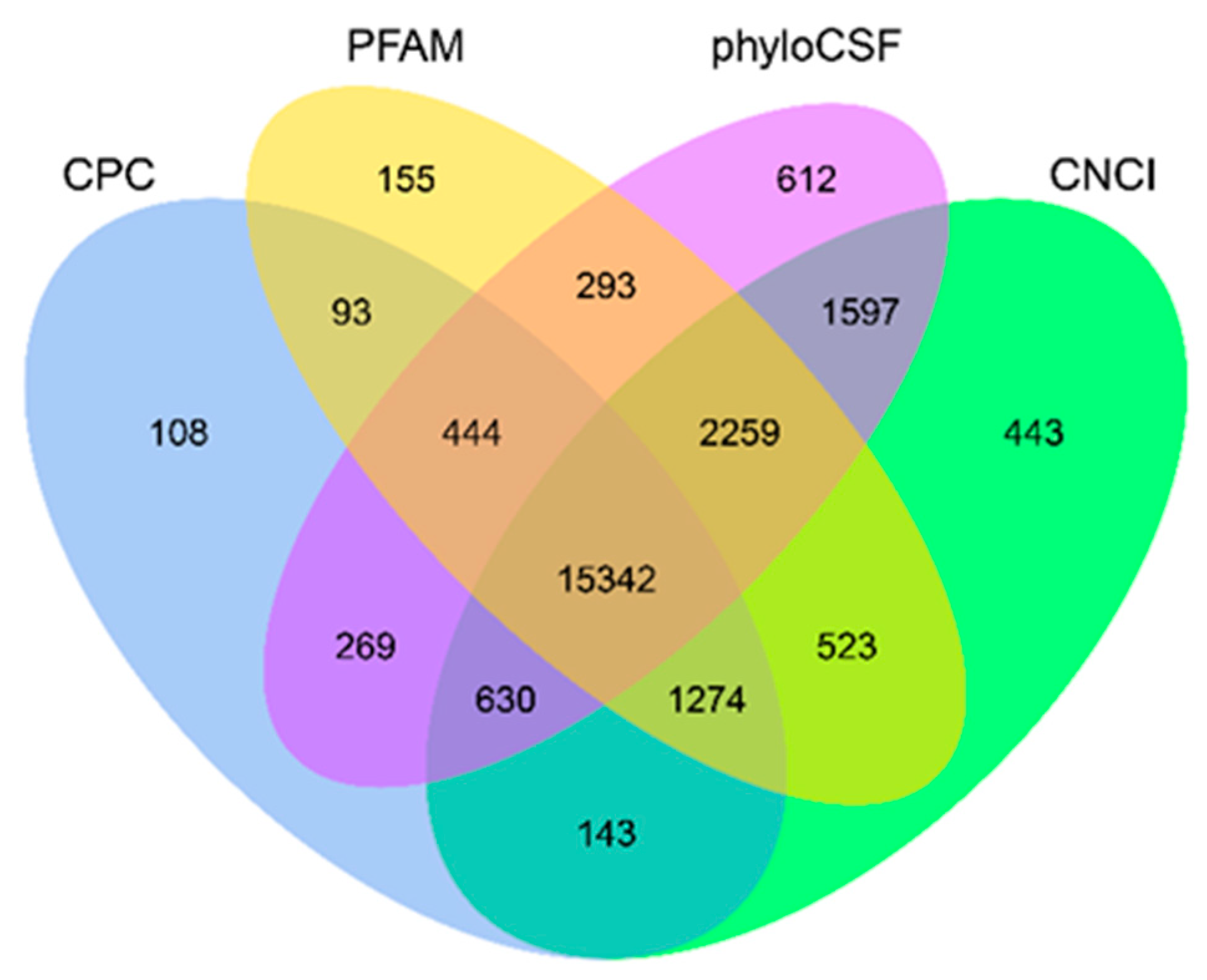
2.2. RNA Sequencing and Identification of Long Non-Coding RNAs (lncRNAs) in All Porcine Placentas
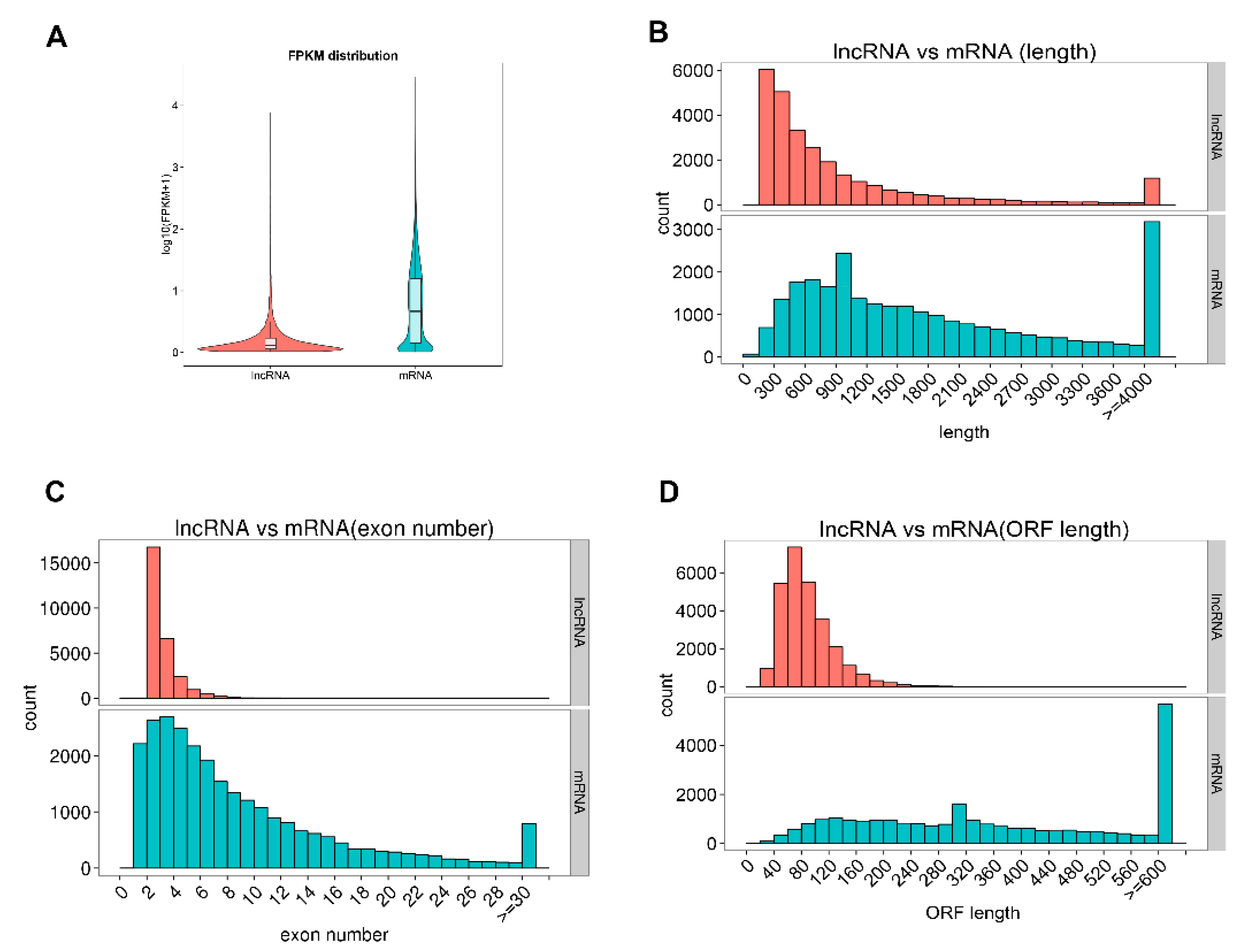
2.3. Comparison of Features of mRNAs and Long Non-Coding RNAs (lncRNAs)
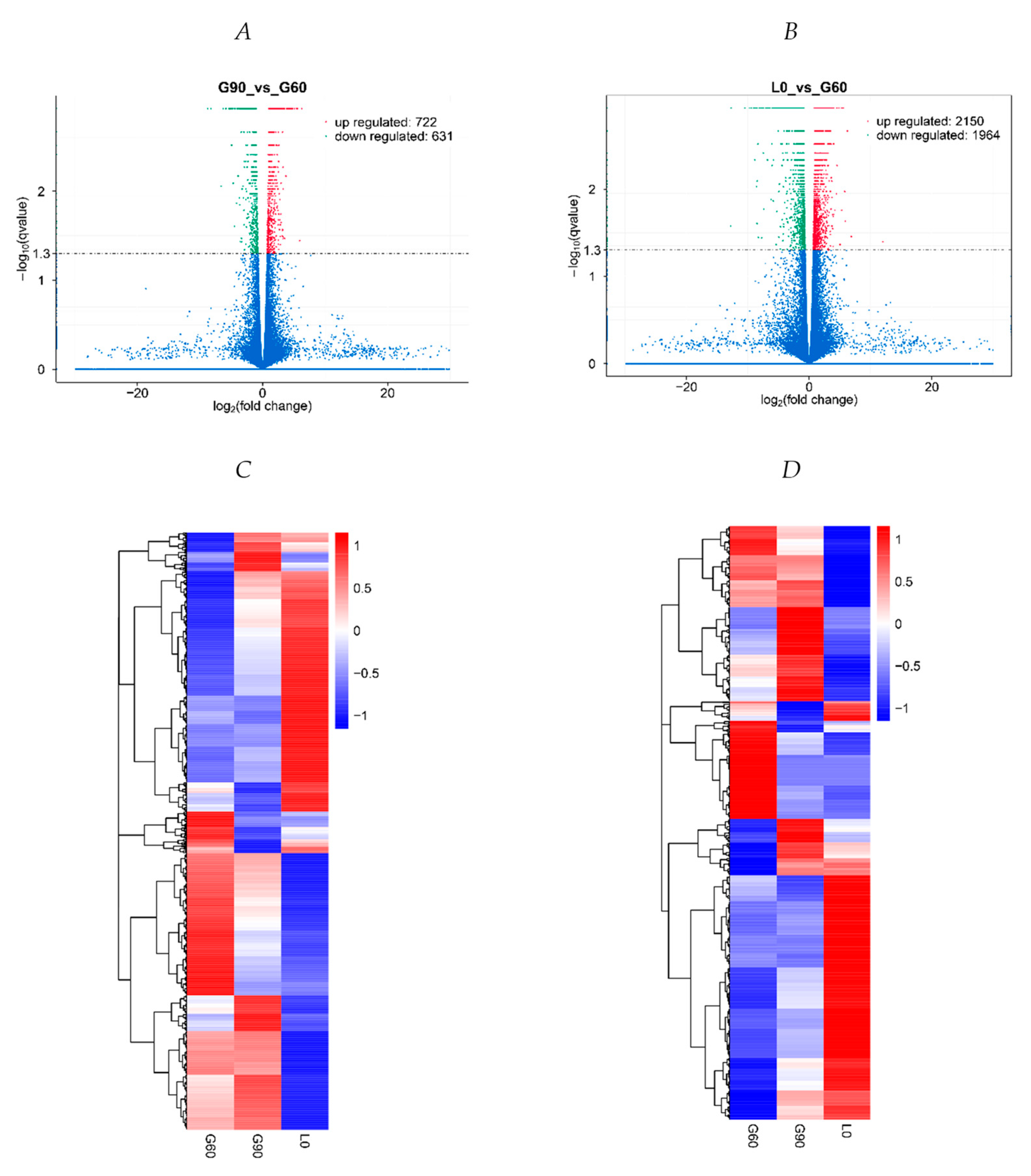
2.4. Placentas at Different Time of Pregnancy Have Distinct Gene Expression Profiles and Cluster Separately
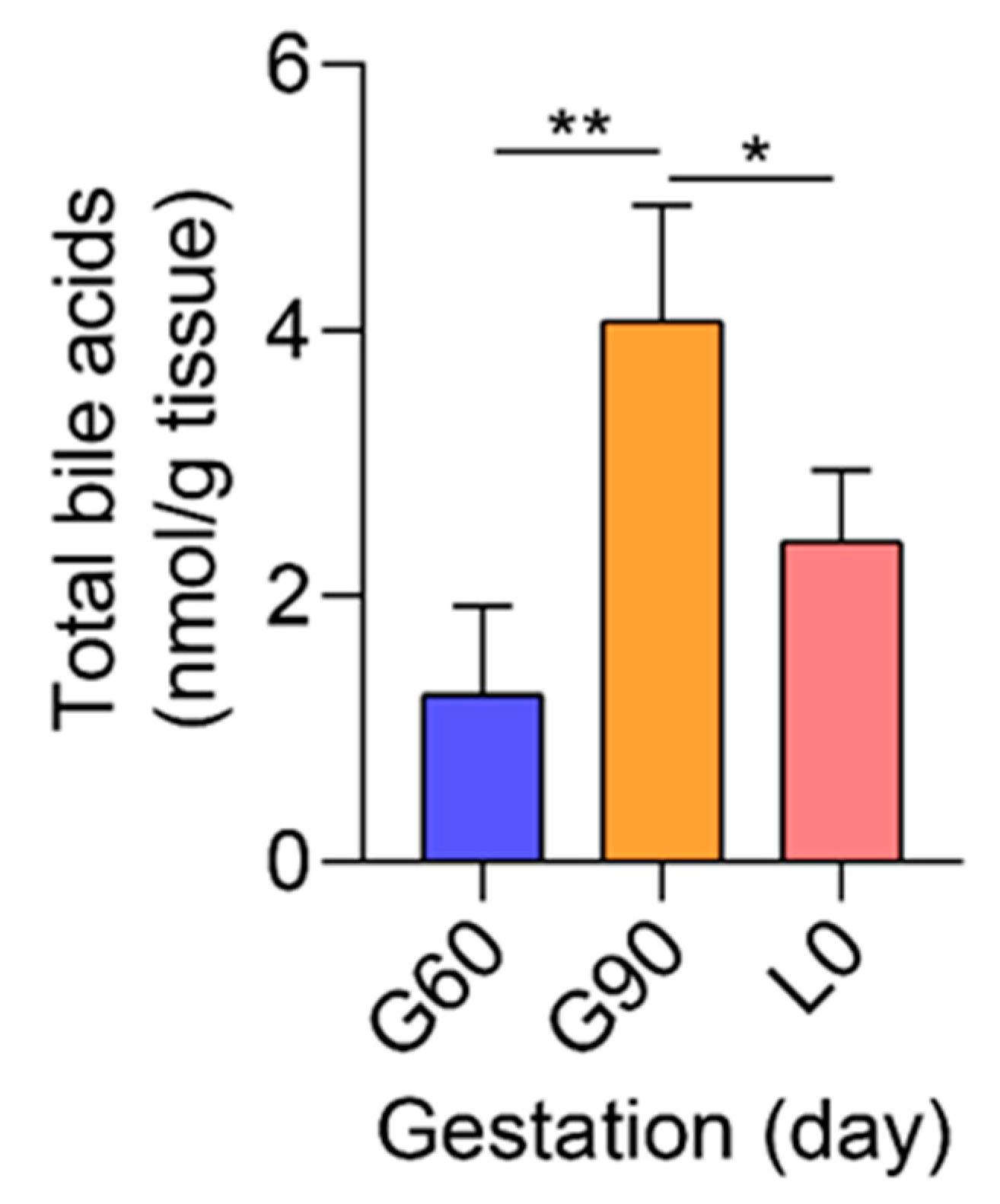
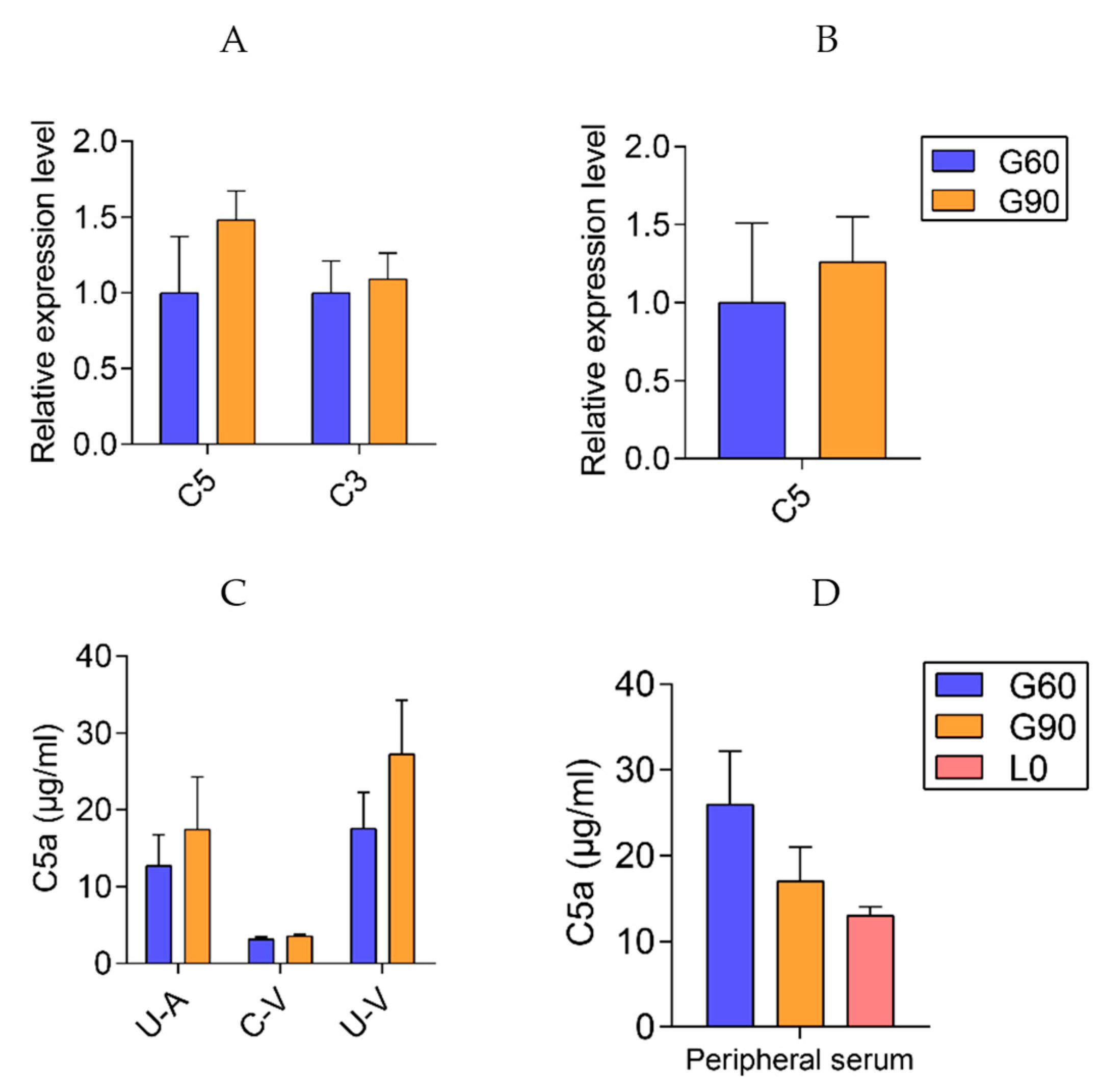
2.5. Dysfunction of Bile Acid Transport and Detoxification Are the Central Characteristics with the Advance of Pregnancy
2.6. Prediction of Long Non-Coded RNAs (LncRNAs) Function
3. Discussion
4. Materials and Methods
4.1. Experimental Design and Sampling
4.2. Total Bile Acids
4.3. RNA Sequencing and Data Analysis of the Porcine Placentas
4.4. Real-Time RT-PCR
4.5. Measurement of C5a
4.6. Data Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ICP | Intrahepatic cholestasis of pregnancy |
| RNA-Seq | RNA sequencing |
| BA | Bile acid |
| LncRNA | Long non-coding RNA |
| ABCB11/BSEP | Bile salt export pump |
| OATP1A2/SLCO1A2 | Organic anion-transporting polypeptide 1A2 |
| TPMa | Apical plasma membrane of the trophoblast |
| TPMb | Basal plasma membrane of the trophoblast |
| NBC1/SLC4A4 | Na+-HCO3− cotransporter |
| SULT2A1 | Hydroxysteroid sulfotransferases |
| CA2 | Carbonate anhydrase 2 |
| FXR/NR1H4 | Farnesoid X receptor |
References
- Battaglia, F.; Meschia, G. Principal substrates of fetal metabolism. Physiol. Rev. 1978, 58, 499–527. [Google Scholar] [CrossRef] [PubMed]
- Carter, A.M. Evolution of placental function in mammals: The molecular basis of gas and nutrient transfer, hormone secretion, and immune responses. Physiol. Rev. 2012, 92, 1543–1576. [Google Scholar] [CrossRef] [PubMed]
- Sood, R.; Zehnder, J.L.; Druzin, M.L.; Brown, P.O. Gene expression patterns in human placenta. Proc. Natl. Acad. Sci. USA 2006, 103, 5478–5483. [Google Scholar] [CrossRef] [PubMed]
- Marin, J.J.; Macias, R.I.; Briz, O.; Perez, M.J.; Blazquez, A.G.; Arrese, M.; Serrano, M.A. Molecular bases of the fetal liver-placenta-maternal liver excretory pathway for cholephilic compounds. Liver Int. 2008, 28, 435–454. [Google Scholar] [CrossRef] [PubMed]
- Barth, A.; Fleck, C.; Klinger, W. Development of organic anion transport in the liver. Exp. Toxicol. Pathol. 1996, 48, 421–432. [Google Scholar] [CrossRef]
- Setchell, K.D.; Dumaswala, R.; Colombo, C.; Ronchi, M. Hepatic bile acid metabolism during early development revealed from the analysis of human fetal gallbladder bile. J. Biol. Chem. 1988, 263, 16637–16644. [Google Scholar]
- Colombo, C.; Zuliani, G.; Ronchi, M.; Breidenstein, J.; Setchell, K.D. Biliary bile acid composition of the human fetus in early gestation. Pediatr. Res. 1987, 21, 197–200. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zhong, H.; Song, Y.; Yuan, P.; Li, Y.; Lin, S.; Zhang, X.; Li, J.; Che, L.; Feng, B.; et al. Targeted metabolomics analysis of maternal-placental-fetal metabolism in pregnant swine reveals links in fetal bile acid homeostasis and sulfation capacity. Am. J. Physiol. Gastrointest. Liver Physiol. 2019, 317, 8–16. [Google Scholar] [CrossRef]
- Macias, R.I.; Marin, J.J.; Serrano, M.A. Excretion of biliary compounds during intrauterine life. World J. Gastroenterol. 2009, 15, 817–828. [Google Scholar] [CrossRef]
- Glantz, A.; Marschall, H.U.; Mattsson, L.A. Intrahepatic cholestasis of pregnancy: Relationships between bile acid levels and fetal complication rates. Hepatology 2004, 40, 467–474. [Google Scholar] [CrossRef]
- Geenes, V.; Williamson, C. Intrahepatic cholestasis of pregnancy. World J. Gastroenterol. 2009, 15, 2049–2066. [Google Scholar] [CrossRef] [PubMed]
- Macias, R.I.; Pascual, M.J.; Bravo, A.; Alcalde, M.P.; Larena, M.G.; St-Pierre, M.V.; Serrano, M.A.; Marin, J.J. Effect of maternal cholestasis on bile acid transfer across the rat placenta-maternal liver tandem. Hepatology 2000, 31, 975–983. [Google Scholar] [CrossRef] [PubMed]
- Gude, N.M.; Roberts, C.T.; Kalionis, B.; King, R.G. Growth and function of the normal human placenta. Thromb. Res. 2004, 114, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Pataia, V.; Dixon, P.H.; Williamson, C. Pregnancy and bile acid disorders. Am. J. Physiol. Gastrointest. Liver Physiol. 2017, 313, 1–6. [Google Scholar] [CrossRef]
- McIlvride, S.; Dixon, P.H.; Williamson, C. Bile acids and gestation. Mol. Asp. Med. 2017, 56, 90–100. [Google Scholar] [CrossRef]
- Tribe, R.M.; Dann, A.T.; Kenyon, A.P.; Seed, P.; Shennan, A.H.; Mallet, A. Longitudinal profiles of 15 serum bile acids in patients with intrahepatic cholestasis of pregnancy. Am. J. Gastroenterol. 2010, 105, 585–595. [Google Scholar] [CrossRef]
- Beuers, U.; Boberg, K.M.; Chapman, R.W.; Chazouilleres, O.; Invernizzi, P.; Jones, D.E.J.; Lammert, F.; Pares, A.; Trauner, M.; European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Management of cholestatic liver diseases. J. Hepatol. 2009, 51, 237–267. [Google Scholar]
- Serrano, M.A.; Brites, D.; Larena, M.G.; Monte, M.J.; Bravo, M.P.; Oliveira, N.; Marin, J.J. Beneficial effect of ursodeoxycholic acid on alterations induced by cholestasis of pregnancy in bile acid transport across the human placenta. J. Hepatol. 1998, 28, 829–839. [Google Scholar] [CrossRef]
- Perez, M.J.; Macias, R.I.; Marin, J.J. Maternal cholestasis induces placental oxidative stress and apoptosis. Protective effect of ursodeoxycholic acid. Placenta 2006, 27, 34–41. [Google Scholar] [CrossRef]
- Majewska, M.; Lipka, A.; Paukszto, L.; Jastrzebski, J.P.; Gowkielewicz, M.; Jozwik, M.; Majewski, M.K. Preliminary RNA-Seq Analysis of Long Non-Coding RNAs Expressed in Human Term Placenta. Int. J. Mol. Sci. 2018, 19, 1894. [Google Scholar] [CrossRef]
- Ackerman, W.; Buhimschi, I.A.; Summerfield, T.; Zhao, G.; Landon, M.B.; Buhimschi, C.S. Global transcriptomic analysis of human placenta in the setting of intrauterine growth restriction (IUGR) using RNA sequencing (RNA-Seq). Am. J. Obstet. Gynecol. 2017, 216, 140. [Google Scholar] [CrossRef]
- Saben, J.; Kang, P.; Zhong, Y.; Thakali, K.M.; Gomez-Acevedo, H.; Borengasser, S.J.; Andres, A.; Badger, T.M.; Shankar, K. RNA-seq analysis of the rat placentation site reveals maternal obesity-associated changes in placental and offspring thyroid hormone signaling. Placenta 2014, 35, 1013–1020. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ananthanarayanan, M. A novel long noncoding RNA regulating cholesterol and bile acid homeostasis: A new kid on the block and a potential therapeutic target? Hepatology 2016, 64, 16–18. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Ruan, X.; Yang, L.; Kiesewetter, K.; Zhao, Y.; Luo, H.; Chen, Y.; Gucek, M.; Zhu, J.; Cao, H. A liver-enriched long non-coding RNA, lncLSTR, regulates systemic lipid metabolism in mice. Cell Metab. 2015, 21, 455–467. [Google Scholar] [CrossRef] [PubMed]
- Machtinger, R.; Zhong, J.; Mansur, A.; Adir, M.; Racowsky, C.; Hauser, R.; Brennan, K.; Karlsson, O.; Baccarelli, A.A. Placental lncRNA Expression Is Associated With Prenatal Phthalate Exposure. Toxicol. Sci. 2018, 163, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Strautnieks, S.S.; Bull, L.N.; Knisely, A.S.; Kocoshis, S.; Dahl, N.; Arnell, H.; Sokal, E.M.; Dahan, K.; Childs, S.; Ling, V.; et al. A gene encoding a liver-specific ABC transporter is mutated in progressive familial intrahepatic cholestasis. Nat. Genet. 1998, 20, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Gerloff, T.; Stieger, B.; Hagenbuch, B.; Madon, J.; Landmann, L.; Roth, J.; Hofmann, A.F.; Meier, P.J. The sister of P-glycoprotein represents the canalicular bile salt export pump of mammalian liver. J. Biol. Chem. 1998, 273, 10046–10050. [Google Scholar] [CrossRef]
- Kullakublick, G.A.; Beuers, U.; Fahney, C.; Hagenbuch, B.; Meier, P.; Paumgartner, G. Identification and functional characterization of the promoter region of the human organic anion transporting polypeptide gene. Hepatology 1997, 26, 991–997. [Google Scholar] [CrossRef]
- Halilbasic, E.; Claudel, T.; Trauner, M. Bile acid transporters and regulatory nuclear receptors in the liver and beyond. J. Hepatol. 2013, 58, 155–168. [Google Scholar] [CrossRef]
- Leuthold, S.; Hagenbuch, B.; Mohebbi, N.; Wagner, C.A.; Meier, P.J.; Stieger, B. Mechanisms of pH-gradient driven transport mediated by organic anion polypeptide transporters. Am. J. Physiol. Cell Physiol. 2008, 296, 570–582. [Google Scholar] [CrossRef]
- Supuran, C.T. Carbonic anhydrases: Novel therapeutic applications for inhibitors and activators. Nat. Rev. Drug Discov. 2008, 7, 168–181. [Google Scholar] [CrossRef] [PubMed]
- Jacob, P.; Christiani, S.; Rossmann, H.; Lamprecht, G.; Vieillardbaron, D.; Muller, R.; Gregor, M.; Seidler, U. Role of Na(+)HCO(3)(-) cotransporter NBC1, Na(+)/H(+) exchanger NHE1, and carbonic anhydrase in rabbit duodenal bicarbonate secretion. Gastroenterology 2000, 119, 406–419. [Google Scholar] [CrossRef] [PubMed]
- Longo, L.D.; Delivoriapapadopoulos, M.; Nd, F.R. Placental CO2 transfer after fetal carbonic anhydrase inhibition. Am. J. Physiol. 1974, 226, 703. [Google Scholar] [CrossRef] [PubMed]
- Milov, D.E.; Jou, W.; Shireman, R.B.; Chun, P.W. The effect of bile salts on carbonic anhydrase. Hepatology 1992, 15, 288–296. [Google Scholar] [CrossRef] [PubMed]
- Alnouti, Y. Bile acid sulfation: A pathway of bile acid elimination and detoxification. Toxicol. Sci. 2009, 108, 225–246. [Google Scholar] [CrossRef] [PubMed]
- Henegouwen, G.P.V.B.; Brandt, K.H.; Eyssen, H.; Parmentier, G. Sulphated and unsulphated bile acids in serum, bile, and urine of patients with cholestasis. Gut 1976, 17, 861–869. [Google Scholar] [CrossRef] [PubMed]
- Radominska, A.; Comer, K.A.; Zimniak, P.; Falany, J.; Iscan, M.; Falany, C.N. Human liver steroid sulphotransferase sulphates bile acids. Biochem. J. 1990, 272, 597–604. [Google Scholar] [CrossRef]
- Huang, J.; Bathena, S.; Tong, J.; Roth, M.; Hagenbuch, B.; Alnouti, Y. Kinetic analysis of bile acid sulfation by stably expressed human sulfotransferase 2A1 (SULT2A1). Xenobiotica 2010, 40, 184–194. [Google Scholar] [CrossRef]
- Makishima, M.; Okamoto, A.Y.; Repa, J.J.; Tu, H.; Learned, R.M.; Luk, A.; Hull, M.V.; Lustig, K.D.; Mangelsdorf, D.J.; Shan, B. Identification of a nuclear receptor for bile acids. Science 1999, 284, 1362–1365. [Google Scholar] [CrossRef]
- Kitada, H.; Miyata, M.; Nakamura, T.; Tozawa, A.; Honma, W.; Shimada, M.; Nagata, K.; Sinal, C.J.; Guo, G.L.; Gonzalez, F.J. Protective role of hydroxysteroid sulfotransferase in lithocholic acid-induced liver toxicity. J. Biol. Chem. 2003, 278, 17838–17844. [Google Scholar] [CrossRef]
- Wu, W.B.; Xu, Y.Y.; Cheng, W.W.; Wang, Y.X.; Liu, Y.; Huang, D.; Zhang, H.J. Agonist of farnesoid X receptor protects against bile acid induced damage and oxidative stress in mouse placenta - A study on maternal cholestasis model. Placenta 2015, 36, 545–551. [Google Scholar] [CrossRef] [PubMed]
- Pickering, M.C.; Cook, H.T.; Warren, J.; Bygrave, A.E.; Moss, J.; Walport, M.J.; Botto, M. Uncontrolled C3 activation causes membranoproliferative glomerulonephritis in mice deficient in complement factor H. Nat. Genet. 2002, 31, 424–428. [Google Scholar] [CrossRef] [PubMed]
- Sakaue, T.; Takeuchi, K.; Maeda, T.; Yamamoto, Y.; Nishi, K.; Ohkubo, I. Factor H in porcine seminal plasma protects sperm against complement attack in genital tracts. J. Biol. Chem. 2010, 285, 2184–2192. [Google Scholar] [CrossRef] [PubMed]
- Tedesco, F.; Narchi, G.; Radillo, O.; Meri, S.; Ferrone, S.; Betterle, C. Susceptibility of human trophoblast to killing by human complement and the role of the complement regulatory proteins. J. Immunol. 1993, 151, 1562–1570. [Google Scholar] [PubMed]
- Huberlang, M.; Sarma, J.V.; Zetoune, F.S.; Rittirsch, D.; Neff, T.A.; Mcguire, S.R.; Lambris, J.D.; Warner, R.L.; Flierl, M.A.; Hoesel, L.M. Generation of C5a in the absence of C3: A new complement activation pathway. Nat. Med. 2006, 12, 682–687. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Jeong, W.I.; Tian, Z. Liver: An organ with predominant innate immunity. Hepatology 2008, 47, 729–736. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef] [PubMed]
- Fatica, A.; Bozzoni, I. Long non-coding RNAs: New players in cell differentiation and development. Nat. Rev. Genet. 2014, 15, 7–21. [Google Scholar] [CrossRef]
- Doyle, W.I.; Dinser, J.A.; Cansler, H.L.; Zhang, X.; Dinh, D.D.; Browder, N.S.; Riddington, I.M.; Meeks, J.P. Faecal bile acids are natural ligands of the mouse accessory olfactory system. Nat. Commun. 2016, 7, 11936. [Google Scholar] [CrossRef]
- Ovadia, C.; Perdones-Montero, A.; Spagou, K.; Smith, A.; Sarafian, M.H.; Gomez Romero, M.; Bellafante, E.; Clarke, L.C.; Sadiq, F.; Nikolova, V.; et al. Enhanced microbial bile acid deconjugation and impaired ileal uptake in pregnancy repress intestinal regulation of bile acid synthesis. Hepatology 2019, 7, 276–293. [Google Scholar] [CrossRef]
- Milona, A.; Owen, B.M.; Cobbold, J.F.L.; Willemsen, E.C.L.; Cox, I.J.; Boudjelal, M.; Cairns, W.; Schoonjans, K.; Taylor-Robinson, S.D.; Klomp, L.W.J.; et al. Raised Hepatic Bile Acid Concentrations During Pregnancy in Mice Are Associated with Reduced Farnesoid X Receptor Function. Hepatology 2010, 52, 1341–1349. [Google Scholar] [CrossRef] [PubMed]
- Pascual, M.J.; Serrano, M.A.; Elmir, M.Y.; Macias, R.I.R.; Jimenez, F.; Marin, J.J.G. Relationship between asymptomatic hypercholanaemia of pregnancy and progesterone metabolism. Clin. Sci. 2002, 102, 587–593. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, F.; Wang, Y.; Pitre, A.; Fang, Z.-Z.; Frank, M.W.; Calabrese, C.; Krausz, K.W.; Neale, G.; Frase, S.; et al. Maternal bile acid transporter deficiency promotes neonatal demise. Nat. Commun. 2015, 6, 8186. [Google Scholar] [CrossRef] [PubMed]
- Estiu, M.C.; Monte, M.J.; Rivas, L.; Moiron, M.; Gomez-Rodriguez, L.; Rodriguez-Bravo, T.; Marin, J.J.G.; Macias, R.I.R. Effect of ursodeoxycholic acid treatment on the altered progesterone and bile acid homeostasis in the mother-placenta-foetus trio during cholestasis of pregnancy. Br. J. Clin. Pharmacol. 2015, 79, 316–329. [Google Scholar] [CrossRef] [PubMed]
- Marin, J.J.; Macias, R.I.; Serrano, M.A. The hepatobiliary-like excretory function of the placenta. A review. Placenta 2003, 24, 431–438. [Google Scholar] [CrossRef] [PubMed]
- Serrano, M.A.; Macias, R.I.R.; Briz, O.; Monte, M.J.; Blazquez, A.G.; Williamson, C.; Kubitz, R.; Marin, J.J.G. Expression in human trophoblast and choriocarcinoma cell lines, BeWo, Jeg-3 and JAr of genes involved in the hepatobiliary-like excretory function of the placenta. Placenta 2007, 28, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Marin, J.J.; Bravo, P.; el-Mir, M.Y.; Serrano, M.A. ATP-dependent bile acid transport across microvillous membrane of human term trophoblast. Am. J. Physiol. 1995, 268, 685–694. [Google Scholar] [CrossRef]
- Bravo, P.; Marin, J.J.; Beveridge, M.J.; Novak, D.A. Reconstitution and characterization of ATP-dependent bile acid transport in human and rat placenta. Biochem. J. 1995, 311, 479–485. [Google Scholar] [CrossRef]
- Shoda, J.; Miura, T.; Utsunomiya, H.; Oda, K.; Yamamoto, M.; Kano, M.; Ikegami, T.; Tanaka, N.; Akita, H.; Ito, K. Genipin enhances Mrp2 (Abcc2)-mediated bile formation and organic anion transport in rat liver. Hepatology 2004, 39, 167–178. [Google Scholar] [CrossRef]
- Cui, Y.; Konig, J.; Buchholz, J.K.; Spring, H.; Leier, I.; Keppler, D. Drug resistance and ATP-dependent conjugate transport mediated by the apical multidrug resistance protein, MRP2, permanently expressed in human and canine cells. Mol. Pharmacol. 1999, 55, 929–937. [Google Scholar]
- Hirohashi, T.; Suzuki, H.; Takikawa, H.; Sugiyama, Y. ATP-dependent transport of bile salts by rat multidrug resistance-associated protein 3 (Mrp3). J. Biol. Chem. 2000, 275, 2905–2910. [Google Scholar] [CrossRef] [PubMed]
- Elmir, M.Y.; Eleno, N.; Serrano, M.A.; Bravo, P.; Marin, J.J. Bicarbonate-induced activation of taurocholate transport across the basal plasma membrane of human term trophoblast. Am. J. Physiol. 1991, 260, 887–894. [Google Scholar]
- Marin, J.J.; Serrano, M.A.; el-Mir, M.Y.; Eleno, N.; Boyd, C.A. Bile acid transport by basal membrane vesicles of human term placental trophoblast. Gastroenterology 1990, 99, 1431–1438. [Google Scholar] [CrossRef]
- Yu, Y.; Liu, X.; Zhang, Z.; Xiao, Y.; Hong, M. Cloning and functional characterization of the pig (Sus scrofa) organic anion transporting polypeptide 1a2. Xenobiotica 2013, 43, 738–744. [Google Scholar] [CrossRef] [PubMed]
- Salomoni, M.; Zuccato, E.; Granelli, P.; Montorsi, W.; Doldi, S.B.; Germiniani, R.; Mussini, E. Effect of bile salts on carbonic anhydrase from rat and human gastric mucosa. Scand. J. Gastroenterol. 1989, 24, 28–32. [Google Scholar] [CrossRef] [PubMed]
- Boone, C.D.; Tu, C.; Mckenna, R. Structural elucidation of the hormonal inhibition mechanism of the bile acid cholate on human carbonic anhydrase II. Acta Crystallogr. Sect. D 2014, 70, 1758–1763. [Google Scholar] [CrossRef] [PubMed]
- Trifunovic, J.; Borcic, V.; Mikov, M. Bile acids and their oxo derivatives: Potential inhibitors of carbonic anhydrase I and II, androgen receptor antagonists and CYP3A4 substrates. Biomed. Chromatogr. 2017, 31, e3870. [Google Scholar] [CrossRef]
- Longo, L.D. Respiratory Gas Exchange in the Placenta. In Handbook of Physiology. The Respiratory System, Gas Exchange; American Physiological Society: Bethesda, MD, USA, 1987; pp. 351–401. [Google Scholar]
- Lan, X.; Yan, J.; Ren, J.; Zhong, B.; Li, J.; Li, Y.; Liu, L.; Yi, J.; Sun, Q.; Yang, X. A novel long noncoding RNA Lnc-HC binds hnRNPA2B1 to regulate expressions of Cyp7a1 and Abca1 in hepatocytic cholesterol metabolism. Hepatology 2016, 64, 58–72. [Google Scholar] [CrossRef]
- Zhang, L.; Yang, Z.; Trottier, J.; Barbier, O.; Wang, L. Long noncoding RNA MEG3 induces cholestatic liver injury by interaction with PTBP1 to facilitate shp mRNA decay. Hepatology 2017, 65, 604–615. [Google Scholar] [CrossRef]
- Zhang, Y.; Pan, Y.; Lin, C.; Zheng, Y.; Sun, H.; Zhang, H.; Wang, J.; Yuan, M.; Duan, T.; Du, Q.; et al. Bile acids evoke placental inflammation by activating Gpbar1/NF-kappaB pathway in intrahepatic cholestasis of pregnancy. J. Mol. Cell Biol. 2016, 8, 530–541. [Google Scholar] [CrossRef]
- Du, Q.; Zhang, Y.; Pan, Y.; Duan, T. Lithocholic acid-induced placental tumor necrosis factor-α upregulation and syncytiotrophoblast cell apoptosis in intrahepatic cholestasis of pregnancy. Hepatol. Res. 2014, 44, 532–541. [Google Scholar] [CrossRef] [PubMed]
- Che, L.; Yang, Z.G.; Xu, M.M.; Xu, S.Y.; Che, L.Q.; Lin, Y.; Fang, Z.F.; Feng, B.; Li, J.; Chen, D.W.; et al. Maternal nutrition modulates fetal development by inducing placental efficiency changes in gilts. BMC Genom. 2017, 18, 14. [Google Scholar] [CrossRef] [PubMed]
- LARusso, N.F.; Korman, M.G.; Hoffman, N.E.; Hofmann, A.F. Dynamics of the enterohepatic circulation of bile acids: Postprandial serum concentrations of conjugates of cholic acid in health, cholecystectomized patients, and patients with bile acid malabsorption. N. Engl. J. Med. 1974, 291, 689–692. [Google Scholar] [CrossRef] [PubMed]
- Sonne, D.P.; Van Nierop, F.S.; Kulik, W.; Soeters, M.R.; Vilsboll, T.; Knop, F.K. Postprandial Plasma Concentrations of Individual Bile Acids and FGF-19 in Patients With Type 2 Diabetes. J. Clin. Endocrinol. Metab. 2016, 101, 3002–3009. [Google Scholar] [CrossRef] [PubMed]
- Takken, A.; Williams, K.C. A simplified procedure for long-term catheterisation of the anterior vena cava in adult pigs. Aust. Vet. J. 1981, 57, 17–20. [Google Scholar] [CrossRef]
- Guttman, M.; Garber, M.; Levin, J.Z.; Donaghey, J.; Robinson, J.T.; Adiconis, X.; Fan, L.; Koziol, M.J.; Gnirke, A.; Nusbaum, C. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat. Biotechnol. 2010, 28, 503–510. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Vandesompele, J.; de, P.K.; Pattyn, F.; Poppe, B.; van, R.N.; de, P.A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 1. [Google Scholar] [CrossRef]
- Zhuo, Y.; Hua, L.; Feng, B.; Jiang, X.M.; Li, J.; Jiang, D.D.; Huang, X.H.; Zhu, Y.G.; Li, Z.; Yan, L.J.; et al. Fibroblast growth factor 21 coordinates adiponectin to mediate the beneficial effects of low-protein diet on primordial follicle reserve. Ebiomedicine 2019, 41, 623–635. [Google Scholar] [CrossRef]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, P.; Song, Y.; Zhong, H.; Lin, S.; Zhang, X.; Li, J.; Che, L.; Feng, B.; Lin, Y.; Xu, S.; et al. Transcriptome Profiling of Placenta through Pregnancy Reveals Dysregulation of Bile Acids Transport and Detoxification Function. Int. J. Mol. Sci. 2019, 20, 4099. https://doi.org/10.3390/ijms20174099
Wang P, Song Y, Zhong H, Lin S, Zhang X, Li J, Che L, Feng B, Lin Y, Xu S, et al. Transcriptome Profiling of Placenta through Pregnancy Reveals Dysregulation of Bile Acids Transport and Detoxification Function. International Journal of Molecular Sciences. 2019; 20(17):4099. https://doi.org/10.3390/ijms20174099
Chicago/Turabian StyleWang, Peng, Yumo Song, Heju Zhong, Sen Lin, Xiaoling Zhang, Jian Li, Lianqiang Che, Bin Feng, Yan Lin, Shengyu Xu, and et al. 2019. "Transcriptome Profiling of Placenta through Pregnancy Reveals Dysregulation of Bile Acids Transport and Detoxification Function" International Journal of Molecular Sciences 20, no. 17: 4099. https://doi.org/10.3390/ijms20174099
APA StyleWang, P., Song, Y., Zhong, H., Lin, S., Zhang, X., Li, J., Che, L., Feng, B., Lin, Y., Xu, S., Zhuo, Y., Wu, D., Burrin, D. G., & Fang, Z. (2019). Transcriptome Profiling of Placenta through Pregnancy Reveals Dysregulation of Bile Acids Transport and Detoxification Function. International Journal of Molecular Sciences, 20(17), 4099. https://doi.org/10.3390/ijms20174099






