Bioactivity and Bactericidal Mechanism of Histidine-Rich β-Hairpin Peptide Against Gram-Negative Bacteria
Abstract
1. Introduction
2. Results
2.1. Design and Characterization of the Peptides
2.2. CD Analysis
2.3. Hemolytic Activity
2.4. Cytotoxicity
2.5. Antimicrobial Activity
2.6. Salt Sensitivity
2.7. Antibacterial Mechanism Study
2.7.1. LPS Binding Assay
2.7.2. Outer Membrane Permeability Assay
2.7.3. Inner Membrane Permeability
2.7.4. Membrane Depolarization
2.7.5. Swimming Motility Analysis
2.7.6. SEM
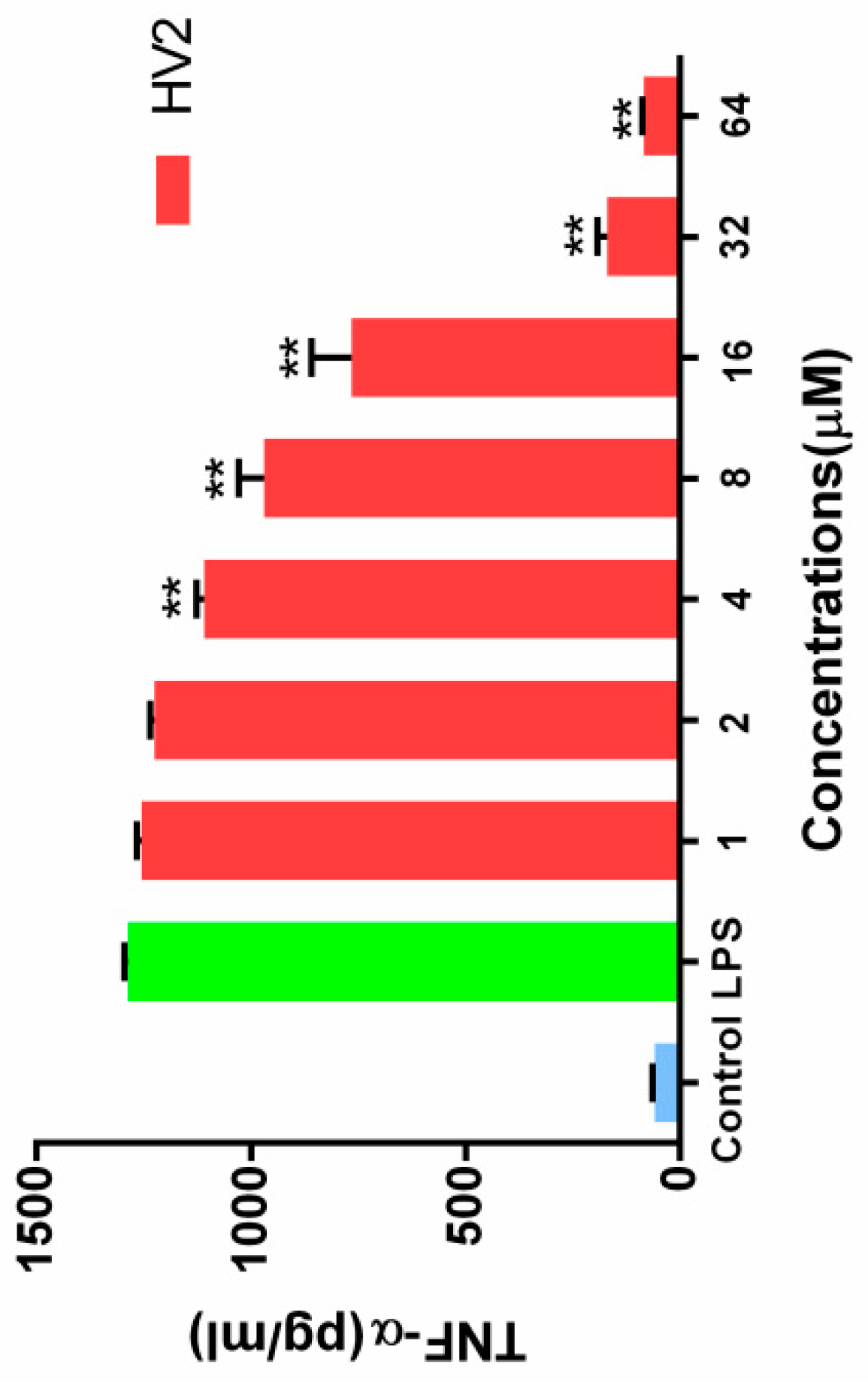
2.7.7. Inhibition of Endotoxin-Induced Inflammation
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Peptide Synthesis and Sequence Analysis
4.3. Circular Dichroism (CD) Measurements
4.4. Measurement of Hemolytic Activity
4.5. Measurement of Cytotoxicity
4.6. Minimum Inhibitory Concentration (MIC) Measurements
4.7. Salt Sensitivity Assays
4.8. Antimicrobial Mechanism Assays
4.8.1. LPS Binding Assay
4.8.2. Outer Membrane Permeability Assay
4.8.3. Inner Membrane Permeability Assay
4.8.4. Membrane Depolarization
4.8.5. Swimming Motility Assay
4.8.6. SEM Characterization
4.8.7. Inhibition of Endotoxin-Induced Inflammation
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Ong, Z.Y.; Cheng, J.; Huang, Y.; Xu, K.; Ji, Z.; Fan, W.; Yang, Y.Y. Effect of stereochemistry, chain length and sequence pattern on antimicrobial properties of short synthetic β-sheet forming peptide amphiphiles. Biomaterials 2014, 35, 1315–1325. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Dou, X.; Song, J.; Lyu, Y.; Zhu, X.; Xu, L.; Li, W.; Shan, A. Antimicrobial peptides: Promising alternatives in the post feeding antibiotic era. Med. Res. Rev. 2019, 39, 831–859. [Google Scholar] [CrossRef] [PubMed]
- Baindara, P.; Korpole, S.; Grover, V. Bacteriocins: Perspective for the development of novel anticancer drugs. Appl. Microbiol. Biotechnol. 2018, 102, 10396–10408. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, T.; Pan, Z.; Sun, X.; Yin, X.; He, M.; Xiao, S.; Liang, H. Theoretical Insights into the Interactions between Star-Shaped Antimicrobial Polypeptides and Bacterial Membranes. Langmuir ACS J. Surf. Colloids 2018, 34, 13438–13448. [Google Scholar]
- Zhu, X.; Dong, N.; Wang, Z.; Ma, Z.; Zhang, L.; Ma, Q.; Shan, A. Design of imperfectly amphipathic α-helical antimicrobial peptides with enhanced cell selectivity. Acta Biomater 2014, 10, 244–257. [Google Scholar] [CrossRef] [PubMed]
- Malanovic, N.; Lohner, K. Gram-positive bacterial cell envelopes: The impact on the activity of antimicrobial peptides. Biochim. Biophys. Acta 2016, 1858, 936–946. [Google Scholar] [CrossRef] [PubMed]
- Dong, N.; Zhu, X.; Chou, S.; Shan, A.; Li, W.; Jiang, J. Antimicrobial potency and selectivity of simplified symmetric-end peptides. Biomaterials 2014, 35, 8028–8039. [Google Scholar] [CrossRef] [PubMed]
- Frecer, V.; Ho, B.; Ding, J.L. De Novo Design of Potent Antimicrobial Peptides. Antimicrob. Agents Chemother. 2004, 48, 3349–3357. [Google Scholar] [CrossRef] [PubMed]
- Shenkarev, Z.O.; Balandin, S.V.; Trunov, K.I.; Paramonov, A.S.; Sukhanov, S.V.; Barsukov, L.I.; Arseniev, A.S.; Ovchinnikova, T.V. Molecular Mechanism of Action of β-Hairpin Antimicrobial Peptide Arenicin: Oligomeric Structure in Dodecylphosphocholine Micelles and Pore Formation in Planar Lipid Bilayers. Biochemistry 2011, 50, 6255–6265. [Google Scholar] [CrossRef]
- Penney, J.; Li, J. Protegrin 1 Enhances Innate Cellular Defense via the Insulin-Like Growth Factor 1 Receptor Pathway. Front. Cell. Infect. Microbiol. 2018, 8, 331. [Google Scholar] [CrossRef]
- Edwards, I.A.; Elliott, A.G.; Kavanagh, A.M.; Zuegg, J.; Blaskovich, M.A.; Cooper, M.A. Contribution of Amphipathicity and Hydrophobicity to the Antimicrobial Activity and Cytotoxicity of β-Hairpin Peptides. ACS Infect. Dis. 2016, 2, 442–450. [Google Scholar] [CrossRef]
- Pereira, A.G.; Jaramillo, M.L.; Remor, A.P.; Latini, A.; Davico, C.E.; da Silva, M.L.; Müller, Y.M.R.; Ammar, D. Low-concentration exposure to glyphosate-based herbicide modulates the complexes of the mitochondrial respiratory chain and induces mitochondrial hyperpolarization in the Danio rerio brain. Chemosphere 2018, 209, 353–362. [Google Scholar] [CrossRef]
- Kharidia, R.; Tu, Z.; Chen, L.; Liang, J.F. Activity and selectivity of histidine-containing lytic peptides to antibiotic-resistant bacteria. Arch. Microbio. 2012, 194, 769–778. [Google Scholar] [CrossRef]
- Lai, Z.; Tan, P.; Zhu, Y.; Shao, C.; Shan, A.; Li, L. Highly Stabilized α-Helical Coiled Coils Kill Gram-Negative Bacteria by Multicomplementary Mechanisms under Acidic Condition. ACS Appl. Mater. Interfaces 2019, 11, 22113–22128. [Google Scholar] [CrossRef]
- Yang, C.; Chen, Y.; Peng, S.; Tsai, A.; Lee, T.; Yen, J.; Liou, J. An engineered arginine-rich α-helical antimicrobial peptide exhibits broad-spectrum bactericidal activity against pathogenic bacteria and reduces bacterial infections in mice. Sci. Rep. 2018, 8, 14602. [Google Scholar] [CrossRef]
- Lai, P.K.; Kaznessis, Y.N. Insights into Membrane Translocation of Protegrin Antimicrobial Peptides by Multistep Molecular Dynamics Simulations. ACS Omega 2018, 3, 6056–6065. [Google Scholar] [CrossRef]
- Papanastasiou, E.; Hua, Q.; Sandouk, A.; Son, U.; Christenson, A.; Van Hoek, M.; Bishop, B. Role of acetylation and charge in antimicrobial peptides based on human beta-defensin-3. APMIS 2009, 117, 492–499. [Google Scholar] [CrossRef]
- Kumar, P.; Kizhakkedathu, J.N.; Straus, S.K. Antimicrobial Peptides: Diversity, Mechanism of Action and Strategies to Improve the Activity and Biocompatibility In Vivo. Biomolecules 2018, 8, 4. [Google Scholar] [CrossRef]
- Hu, H.; Kofoed, C.; Li, M.; Gonçalves, J.P.L.; Hansen, J.; Wolfram, M.; Hansen, A.K.; Friis Hansen, C.H.; Diness, F.; Schoffelen, S.; et al. Computational Evolution of Threonine-Rich β-Hairpin Peptides Mimicking Specificity and Affinity of Antibodies. ACS Cent. Sci. 2019, 5, 259–269. [Google Scholar] [CrossRef]
- Tang, M.; Waring, A.; Hong, M. Effects of arginine density on the membrane-bound structure of a cationic antimicrobial peptide from solid-state NMR. Biochim. Biophys. Acta 2009, 1788, 514–521. [Google Scholar] [CrossRef]
- Jin, Y.; Hammer, J.; Pate, M.; Zhang, Y.; Zhu, F.; Zmuda, E.; Blazyk, J. Antimicrobial activities and structures of two linear cationic peptide families with various amphipathic beta-sheet and alpha-helical potentials. Antimicrob. Agents Chemother. 2005, 49, 4957–4964. [Google Scholar] [CrossRef]
- Varkey, J.; Nagaraj, R. Antibacterial activity of human neutrophil defensin HNP-1 analogs without cysteines. Antimicrob. Agents Chemother. 2005, 49, 4561–4566. [Google Scholar] [CrossRef]
- Rydengård, V.; Andersson Nordahl, E.; Schmidtchen, A. Zinc potentiates the antibacterial effects of histidine-rich peptides against Enterococcus faecalis. FEBS J. 2006, 273, 2399–2406. [Google Scholar] [CrossRef]
- Zhang, W.; He, J.; Wu, J.; Schmuck, C. In Vivo Detoxification of Lipopolysaccharide by Antimicrobial Peptides. Bioconjugate Chem. 2017, 28, 319–324. [Google Scholar] [CrossRef]
- Kwon, S.Y.; Ro, M.; Kim, J.H. Mediatory roles of leukotriene B receptors in LPS-induced endotoxic shock. Sci. Rep. 2019, 9, 5936. [Google Scholar] [CrossRef]
- Balleza, D.; Alessandrini, A.; Beltrán García, M.J. Role of Lipid Composition, Physicochemical Interactions, and Membrane Mechanics in the Molecular Actions of Microbial Cyclic Lipopeptides. J. Membr. Bio. 2019, 252, 131–157. [Google Scholar] [CrossRef]
- Sato, H.; Feix, J. Peptide-membrane interactions and mechanisms of membrane destruction by amphipathic alpha-helical antimicrobial peptides. Biochim. Biophys. Acta 2006, 1758, 1245–1256. [Google Scholar] [CrossRef]
- Farha, M.; Verschoor, C.; Bowdish, D.; Brown, E. Collapsing the proton motive force to identify synergistic combinations against Staphylococcus aureus. Chem. Biol. 2013, 20, 1168–1178. [Google Scholar] [CrossRef]
- Campoccia, D.; Montanaro, L.; Speziale, P.; Arciola, C. Antibiotic-loaded biomaterials and the risks for the spread of antibiotic resistance following their prophylactic and therapeutic clinical use. Biomaterials 2010, 31, 6363–6377. [Google Scholar] [CrossRef]
- Kim, S.; Kim, S.; Lee, B. Correlation between the activities of alpha-helical antimicrobial peptides and hydrophobicities represented as RP HPLC retention times. Peptides 2005, 26, 2050–2056. [Google Scholar] [CrossRef]
- Zelezetsky, I.; Tossi, A. Alpha-helical antimicrobial peptides--using a sequence template to guide structure-activity relationship studies. Biochim. Biophys. Acta 2006, 1758, 1436–1449. [Google Scholar] [CrossRef]
- Arouri, A.; Dathe, M.; Blume, A. The helical propensity of KLA amphipathic peptides enhances their binding to gel-state lipid membranes. Biophys. Chem. 2013, 180, 10–21. [Google Scholar] [CrossRef]
- Huang, J.; Hao, D.; Chen, Y.; Xu, Y.; Tan, J.; Huang, Y.; Li, F.; Chen, Y. Inhibitory effects and mechanisms of physiological conditions on the activity of enantiomeric forms of an α-helical antibacterial peptide against bacteria. Peptides 2011, 32, 1488–1495. [Google Scholar] [CrossRef]
- Fjell, C.; Hiss, J.; Hancock, R.; Schneider, G. Designing antimicrobial peptides: Form follows function. Nat. Rev. Drug Discov. 2011, 11, 37–51. [Google Scholar] [CrossRef]
- Ong, Z.; Wiradharma, N.; Yang, Y. Strategies employed in the design and optimization of synthetic antimicrobial peptide amphiphiles with enhanced therapeutic potentials. Adv. Drug Deliv. Rev. 2014, 78, 28–45. [Google Scholar] [CrossRef]
- Epand, R.; Pollard, J.; Wright, J.; Savage, P.; Epand, R. Depolarization, bacterial membrane composition, and the antimicrobial action of ceragenins. Antimicrob. Agents Chemother. 2010, 54, 3708–3713. [Google Scholar] [CrossRef]
- Paul, K.; Erhardt, M.; Hirano, T.; Blair, D.; Hughes, K. Energy source of flagellar type III secretion. Nature 2008, 451, 489–492. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, X.; Wang, J. Recent Advances in Antibacterial and Antiendotoxic Peptides or Proteins from Marine Resources. Mar. Drugs 2018, 16, 57. [Google Scholar] [CrossRef]
- Hancock, R.; Haney, E.; Gill, E. The immunology of host defence peptides: Beyond antimicrobial activity. Nat. Rev. Immunol. 2016, 16, 321–334. [Google Scholar] [CrossRef]
- Rosenfeld, Y.; Shai, Y. Lipopolysaccharide (Endotoxin)-host defense antibacterial peptides interactions: Role in bacterial resistance and prevention of sepsis. Biochim. Biophys. Acta 2006, 1758, 1513–1522. [Google Scholar] [CrossRef]
- Hagar, J.; Powell, D.; Aachoui, Y.; Ernst, R.; Miao, E. Cytoplasmic LPS activates caspase-11: Implications in TLR4-independent endotoxic shock. Science 2013, 341, 1250–1253. [Google Scholar] [CrossRef]
- Lee, E.; Kim, Y.; Nan, Y.; Shin, S. Cell selectivity, mechanism of action and LPS-neutralizing activity of bovine myeloid antimicrobial peptide-18 (BMAP-18) and its analogs. Peptides 2011, 32, 1123–1130. [Google Scholar] [CrossRef]
- Fang, L.; Xu, Z.; Wang, G.; Ji, F.; Mei, C.; Liu, J.; Wu, G. Directed evolution of an LBP/CD14 inhibitory peptide and its anti-endotoxin activity. PLoS ONE 2014, 9, e101406. [Google Scholar] [CrossRef]
- Gutsmann, T.; Razquin-Olazarán, I.; Kowalski, I.; Kaconis, Y.; Howe, J.; Bartels, R.; Hornef, M.; Schürholz, T.; Rössle, M.; Sanchez-Gómez, S.; et al. New antiseptic peptides to protect against endotoxin-mediated shock. Antimicrob. Agents Chemother. 2010, 54, 3817–3824. [Google Scholar] [CrossRef]
- Heinbockel, L.; Sánchez-Gómez, S.; Martinez de Tejada, G.; Dömming, S.; Brandenburg, J.; Kaconis, Y.; Hornef, M.; Dupont, A.; Marwitz, S.; Goldmann, T.; et al. Preclinical investigations reveal the broad-spectrum neutralizing activity of peptide Pep19–2.5 on bacterial pathogenicity factors. Antimicrob. Agents Chemother. 2013, 57, 1480–1487. [Google Scholar] [CrossRef]
- Proft, T.; Baker, E. Pili in Gram-negative and Gram-positive bacteria—structure, assembly and their role in disease. Cell. Mol. Life Sci. 2009, 66, 613. [Google Scholar] [CrossRef]
- Xu, W.; Zhu, X.; Tan, T.; Li, W.; Shan, A. Design of embedded-hybrid antimicrobial peptides with enhanced cell selectivity and anti-biofilm activity. PLoS ONE 2014, 9, e98935. [Google Scholar] [CrossRef]
- Chou, S.; Wang, J.; Shang, L.; Akhtar, M.U.; Wang, Z.; Shi, B.; Feng, X.; Shan, A. Short, symmetric-helical peptides have narrow-spectrum activity with low resistance potential and high selectivity. Biomater. Sci. 2019, 7, 2394–2409. [Google Scholar] [CrossRef]
- Ejim, L.; Farha, M.; Falconer, S.; Wildenhain, J.; Coombes, B.; Tyers, M.; Brown, E.; Wright, G. Combinations of antibiotics and nonantibiotic drugs enhance antimicrobial efficacy. Nat. Chem. Biol. 2011, 7, 348–350. [Google Scholar] [CrossRef]
- Sun, E.; Liu, S.; Hancock, R.E.W. Surfing Motility: A Conserved yet Diverse Adaptation among Motile Bacteria. J. Bacteriol. 2018, 200, e00394. [Google Scholar] [CrossRef]
- Ma, Z.; Yang, J.; Han, J.; Gao, L.; Liu, H.; Lu, Z.; Zhao, H.; Bie, X. Insights into the Antimicrobial Activity and Cytotoxicity of Engineered α-Helical Peptide Amphiphiles. J. Med. Chem. 2016, 59, 10946–10962. [Google Scholar] [CrossRef]
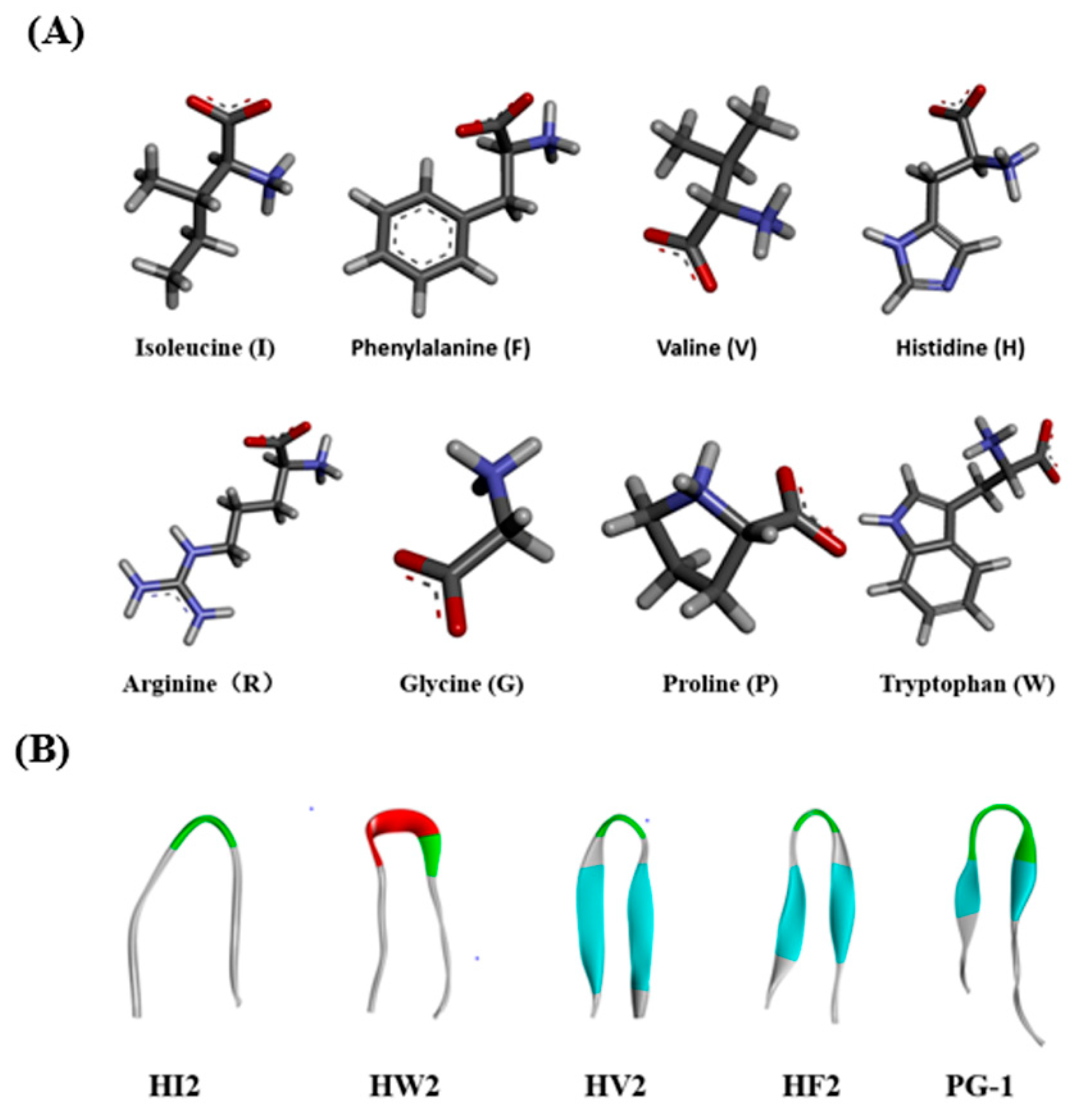

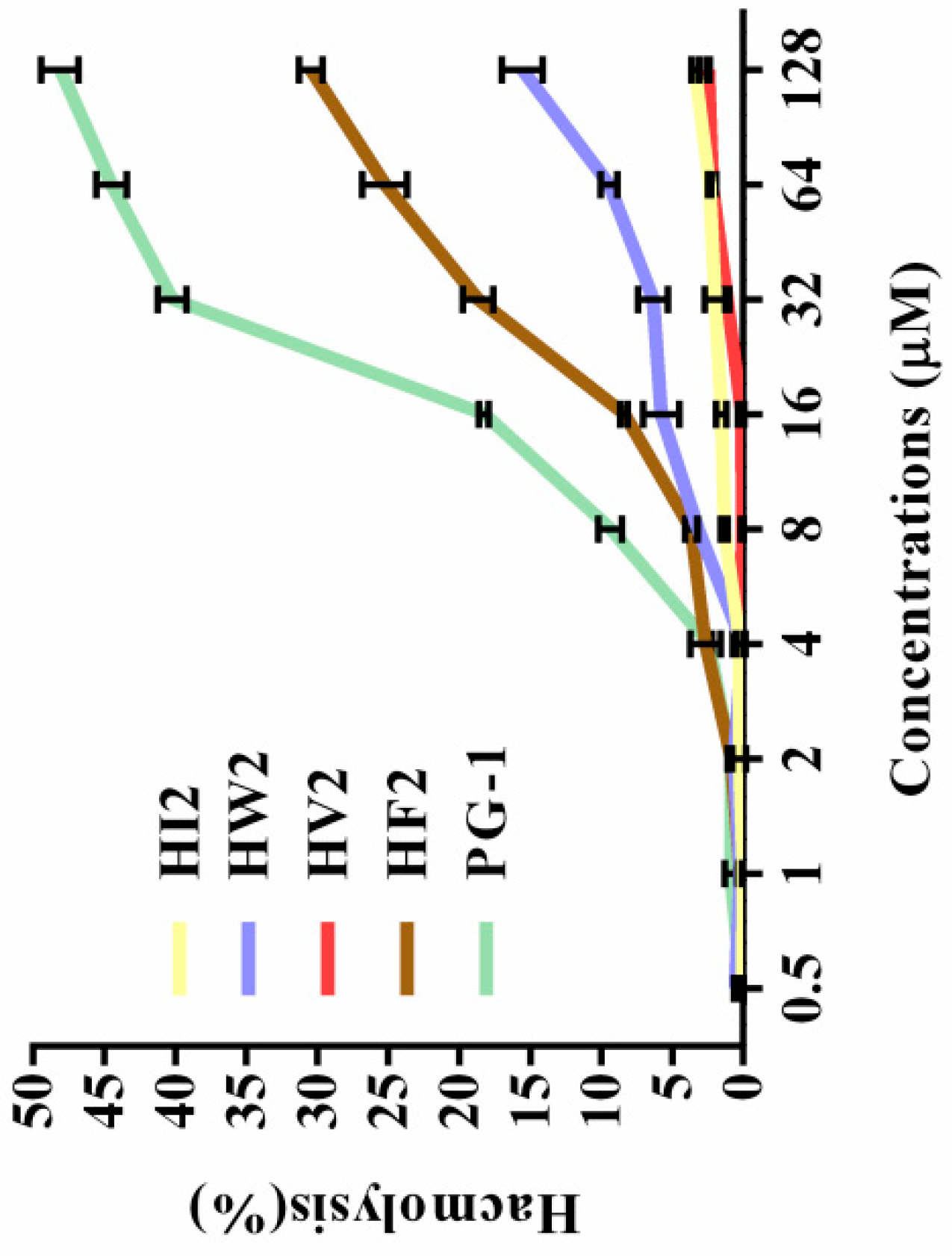
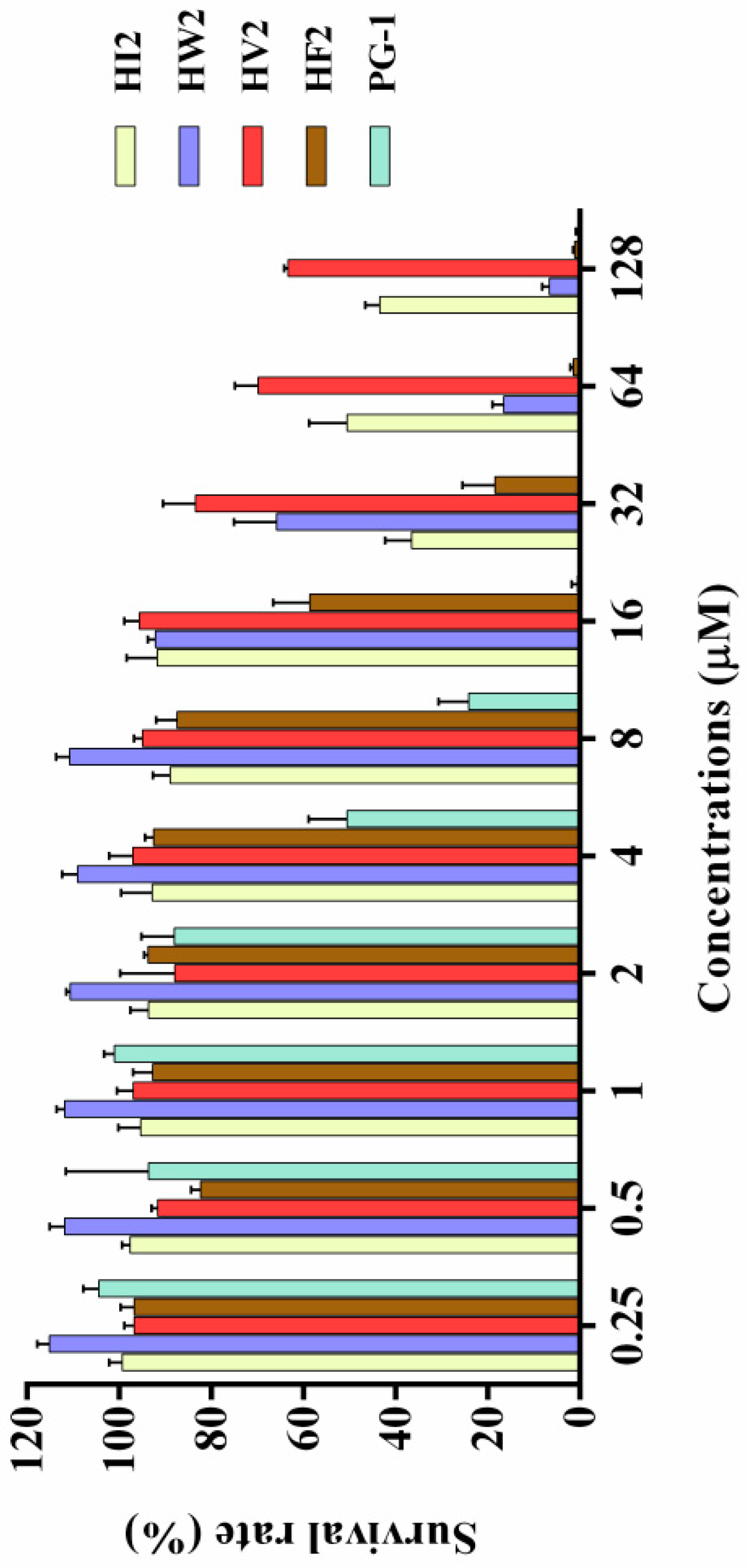
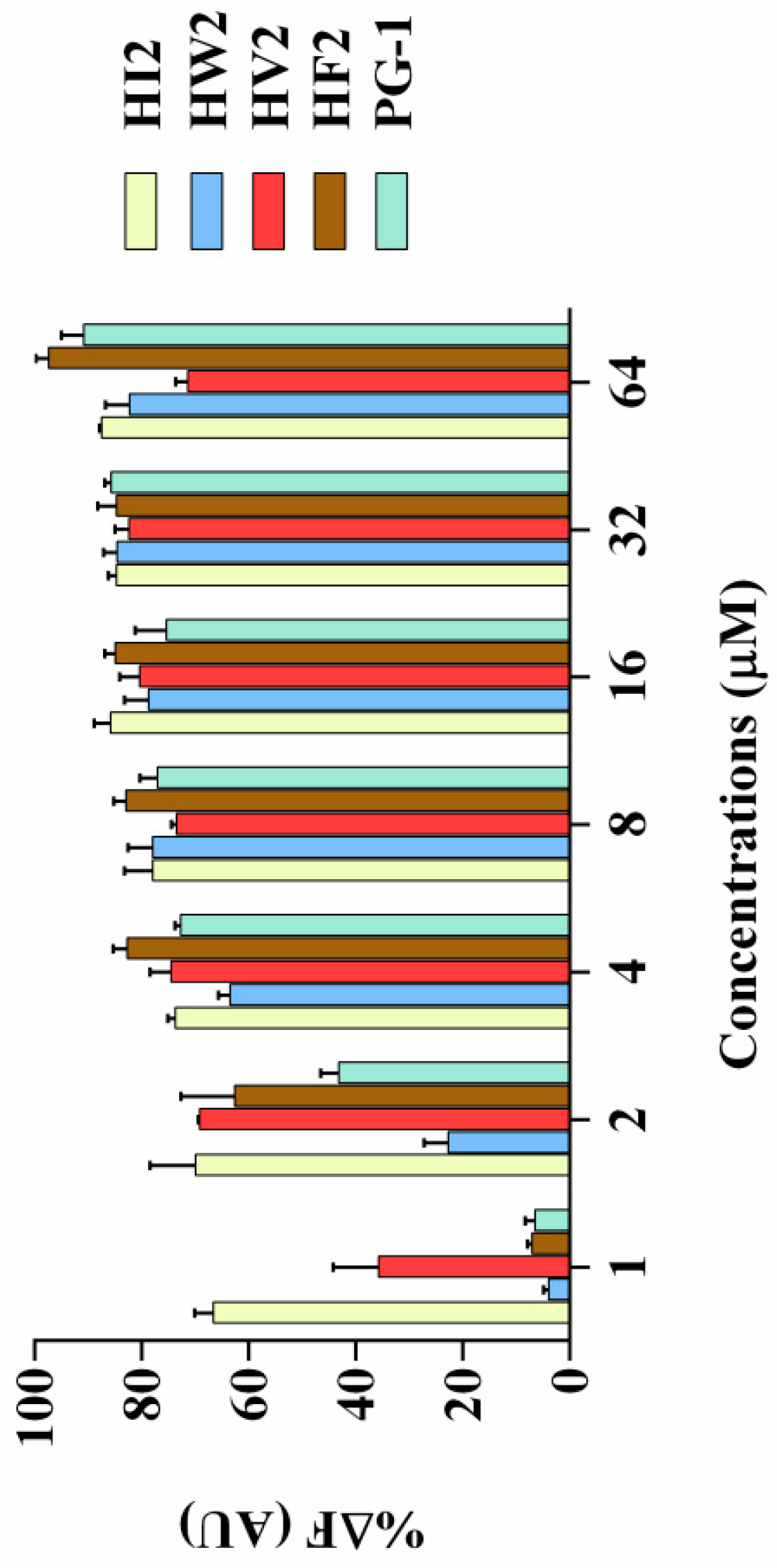
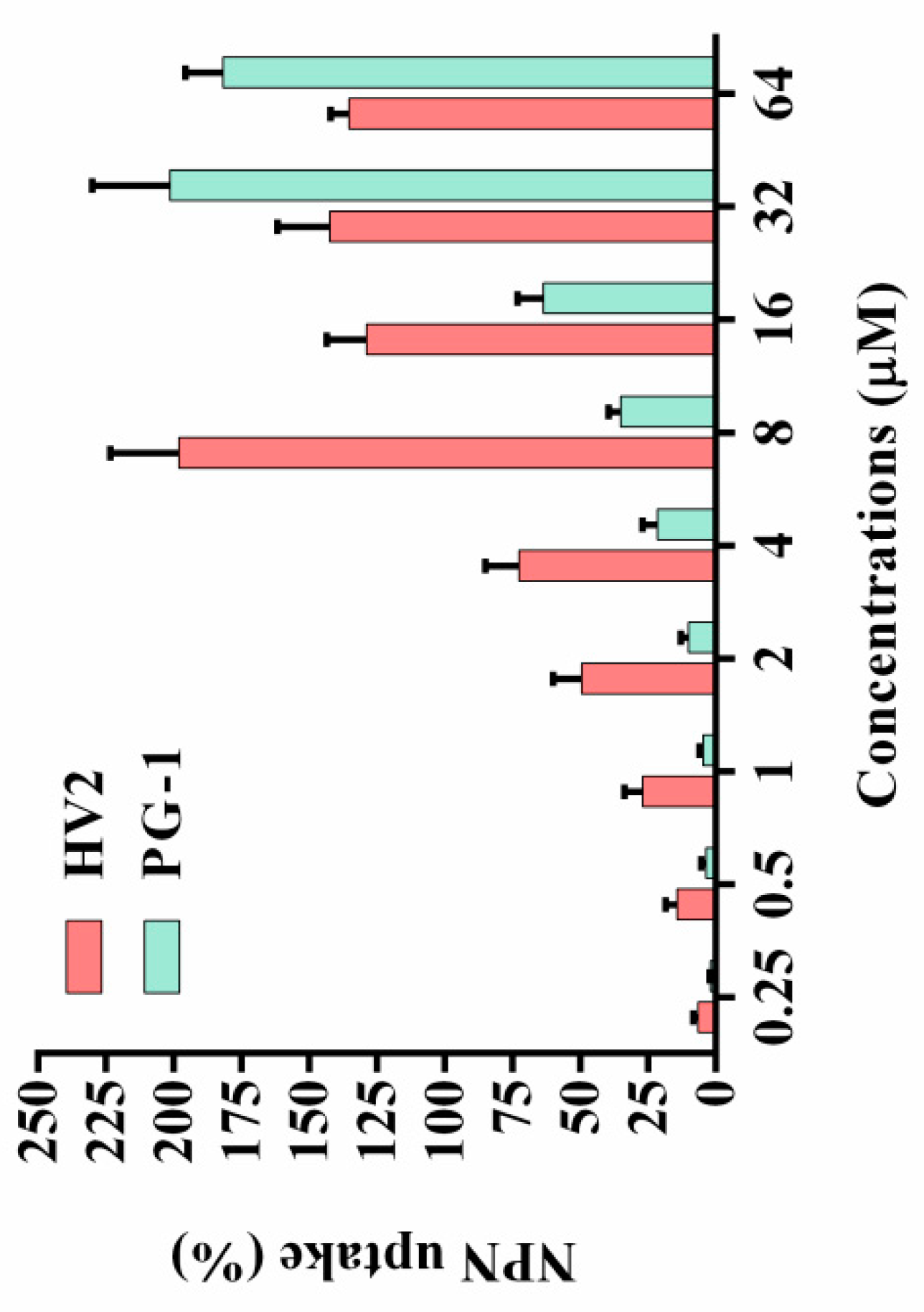
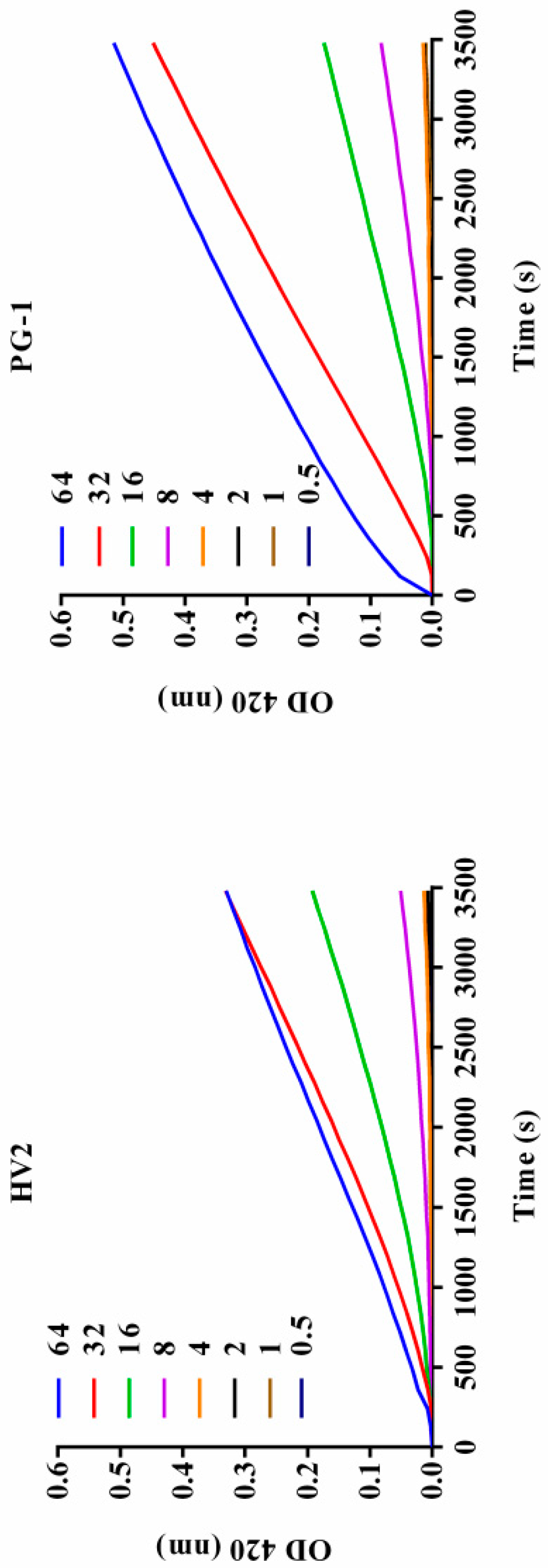
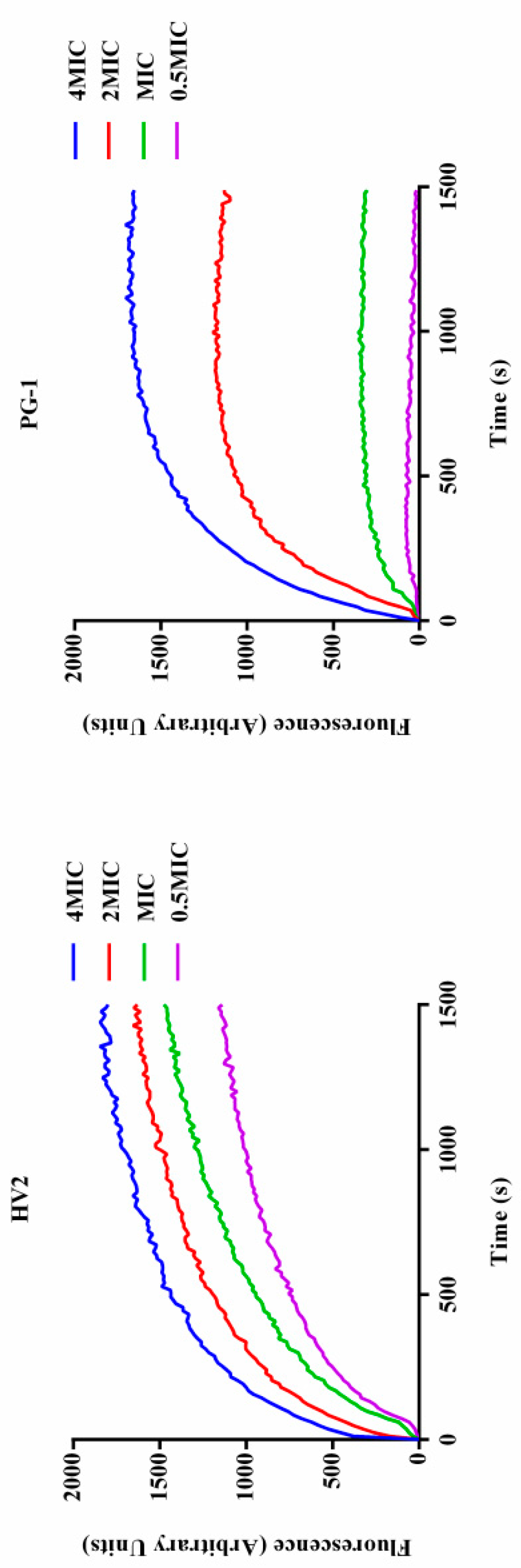
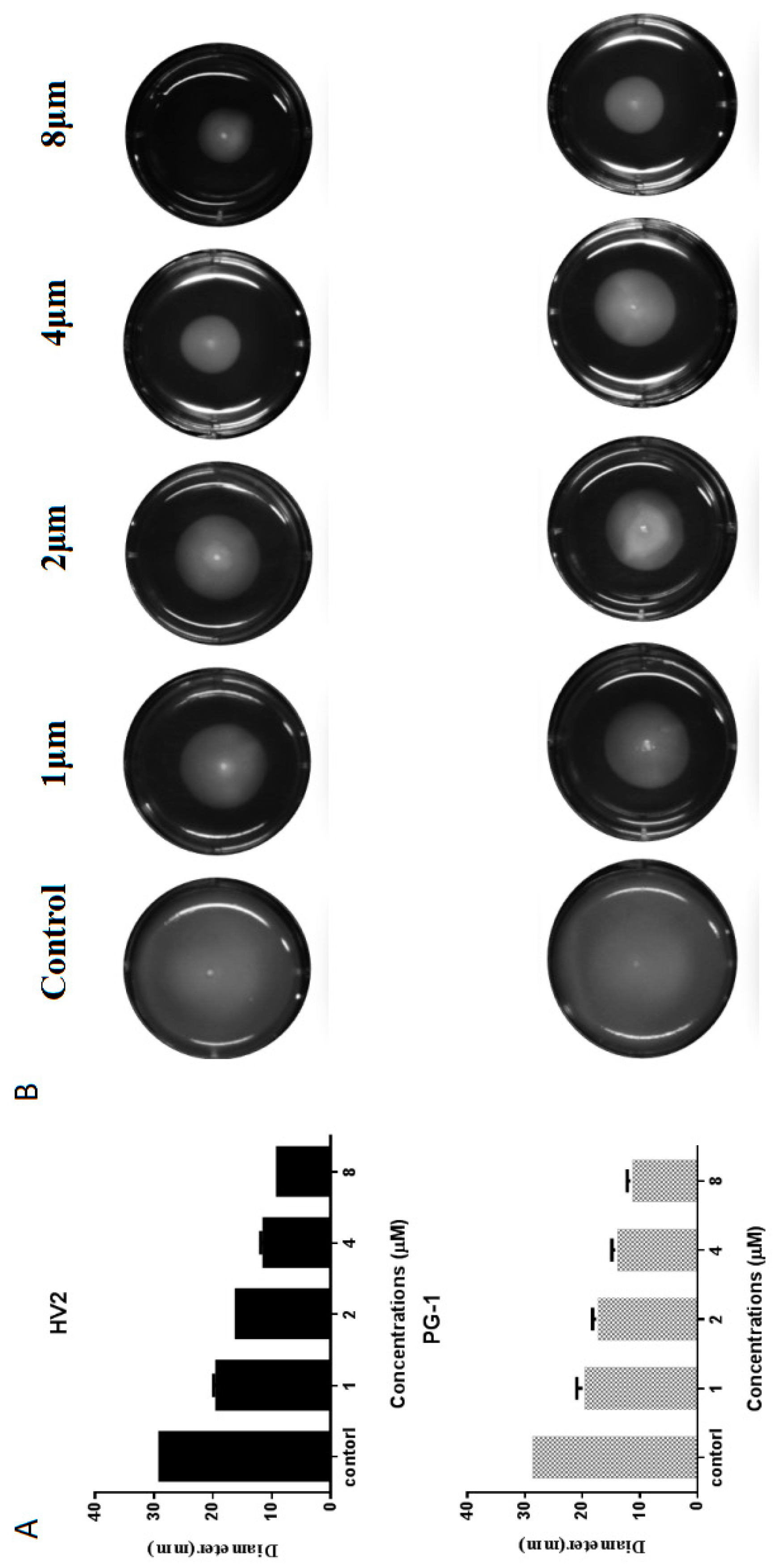

| Peptides | Sequence | Theoretical MWa | Measured MW | Retention Time (min) | Hb | μHrelc | Net Charge |
|---|---|---|---|---|---|---|---|
| HI2 | RRIHIHIDPGIHIHIRR-NH2 | 2023.46 | 2024.47 | 11.13 | −0.35 | 2.39 | +9 |
| HW2 | RRWHWHWDPGWHWHWRR-NH2 | 2461.78 | 2461.82 | 10.55 | 0.02 | 2.35 | +9 |
| HV2 | RRVHVHVDPGVHVHVRR-NH2 | 1939.29 | 1939.33 | 12.20 | −2.07 | 1.89 | +9 |
| HF2 | RRFHFHFDPGFHFHFRR-NH2 | 2227.56 | 2227.59 | 18.06 | 0.13 | 2.53 | +9 |
| PG-1 | RGGRLCYCRRFCVCVGR-NH2 | 2155.61 | 2156.35 | 12.56 | −2.42 | 1.37 | +7 |
| HI2 | HW2 | HV2 | HF2 | PG-1 | |
|---|---|---|---|---|---|
| MICa (μM) | |||||
| Gram- | |||||
| E. coli 25922 | 32 | 4 | 8 | 4 | 4 |
| E. coli K88 | 32 | 4 | 4 | 2 | 2 |
| E. coli K99 | >128 | 4 | 8 | 4 | 2 |
| E. coli UB1005 | 16 | 2 | 8 | 8 | 2 |
| S.pullorum C7913 | 16 | 2 | 8 | 4 | 4 |
| P. auruginosa 27853 | >128 | 8 | 8 | >128 | 8 |
| Gram+ | |||||
| S.aureus 29213 | >128 | 4 | >128 | 64 | 2 |
| S.aureus 43300 | >128 | 2 | >128 | 4 | 8 |
| S.epidermidis 12228 | >128 | 2 | >128 | 4 | 8 |
| MBCb (GM) | |||||
| Gram (−) | 50.80 | 3.56 | 7.13 | 8.00 | 3.17 |
| Gram (+) | 256 | 2.52 | 256 | 10.08 | 5.04 |
| Gram (+,−) | 87.09 | 3.17 | 23.52 | 8.64 | 3.70 |
| MHCc(µM) | >128 | 8 | >128 | 16 | 4 |
| TId | |||||
| TI(−) | 5.04 | 2.25 | 35.90 | 2.00 | 1.26 |
| TI(+) | 1 | 3.17 | 1 | 1.59 | 0.79 |
| TI(+,−) | 2.94 | 2.52 | 10.88 | 1.85 | 1.08 |
| Peptides | Control b | NaCl b | KCl b | NH4Cl b | MgCl2 b | ZnCl2 b | FeCl3 b |
|---|---|---|---|---|---|---|---|
| Gram-negative strain E. coli ATCC25922 | |||||||
| HV2 | 8 | 4 | 4 | 4 | 8 | 1 | 4 |
| PG-1 | 4 | 4 | 4 | 4 | 4 | 2 | 4 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, N.; Wang, C.; Zhang, T.; Zhang, L.; Xue, C.; Feng, X.; Bi, C.; Shan, A. Bioactivity and Bactericidal Mechanism of Histidine-Rich β-Hairpin Peptide Against Gram-Negative Bacteria. Int. J. Mol. Sci. 2019, 20, 3954. https://doi.org/10.3390/ijms20163954
Dong N, Wang C, Zhang T, Zhang L, Xue C, Feng X, Bi C, Shan A. Bioactivity and Bactericidal Mechanism of Histidine-Rich β-Hairpin Peptide Against Gram-Negative Bacteria. International Journal of Molecular Sciences. 2019; 20(16):3954. https://doi.org/10.3390/ijms20163954
Chicago/Turabian StyleDong, Na, Chensi Wang, Tingting Zhang, Lei Zhang, Chenyu Xue, Xinjun Feng, Chongpeng Bi, and Anshan Shan. 2019. "Bioactivity and Bactericidal Mechanism of Histidine-Rich β-Hairpin Peptide Against Gram-Negative Bacteria" International Journal of Molecular Sciences 20, no. 16: 3954. https://doi.org/10.3390/ijms20163954
APA StyleDong, N., Wang, C., Zhang, T., Zhang, L., Xue, C., Feng, X., Bi, C., & Shan, A. (2019). Bioactivity and Bactericidal Mechanism of Histidine-Rich β-Hairpin Peptide Against Gram-Negative Bacteria. International Journal of Molecular Sciences, 20(16), 3954. https://doi.org/10.3390/ijms20163954





