Identification and Characterization of MiRNAs in Coccomyxa subellipsoidea C-169
Abstract
1. Introduction
2. Results and Discussion
2.1. High-Throughput Sequencing of Small RNAs in C-169
2.2. Identification of Mirnas in C-169
2.3. Validation of miRNAs in C-169 Through qRT-PCR
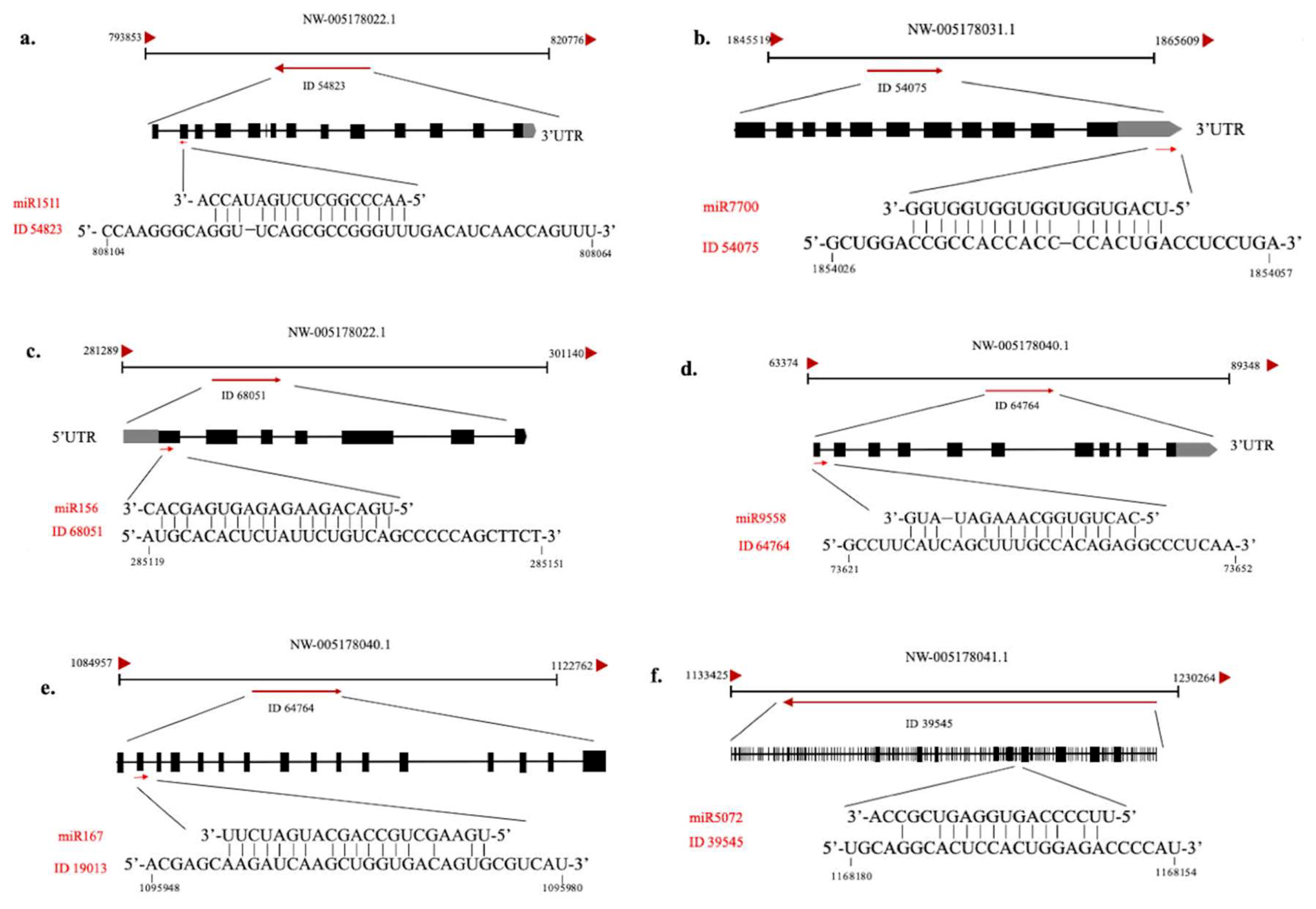
2.4. Functional Analysis of the Putative Target Genes of C-169 miRNAs
2.5. Functional Analysis of the Putative Target Genes in Metabolism and Transcription and Translation Process
3. Materials and Methods
3.1. Algal Strain and Culture Conditions
3.2. RNA Extraction, Library Construction and Sequencing
3.3. Sequencing Data Analysis
3.4. Target Gene Prediction and Function Analysis
3.5. Quantitative RT-PCR
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Chisti, Y. Biodiesel from microalgae. Biotechnol. Adv. 2007, 25, 294–306. [Google Scholar] [CrossRef] [PubMed]
- Chisti, Y. Biodiesel from microalgae beats bioethanol. Trends Biotechnol. 2008, 26, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Miao, X.L.; Wu, Q.Y.; Yang, C.Y. Fast pyrolysis of microalgae to produce renewable fuels. J. Anal. Appl. Pyrol. 2004, 71, 855–863. [Google Scholar] [CrossRef]
- Huang, G.; Chen, F.; Wei, D.; Zhang, X.; Chen, G. Biodiesel production by microalgal biotechnology. Applied. Energy 2010, 87, 38–46. [Google Scholar] [CrossRef]
- Khan, S.A.; Hussain, M.Z.; Prasad, S.; Banerjee, U.C. Prospects of biodiesel production from microalgae in India. Renew. Sust. Energy Rev. 2009, 13, 2361–2372. [Google Scholar] [CrossRef]
- Sharma, T.; Gour, R.S.; Kant, A.; Chauhan, R.S. Lipid content in Scenedesmus species correlates with multiple genes of fatty acid and triacylglycerol biosynthetic pathways. Algal Res. 2015, 12, 341–349. [Google Scholar] [CrossRef]
- Biswas, B.; Singh, R.; Krishna, B.B.; Kumar, J.; Bhaskar, T. Pyrolysis of azolla, sargassum tenerrimum and water hyacinth for production of bio-oil. Bioresour. Technol. 2017, 242, 139–145. [Google Scholar] [CrossRef]
- Blanc, G.; Agarkova, I.; Grimwood, J.; Kuo, A.; Brueggeman, A.; Dunigan, D.D.; Gurnon, J.; Ladunga, I.; Lindquist, E.; Lucas, S.; et al. The genome of the polar eukaryotic microalga Coccomyxa subellipsoidea reveals traits of cold adaptation. Genome Biol. 2012, 13, R39. [Google Scholar] [CrossRef]
- Peng, H.; Wei, D.; Chen, G.; Chen, F. Transcriptome analysis reveals global regulation in response to CO2 supplementation in oleaginous microalga Coccomyxa subellipsoidea C-169. Biotechnol. Biofuels 2016, 9, 151. [Google Scholar] [CrossRef]
- Achkar, N.P.; Cambiagno, D.A.; Manavella, P.A. MiRNA Biogenesis: A Dynamic Pathway. Trends Plant Sci. 2016, 21, 1034–1044. [Google Scholar] [CrossRef]
- Hausser, J.; Zavolan, M. Identification and consequences of miRNA-target interactions—Beyond repression of gene expression. Nat. Rev. Genet. 2014, 15, 599–612. [Google Scholar] [CrossRef] [PubMed]
- Naqvi, A.R.; Shango, J.; Seal, A.; Shukla, D.; Nares, S. Herpesviruses and MicroRNAs: New Pathogenesis Factors in Oral Infection and Disease? Front. Immunol. 2018, 9, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Tuddenham, L.; Jung, J.S.; Chane-Woon-Ming, B.; Doelken, L.; Pfeffer, S. Small RNA Deep Sequencing Identifies MicroRNAs and Other Small Noncoding RNAs from Human Herpesvirus 6B. J. Virol. 2012, 86, 1638–1649. [Google Scholar] [CrossRef] [PubMed]
- Moran, Y.; Agron, M.; Praher, D.; Technau, U. The evolutionary origin of plant and animal microRNAs. Nat. Ecol. Evolut. 2017, 1, 0027. [Google Scholar] [CrossRef] [PubMed]
- Murchison, E.P.; Hannon, G.J. MiRNAs on the move: MiRNA biogenesis and the RNAi machinery. Curr. Opin. Cell Biol. 2004, 16, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Ameres, S.L.; Zamore, P.D. Diversifying microRNA sequence and function. Nat. Rev. Mol. Cell Bio. 2013, 14, 475–488. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Koyama, T.; Sato, F.; Ohme-Takagi, M. Roles of miR319 and TCP Transcription Factors in Leaf Development. Plant Physiol. 2017, 175, 874–885. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; He, H.; Li, Y.; Wang, F.; Yu, D. Molecular mechanism of microRNA396 mediating pistil development in Arabidopsis. Plant Physiol. 2014, 164, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Bi, F.C.; Meng, X.C.; Ma, C.; Yi, G.J. Identification of miRNAs involved in fruit ripening in Cavendish bananas by deep sequencing. BMC Genom. 2015, 16, 776. [Google Scholar] [CrossRef] [PubMed]
- Gupta, O.P.; Karkute, S.G.; Banerjee, S.; Meena, N.L.; Dahuja, A. Contemporary Understanding of miRNA-Based Regulation of Secondary Metabolites Biosynthesis in Plants. Front. Plant Sci. 2017, 8, 374. [Google Scholar] [CrossRef] [PubMed]
- Rupaimoole, R.; Calin, G.A.; Lopez-Berestein, G.; Sood, A.K. MiRNA Deregulation in Cancer Cells and the Tumor Microenvironment. Cancer Discov. 2016, 6, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.; He, L.; Wang, G. Identification and characterization of microRNAs from Phaeodactylum tricornutum by high-throughput sequencing and bioinformatics analysis. BMC Genom. 2011, 12, 337. [Google Scholar] [CrossRef] [PubMed]
- Voshall, A.; Kim, E.-J.; Ma, X.; Moriyama, E.N.; Cerutti, H. Identification of AGO3-Associated miRNAs and Computational Prediction of Their Targets in the Green Alga Chlamydomonas reinhardtii. Genetics 2015, 200, 105–121. [Google Scholar] [CrossRef]
- Voshall, A.; Kim, E.J.; Ma, X.; Yamasaki, T.; Moriyama, E.N.; Cerutti, H. MiRNAs in the alga Chlamydomonas reinhardtii are not phylogenetically conserved and play a limited role in responses to nutrient deprivation. Sci. Rep.-UK 2017, 7, 5462. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wu, Y.; Qi, Y. MicroRNAs in a multicellular green alga Volvox carteri. Sci. China-Life Sci. 2014, 57, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Dueck, A.; Evers, M.; Henz, S.R.; Unger, K.; Eichner, N.; Merkl, R.; Berezikov, E.; Engelmann, J.C.; Weigel, D.; Wenzl, S.; et al. Gene silencing pathways found in the green alga Volvox carteri reveal insights into evolution and origins of small RNA systems in plants. BMC Genom. 2016, 17, 853. [Google Scholar] [CrossRef]
- Deng, X.-Y.; Hu, X.-L.; Li, D.; Wang, L.; Cheng, J.; Gao, K. Identification and analysis of microRNAs in Botryococcus braunii using high-throughput sequencing. Aquat. Biol. 2017, 26, 41–48. [Google Scholar] [CrossRef]
- Gao, X.N.; Cong, Y.T.; Yue, J.R.; Xing, Z.Y.; Wang, Y.; Chai, X.J. Small RNA, transcriptome, and degradome sequencing to identify salinity stress responsive miRNAs and target genes in Dunaliella salina. J. Appl. Phycol. 2019, 31, 1175–1183. [Google Scholar] [CrossRef]
- Gao, F.; Nan, F.R.; Song, W.; Feng, J.; Lv, J.P.; Xie, S.L. Identification and Characterization of miRNAs in Chondrus crispus by High-Throughput Sequencing and Bioinformatics Analysis. Sci. Rep.-UK 2016, 6, 26397. [Google Scholar] [CrossRef]
- Gao, F.; Nan, F.; Feng, J.; Lv, J.; Liu, Q.; Xie, S. Identification and characterization of microRNAs in Eucheuma denticulatum by high-throughput sequencing and bioinformatics analysis. RNA Biol. 2016, 13, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Nan, F.; Feng, J.; Lv, J.; Liu, Q.; Xie, S. Identification of conserved and novel microRNAs in Porphyridium purpureum via deep sequencing and bioinformatics. BMC Genom. 2016, 17, 612. [Google Scholar] [CrossRef] [PubMed]
- Barozai, M.Y.K.; Qasim, M.; Din, M.; Achakzai, A.K.K. An update on the microRNAs and their targets in unicellular red alga Porphyridium cruentum. Pak. J. Bot. 2018, 50, 817–825. [Google Scholar]
- Billoud, B.; Nehr, Z.; Le Bail, A.; Charrier, B. Computational prediction and experimental validation of microRNAs in the brown alga Ectocarpus siliculosus. Nucleic Acids Res. 2014, 42, 417–429. [Google Scholar] [CrossRef] [PubMed]
- Cock, J.M.; Liu, F.; Duan, D.; Bourdareau, S.; Lipinska, A.P.; Coelho, S.M.; Tarver, J.E. Rapid Evolution of microRNA Loci in the Brown Algae. Genome Biol. Evol. 2017, 9, 740–749. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhuang, X.; Chen, M.; Zeng, Z.; Cai, X.; Li, H.; Hu, Z. An endogenous microRNA (miRNA1166.1) can regulate photobio-H2 production in eukaryotic green alga Chlamydomonas reinhardtii. Biotechnol. Biofuels 2018, 11, 126. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jiang, X.; Hu, C.; Sun, T.; Zeng, Z.; Cai, X.; Li, H.; Hu, Z. Optogenetic regulation of artificial microRNA improves H2 production in green alga Chlamydomonas reinhardtii. Biotechnol. Biofuels 2017, 10, 257. [Google Scholar] [CrossRef]
- Li, H.; Liu, Y.; Wang, Y.; Chen, M.; Zhuang, X.; Wang, C.; Wang, J.; Hu, Z. Improved photobio-H2 production regulated by artificial miRNA targeting psbA in green microalga Chlamydomonas reinhardtii. Biotechnol. Biofuels 2018, 11, 36. [Google Scholar] [CrossRef]
- Chung, B.Y.W.; Deery, M.J.; Groen, A.J.; Howard, J.; Baulcombe, D.C. Endogenous miRNA in the green alga Chlamydomonas regulates gene expression through CDS-targeting. Nat. Plants 2017, 3, 787–794. [Google Scholar] [CrossRef]
- Lou, S.; Sun, T.; Li, H.; Hu, Z. Mechanisms of microRNA-mediated gene regulation in unicellular model alga Chlamydomonas reinhardtii. Biotechnol. Biofuels 2018, 11, 244. [Google Scholar] [CrossRef]
- Samad, A.F.A.; Sajad, M.; Nazaruddin, N.; Fauzi, I.A.; Murad, A.M.A.; Zainal, Z.; Ismail, I. MicroRNA and Transcription Factor: Key Players in Plant Regulatory Network. Front. Plant Sci. 2017, 8, 565. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Bao, Y.; Shan, D.; Wang, Z.; Song, X.; Wang, Z.; Wang, J.; He, L.; Wu, L.; Zhang, Z.; et al. Magnaporthe oryzae Induces the Expression of a MicroRNA to Suppress the Immune Response in Rice. Plant Physiol. 2018, 177, 352–368. [Google Scholar] [PubMed]
- Sun, X.H.; Zhao, L.P.; Zou, Q.; Wang, Z.B. Identification of MicroRNA Genes and their mRNA Targets in Festuca arundinacea. Appl. Biochem. Biotech. 2014, 172, 3875–3887. [Google Scholar] [CrossRef]
- Shi, T.; Wang, K.; Yang, P.F. The evolution of plant microRNAs: Insights from a basal eudicot sacred lotus. Plant J. 2017, 89, 442–457. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.-L.; Li, Q.; Lv, W.-Q.; Zhang, G.-H.; Yu, Y.-H. MicroRNA profiling analysis of developing berries for ‘Kyoho’ and its early-ripening mutant during berry ripening. BMC Plant Biol. 2018, 18, 285. [Google Scholar] [CrossRef]
- Matthijs, M.; Fabris, M.; Obata, T.; Foubert, I.; Franco-Zorrilla, J.M.; Solano, R.; Fernie, A.R.; Vyverman, W.; Goossens, A. The transcription factor bZIP14 regulates the TCA cycle in the diatom Phaeodactylum tricornutum. EMBO J. 2017, 36, 1559–1576. [Google Scholar] [CrossRef]
- Thiriet-Rupert, S.; Carrier, G.; Trottier, C.; Eveillard, D.; Schoefs, B.; Bougaran, G.; Cadoret, J.P.; Chenais, B.; Saint-Jean, B. Identification of transcription factors involved in the phenotype of a domesticated oleaginous microalgae strain of Tisochrysis lutea. Algal Res. 2018, 30, 59–72. [Google Scholar] [CrossRef]
- Kwon, S.; Kang, N.K.; Koh, H.G.; Shin, S.E.; Lee, B.; Jeong, B.R.; Chang, Y.K. Enhancement of biomass and lipid productivity by overexpression of a bZIP transcription factor in Nannochloropsis salina. Biotechnol. Bioeng. 2018, 115, 331–340. [Google Scholar] [CrossRef]
- Tang, J.Y.; Chu, C.C. MicroRNAs in crop improvement: Fine-tuners for complex traits. Nat. Plants 2017, 3, 17077. [Google Scholar] [CrossRef]
- Wang, C.G.; Chen, X.; Li, H.; Wang, J.X.; Hu, Z.L. Artificial miRNA inhibition of phosphoenolpyruvate carboxylase increases fatty acid production in a green microalga Chlamydomonas reinhardtii. Biotechnol. Biofuels 2017, 10, 91. [Google Scholar] [CrossRef]
- Franco-Zorrilla, J.M.; Valli, A.; Todesco, M.; Mateos, I.; Puga, M.I.; Rubio-Somoza, I.; Leyva, A.; Weigel, D.; Garcia, J.A.; Paz-Ares, J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007, 39, 1033–1037. [Google Scholar] [CrossRef] [PubMed]
- Todesco, M.; Rubio-Somoza, I.; Paz-Ares, J.; Weigel, D. A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet. 2010, 6, e1001031. [Google Scholar] [CrossRef] [PubMed]
- Bazin, J.; Khan, G.A.; Combier, J.P.; Bustos-Sanmamed, P.; Debernardi, J.M.; Rodriguez, R.; Sorin, C.; Palatnik, J.; Hartmann, C.; Crespi, M.; et al. MiR396 affects mycorrhization and root meristem activity in the legume Medicago truncatula. Plant J. 2013, 74, 920–934. [Google Scholar] [CrossRef] [PubMed]
- Kielbasa, S.M.; Bluthgen, N.; Fahling, M.; Mrowka, R. Targetfinder.org: A resource for systematic discovery of transcription factor target genes. Nucleic Acids Res. 2010, 38, W233–W238. [Google Scholar] [CrossRef] [PubMed]
- Harris, M.A.; Clark, J.; Ireland, A.; Lomax, J.; Ashburner, M.; Foulger, R.; Eilbeck, K.; Lewis, S.; Marshall, B.; Mungall, C.; et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004, 32, D258–D261. [Google Scholar] [PubMed]
- Kanehisa, M. The KEGG database. Novart. Fdn. Symp. 2002, 247, 91–103. [Google Scholar]
- Li, H.; Zhang, L.; Shu, L.; Zhuang, X.; Liu, Y.; Chen, J.; Hu, Z. Sustainable photosynthetic H2-production mediated by artificial miRNA silencing of OEE2 gene in green alga Chlamydomonas reinhardtii. Int. J. Hydrog. Energy 2015, 40, 5609–5616. [Google Scholar] [CrossRef]
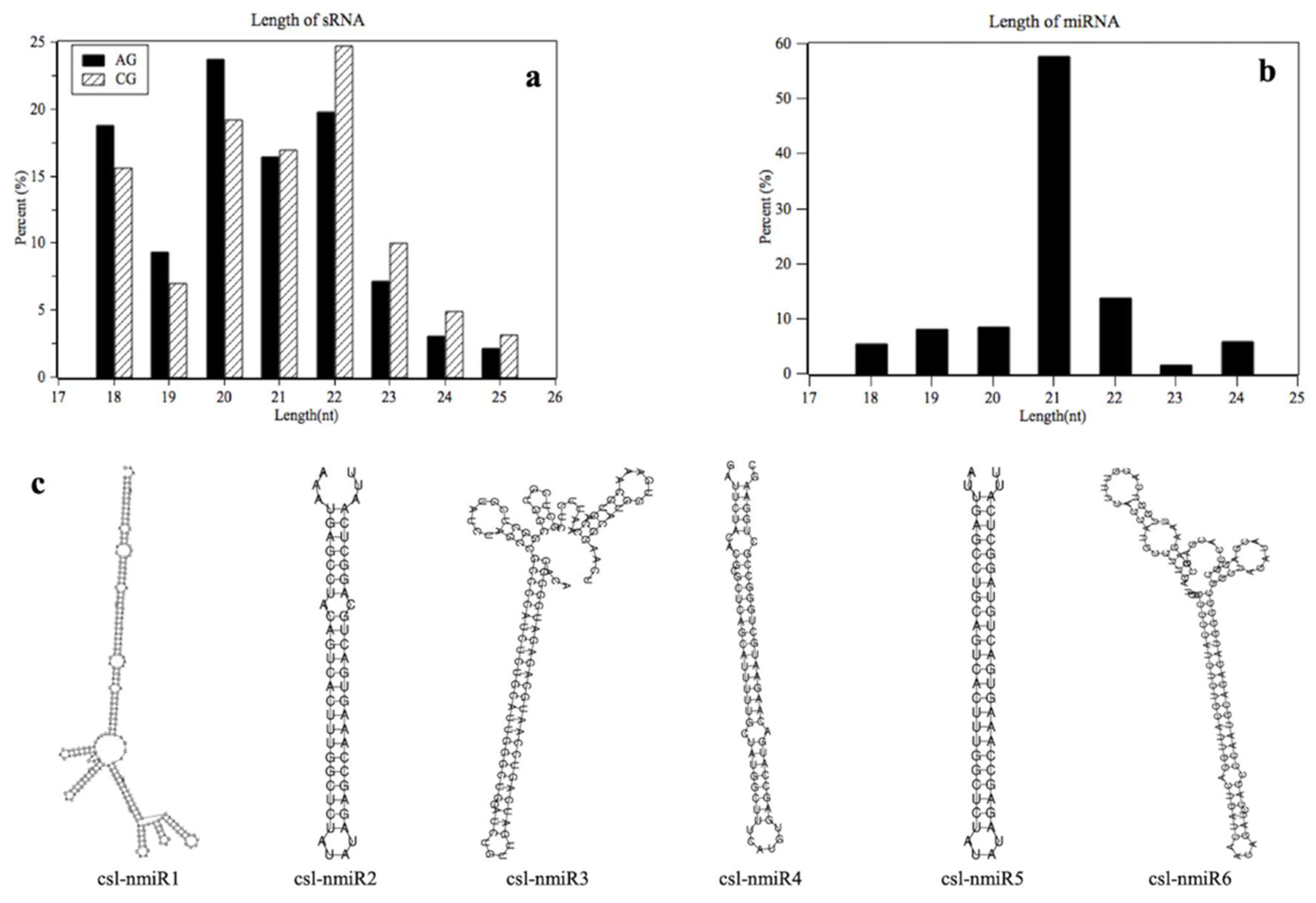

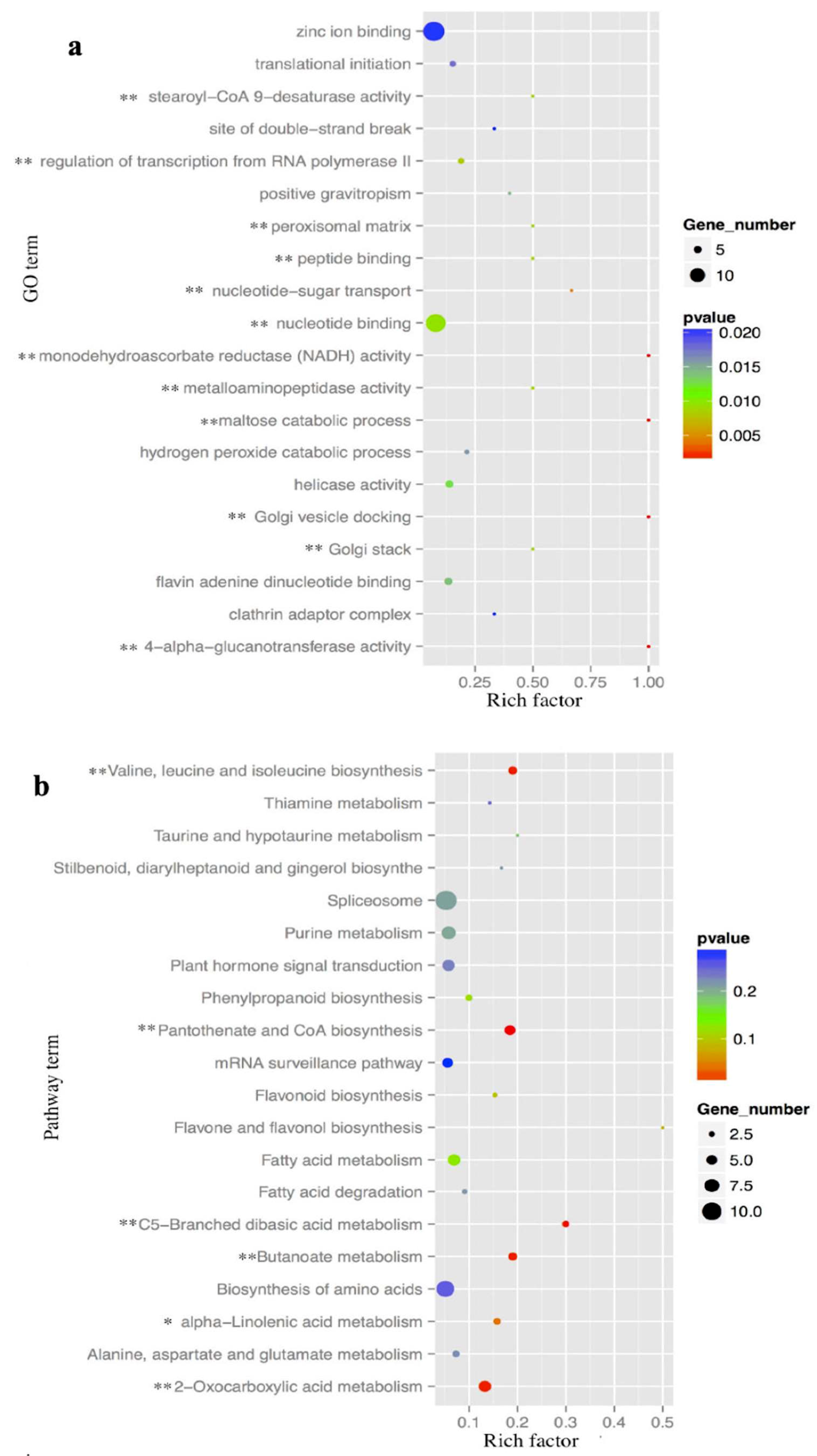
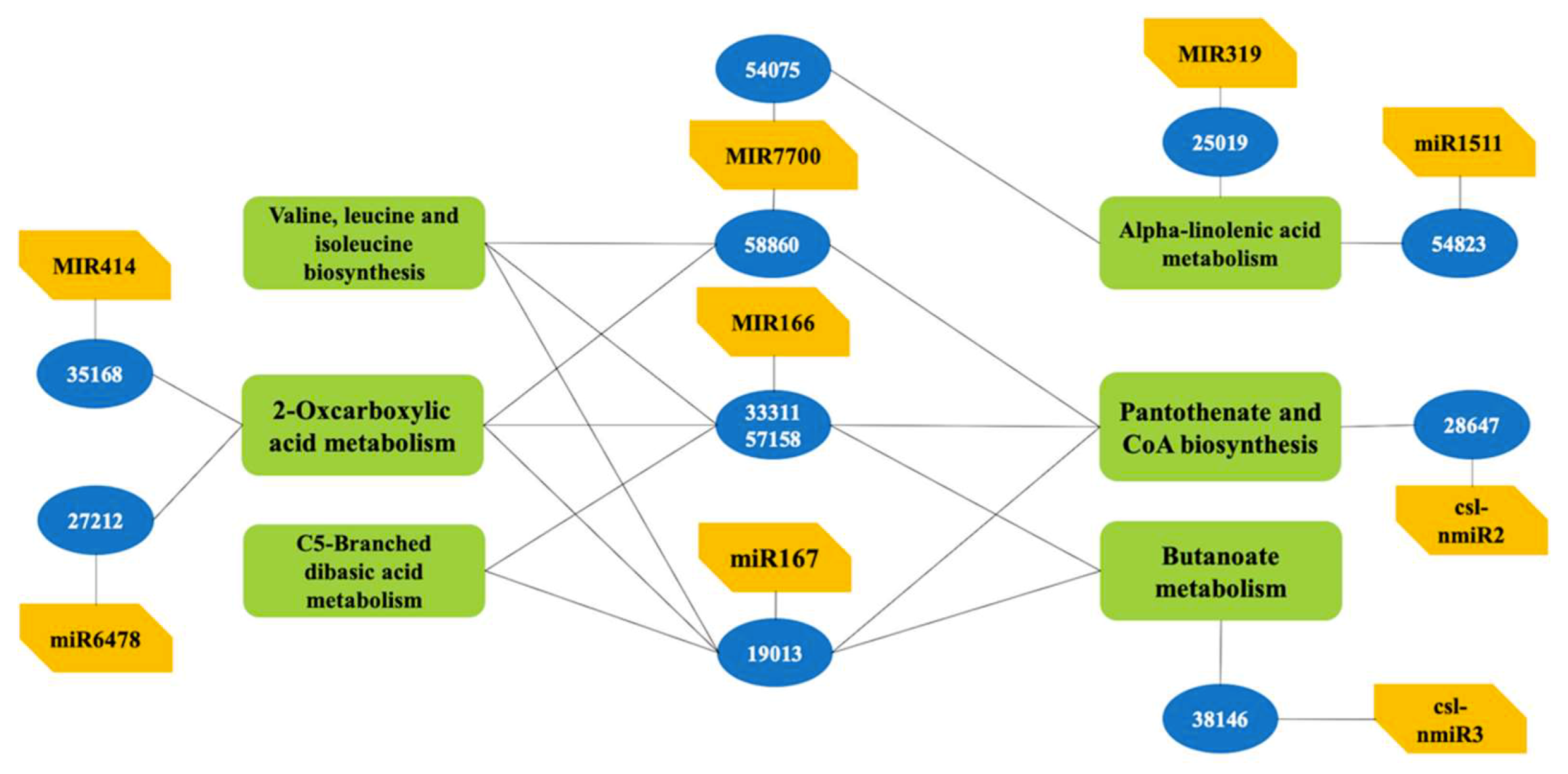
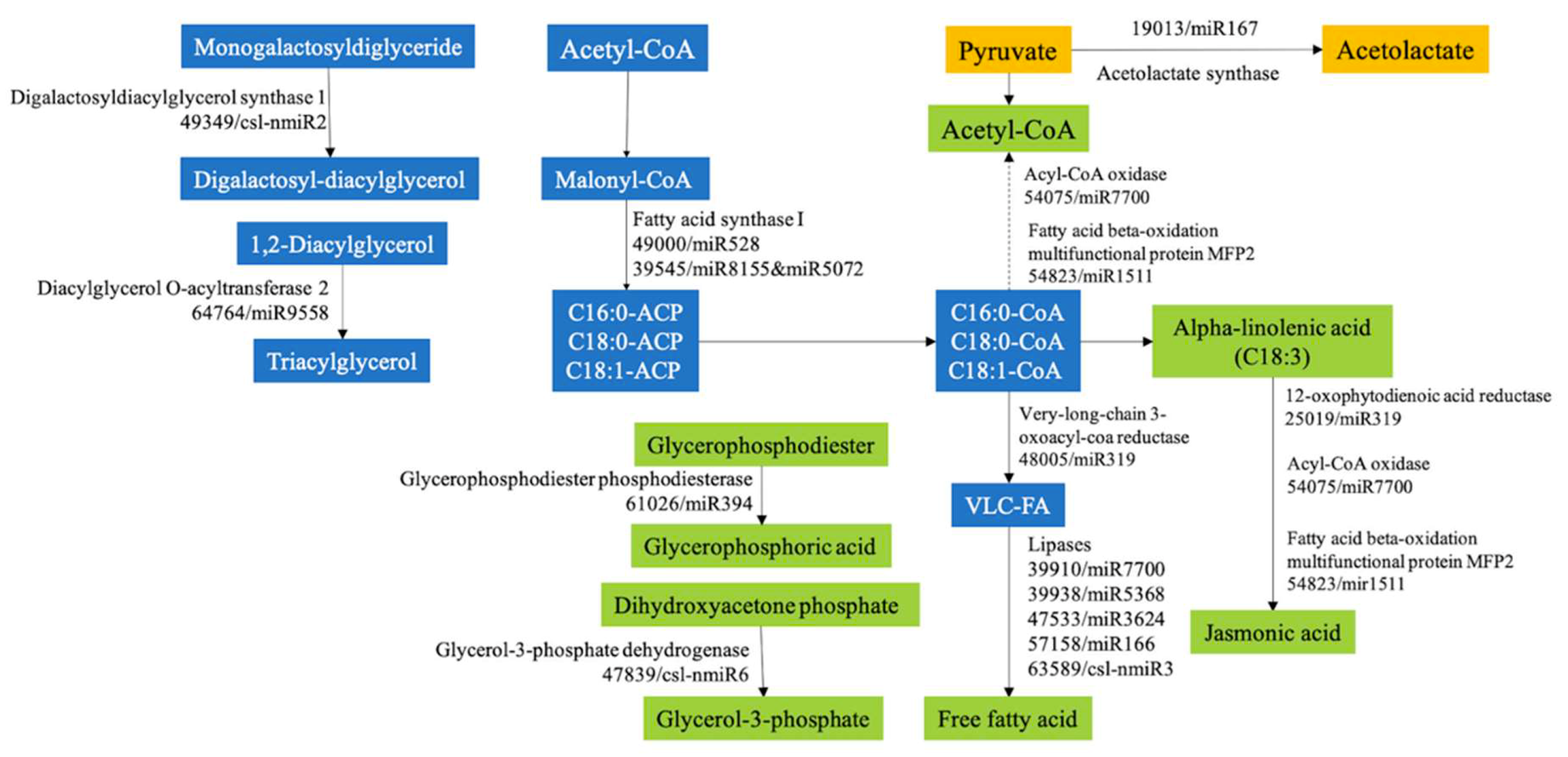
| Read | AG | CG |
|---|---|---|
| Raw data | 8409893 | 8102757 |
| 3′ adaptor sequence & length filter | 2627653 | 2254543 |
| Junk reads | 5554 | 7345 |
| Valid reads | 5142305 | 4856938 |
| Genome mapped | 2913297 | 3251099 |
| Genome mapped (%) | 56.65% | 66.94% |
| Unique reads | 137160 | 228512 |
| miRNA mapped | 24974 | 27025 |
| miRNA mapped (%) | 0.49% | 0.55% |
| Family | No of Members | miRNA Name | miRNA Sequence (5′–3′) |
|---|---|---|---|
| Conserved miRNA | 133 | ||
| miR156 | 1 | csl-miR156a | TGACAGAAGAGAGTGAGCAC |
| miR157 | 1 | csl-miR157a-5p | TTGACAGAAGATAGAGAGCAC |
| miR159 | 4 | csl-miR159a | TTTGGATTGAAGGGAGCTCTA |
| csl-miR159a-5p | AGCTCCCTTCGATCCAATC | ||
| csl-miR159a-3p | CTTGGATTGAAGGTAGCTCT | ||
| csl-miR159b | TTTGGATTGAAGGGAGCTCTATT | ||
| miR160 | 1 | csl-miR160a-3p | GCGTATGAGGAGCGAAGCATA |
| miR162 | 1 | csl-miR162a-3p | ATCGATAAACCTCTGCATCCAG |
| miR164 | 2 | csl-miR164a | TGGAGAAGCAGGGCACGTGCA |
| csl-miR164a-1 | TGTAGAAGCAGGGCACATGCC | ||
| miR166 | 11 | csl-miR166a-3p | TCGGACCAGGCTTCATTCCCC |
| csl-miR166a-5p-1 | GGAATGTTGTCTGGTTCAAGG | ||
| csl-miR166a-5p | GGAATGTTGTCTGGCTCGGGG | ||
| csl-MIR166b-3p | GATAATGATAATGATAATG | ||
| csl-MIR166b-5p | TAATGATAATGATAATGATAAT | ||
| csl-miR166e-3p | TGGAACCAGGCTTCATTCCCC | ||
| csl-miR166h-5p | GGAATGTTGGCTGGCTCGAGG | ||
| csl-miR166i-3p | TCTCGGATCAGGCTTCATTCC | ||
| csl-miR166j | CCGGACAAGGCTTCATTCCCC | ||
| csl-miR166k-5p | GGTTTGTTGTCTGGCTCGAGG | ||
| csl-miR166m | GCTGACCAGGCTTCATTCCCC | ||
| miR167 | 1 | csl-miR167e | TGAAGCTGCCAGCATGATCTT |
| miR168 | 4 | csl-miR168 | TCTCTTGGTGCAGGTCGGGAA |
| csl-miR168a-3p-1 | CCCGCCTTGCATCAACTGAAT | ||
| csl-miR168a-3p | CCCGCCTTGCACCAAGTGAAT | ||
| csl-miR168a-5p | TCGCTTGGTGCAGGTCGGGAA | ||
| miR171 | 3 | csl-miR171a | TTGAGCCGCGTCAATATCTCC |
| csl-miR171h | TGAGCCGAACCAATATCACTC | ||
| csl-miR171k-3p | TTGAGCCGCGCCAATATCACT | ||
| miR172 | 1 | csl-miR172a | AGAATCTTGATGATGCTGCAT |
| miR319 | 6 | csl-miR319 | GTGGGACTGAAGGGAGCTCCC |
| csl-MIR319a-5p | TAACACGTCCGGGTTGCT | ||
| csl-miR319a-3p | TTGGACTGAAGGGTGCTCCCT | ||
| csl-miR319a-5p | AGAGCTTCCTTCAGTCCACTCA | ||
| csl-miR319b | GCTTGGACTGAAGGGAGCTCCTTC | ||
| csl-miR319b-5p | AGAGCGTCCTTCAGTCCACTC | ||
| miR390 | 2 | csl-miR390-3p | CGCTATCTATCCTGAGCTCC |
| csl-miR390-5p | AAGCTCAGGAGGGATAGCGCC | ||
| miR393 | 1 | csl-miR393a | TCCAAAGGGATCGCATTGATT |
| miR394 | 1 | csl-miR394a-5p | TTGGCATTCTGTCCACCTCC |
| miR396 | 6 | csl-miR396a-3p-1 | GTTCAATAAAGTTGTGGGAAG |
| csl-miR396a-3p | CTCAAGAAAGCTGTGGGAGA | ||
| csl-MIR396a-5p | TTGATGATCTTCTTCAAAA | ||
| csl-miR396a-5p-1 | TTCCACAGCTTTCTTGAACTG | ||
| csl-miR396a-5p | TCCACAGGCTTTCTTGAACTG | ||
| csl-miR396b | TTCCACAGCTTTCTTGAACTTTT | ||
| miR397 | 2 | csl-miR397a | TCATTGAGTGCAGCGTTGATG |
| csl-miR397b-5p | TTGAGTGCAGCGTTGATGAACC | ||
| miR398 | 1 | csl-miR398c | CATGTGTTCTCAGGTCGCCCC |
| miR403 | 1 | csl-miR403-3p | TTAGATTCACGCACAAACTCG |
| miR408 | 1 | csl-miR408-3p | TGCACTGCCTCTTCCCTGGCT |
| miR414 | 1 | csl-MIR414-5p | ACATCATCATCGTCATCATC |
| miR444 | 1 | csl-miR444c | TGCAGTTGTTGTCTCAAGCTT |
| miR477 | 1 | csl-miR477a | ACTCTCCCTCAAGGGCTTCTGA |
| miR482 | 6 | csl-miR482-3p | TTCCCAATTCCGCCCATTCCTA |
| csl-miR482-5p | GGAATGGGCTGATTGGGAAGC | ||
| csl-miR482a-3p | TTCCCAAGCCCGCCCATTCCAA | ||
| csl-miR482a-5p | GGAATGGGCTGTTTGGGATG | ||
| csl-miR482b-3p | TCTTCCCTACACCTCCCATACC | ||
| csl-miR482c | TCTTTCCTACTCCACCCATTCC | ||
| miR528 | 1 | csl-miR528-5p | TGGAAGGGGCATGCAGAGGAG |
| miR529 | 1 | csl-miR529-5p | AGAAGAGAGAGAGTACAGCCT |
| miR535 | 1 | csl-miR535-5p | TGACAACGAGAGAGAGCACGC |
| miR812 | 1 | csl-miR812a | GACGGACGGTCAAACGTTGGACAC |
| miR827 | 1 | csl-miR827 | TTAGATGACCATCAGCAAACA |
| miR894 | 1 | csl-miR894 | CGTTTCACGTCGGGTTCACCA |
| miR946 | 1 | csl-miR946 | CAGCCCTTCTCCTATCCACAAT |
| miR1314 | 1 | csl-miR1314 | CCGGCCTCGAATGTTAGGAGAA |
| miR1423 | 1 | csl-miR1423-5p | GCAACTACACGTTGGGCGCTCGAT |
| miR1425 | 1 | csl-miR1425-5p | TAGGATTCAATCCTTGCTGCT |
| miR1448 | 1 | csl-miR1448 | TCTTTCCAACGCCTCCCATACC |
| miR1507 | 1 | csl-miR1507a | TCTCATTCCATACATCGTCTGA |
| miR1509 | 1 | csl-miR1509a | TTAATCAAGGAAATCACGGTCG |
| miR1510 | 2 | csl-miR1510b-3p | TGTTGTTTTACCTATTCCACCT |
| csl-miR1510b-5p | AGGGATAGGTAAAACAACTAC | ||
| miR1511 | 1 | csl-miR1511 | AACCCGGCTCTGATACCA |
| miR1850 | 1 | csl-miR1850.1 | TGGAAAGTTGGGAGATTGGGG |
| miR1862 | 1 | csl-miR1862e | CTAGATTTGTTTATTTTGGGACGG |
| miR1876 | 1 | csl-miR1876 | ATAAGTGGGTTTGTGGGCTGGCCC |
| miR2109 | 1 | csl-miR2109-3p | GGAGGCGTAGATACTCACACC |
| miR2118 | 4 | csl-miR2118a-3p | TTGCCGATTCCACCCATTCCTA |
| csl-miR2118b | TTCCCAATGCCTCCCATTCCTA | ||
| csl-miR2118f | TCCTGATGCCTCCCATTCCTA | ||
| csl-miR2118o | CTCCTGATGCCTCCCAAGCCTA | ||
| miR2275 | 3 | csl-miR2275b-3p | TTCAGTTTCCTCTAATATCTCG |
| csl-miR2275b-5p | AGGATTAGAGGGAACTGAACC | ||
| csl-miR2275c | AGAATTGGAGGAAAACAAACT | ||
| miR3522 | 1 | csl-miR3522 | TGAGACCAAATGAGCAGCTGA |
| miR3624 | 1 | csl-miR3624-3p | TCAGGGCAGCAGCATACTA |
| miR3630 | 1 | csl-miR3630-3p | TGGGAATCTCTCTGACGCT |
| miR4412 | 1 | csl-miR4412-5p | TGTTGCGGGTATCTTTGCCTC |
| miR4413 | 1 | csl-miR4413a | TAAGAGAATTGTAAGTCACTG |
| miR4995 | 1 | csl-miR4995 | GGCAGTGGCTTGGTTAAGGGAACC |
| miR4996 | 1 | csl-miR4996 | TAGAAGCTCCCCATGTTCTCA |
| miR5054 | 1 | csl-miR5054 | TCCCCACGGACGGCGCCA |
| miR5072 | 1 | csl-miR5072 | TTCCCCAGTGGAGTCGCCA |
| miR5077 | 1 | csl-miR5077 | GTTCACGTCGGGTTCACCA |
| miR5139 | 1 | csl-miR5139 | CGAAACCTGGCTCTGATACCA |
| miR5368 | 1 | csl-MIR5368-3p | CTGGGATTGGCTTTGGGC |
| miR5372 | 1 | csl-miR5372 | TTGTTCGATAAAACTGTTGTG |
| miR5527 | 1 | csl-miR5527 | TCTCAGCCAGGGCAGTAACAG |
| miR5530 | 1 | csl-miR5530 | AGTGGTGTCGTATTACCTGCC |
| miR5724 | 1 | csl-miR5724 | AACCGCCGGTTCGATAAT |
| miR5770 | 1 | csl-miR5770a | TTAGGACTATGGTTTGGACGA |
| miR5792 | 1 | csl-miR5792 | GATGACAGCGGTGGTTCGGACCTC |
| miR5796 | 1 | csl-miR5796 | TCATTCAGGATTGAAGCCGCC |
| miR5818 | 1 | csl-miR5818 | TCGAACTAGAAGGGCCAGGTT |
| miR6300 | 1 | csl-miR6300 | GTCGTTGTAGTATAGTGGT |
| miR6478 | 3 | csl-miR6478-2 | CCGACCTTAGCTCAGTTGGTAGA |
| csl-miR6478-1 | CCGGCCTTAGCTCAGTTGG | ||
| csl-miR6478 | CCGACTTTAGCTCAGTTGG | ||
| miR7700 | 1 | csl-MIR7700-5p | TCAGTGGTGGTGGTGGTGG |
| miR8005 | 1 | csl-MIR8005c-3p | TTTAGGGTTTAGGGTTTAGG |
| miR7972 | 1 | csl-miR7972 | TTGTCAGGCTTGTTATTCTCC |
| miR8155 | 2 | csl-miR8155-1 | GTAACCTGGCTCTGATACCA |
| csl-miR8155 | AACCTCGCTCTGATACCA | ||
| miR8175 | 1 | csl-miR8175 | TCGTTCCCCGGCAACGGCGCCA |
| miR9558 | 1 | csl-MiR9558-5p | CACTGTGGCAAAGATATG |
| Novel | 6 | ||
| csl-nmiR1 | 1 | csl-nmiR1 | AGGGCGTTCCGTCGGACGGGTT |
| csl-nmiR2 | 1 | csl-nmiR2 | GCCAAAGTGACTGCAGGCT |
| csl-nmiR3 | 1 | csl-nmiR3 | TCTCTGCATTGGGCTGGATCT |
| csl-nmiR4 | 1 | csl-nmiR4 | GCCATGACAAGAATGCTGGGCC |
| csl-nmiR5 | 1 | csl-nmiR5 | GCCTACAGTCACTTTGGCT |
| csl-nmiR6 | 1 | csl-nmiR6 | TGCAGAGATGGGACGGCT |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, R.; Chen, G.; Peng, H.; Wei, D. Identification and Characterization of MiRNAs in Coccomyxa subellipsoidea C-169. Int. J. Mol. Sci. 2019, 20, 3448. https://doi.org/10.3390/ijms20143448
Yang R, Chen G, Peng H, Wei D. Identification and Characterization of MiRNAs in Coccomyxa subellipsoidea C-169. International Journal of Molecular Sciences. 2019; 20(14):3448. https://doi.org/10.3390/ijms20143448
Chicago/Turabian StyleYang, Runqing, Gu Chen, Huifeng Peng, and Dong Wei. 2019. "Identification and Characterization of MiRNAs in Coccomyxa subellipsoidea C-169" International Journal of Molecular Sciences 20, no. 14: 3448. https://doi.org/10.3390/ijms20143448
APA StyleYang, R., Chen, G., Peng, H., & Wei, D. (2019). Identification and Characterization of MiRNAs in Coccomyxa subellipsoidea C-169. International Journal of Molecular Sciences, 20(14), 3448. https://doi.org/10.3390/ijms20143448





