Comprehensive Transcriptome Reveals an Opposite Regulatory Effect of Plant Growth Retardants in Controlling Seedling Overgrowth between Roots and Shoots
Abstract
1. Introduction
2. Results
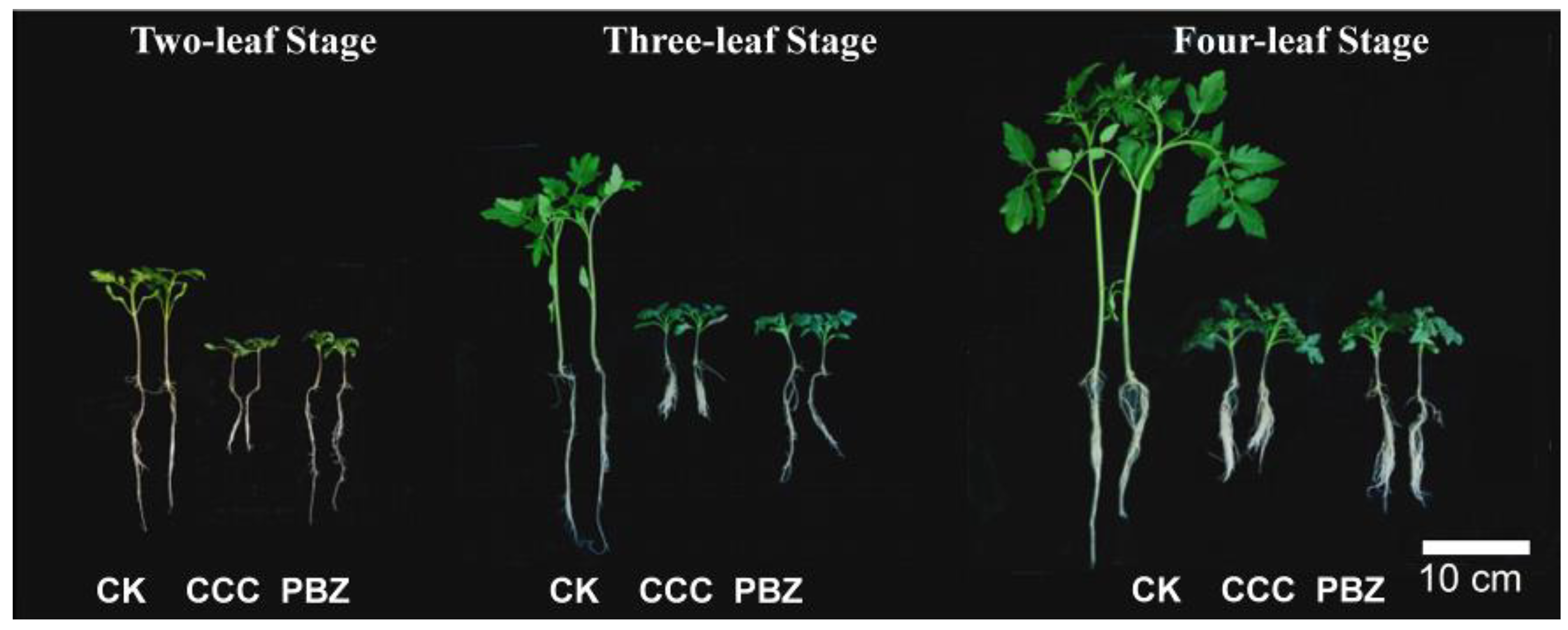
2.1. CCC and PBZ Treatments Restrain Tomato Seedlings Overgrowth
2.2. CCC and PBZ Increased the Root Diameter in Tomato Seedlings
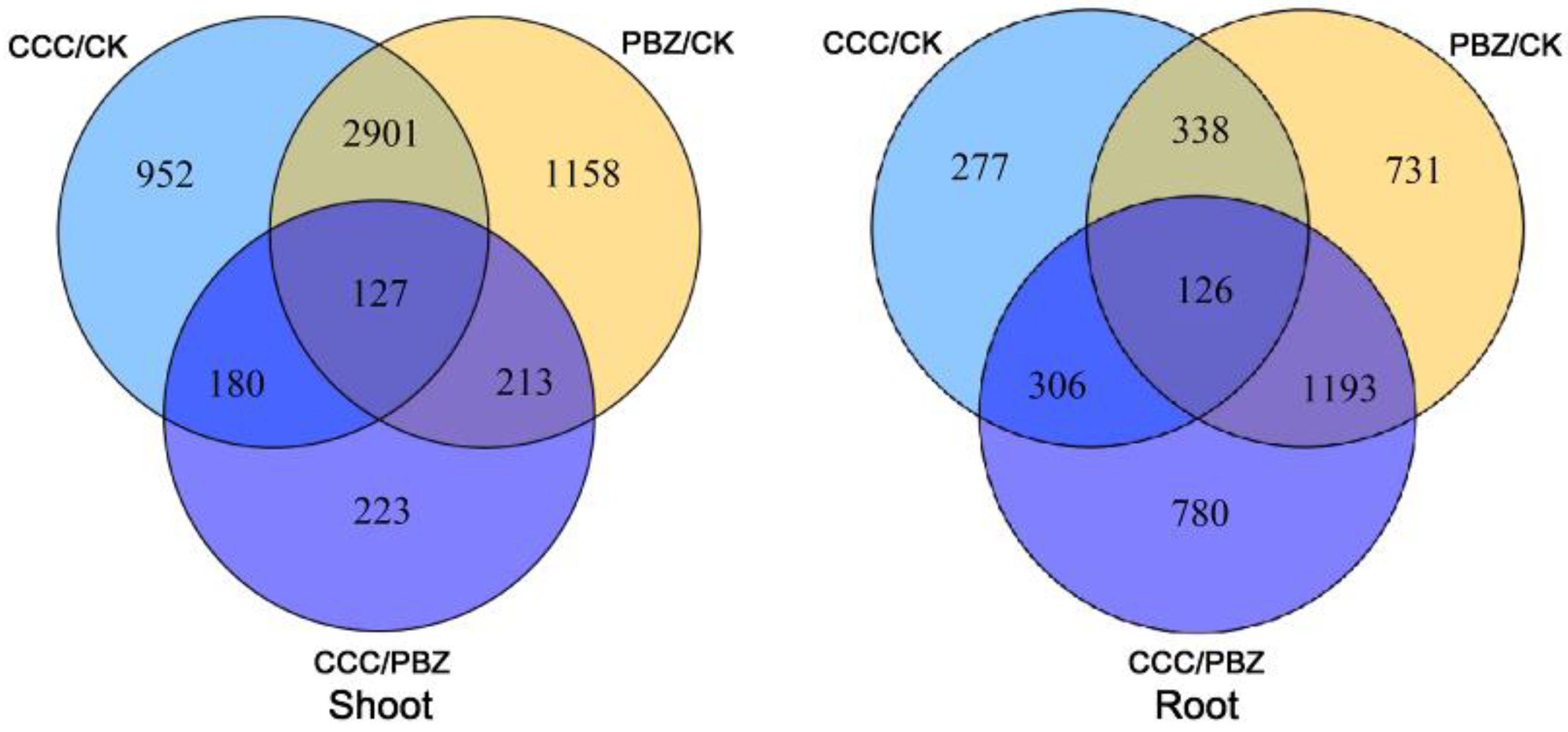
2.3. Transcriptome Profiling Analysis in CCC/PBZ Controlling Tomato Overgrowth
2.4. Plant Growth Retardants Induced Differentially Expressed Genes in GA Metabolism and the Signaling Pathway
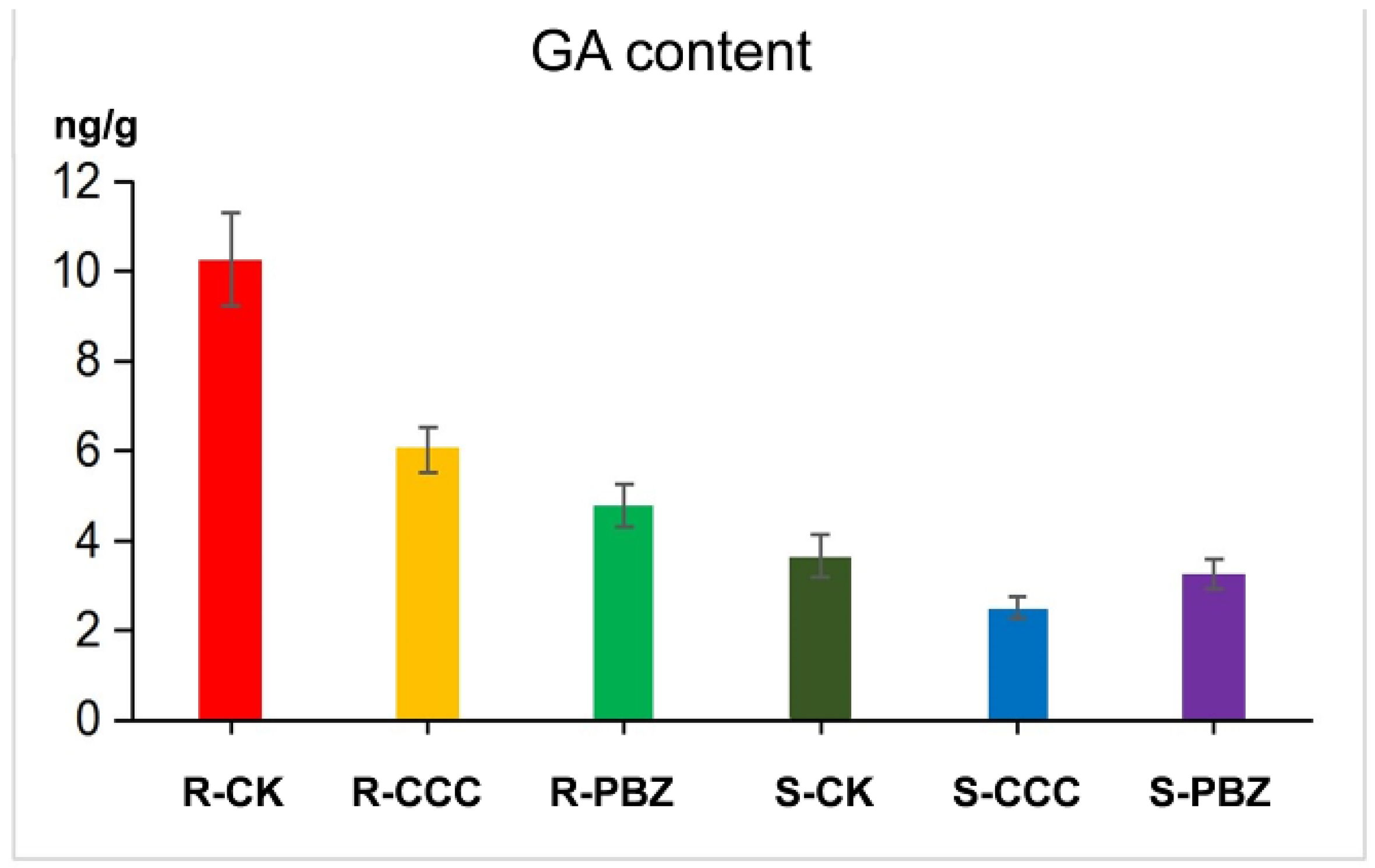
2.5. Cross Talk of Plant Growth Retardants and Plant Hormone GA in Controlling Overgrowth
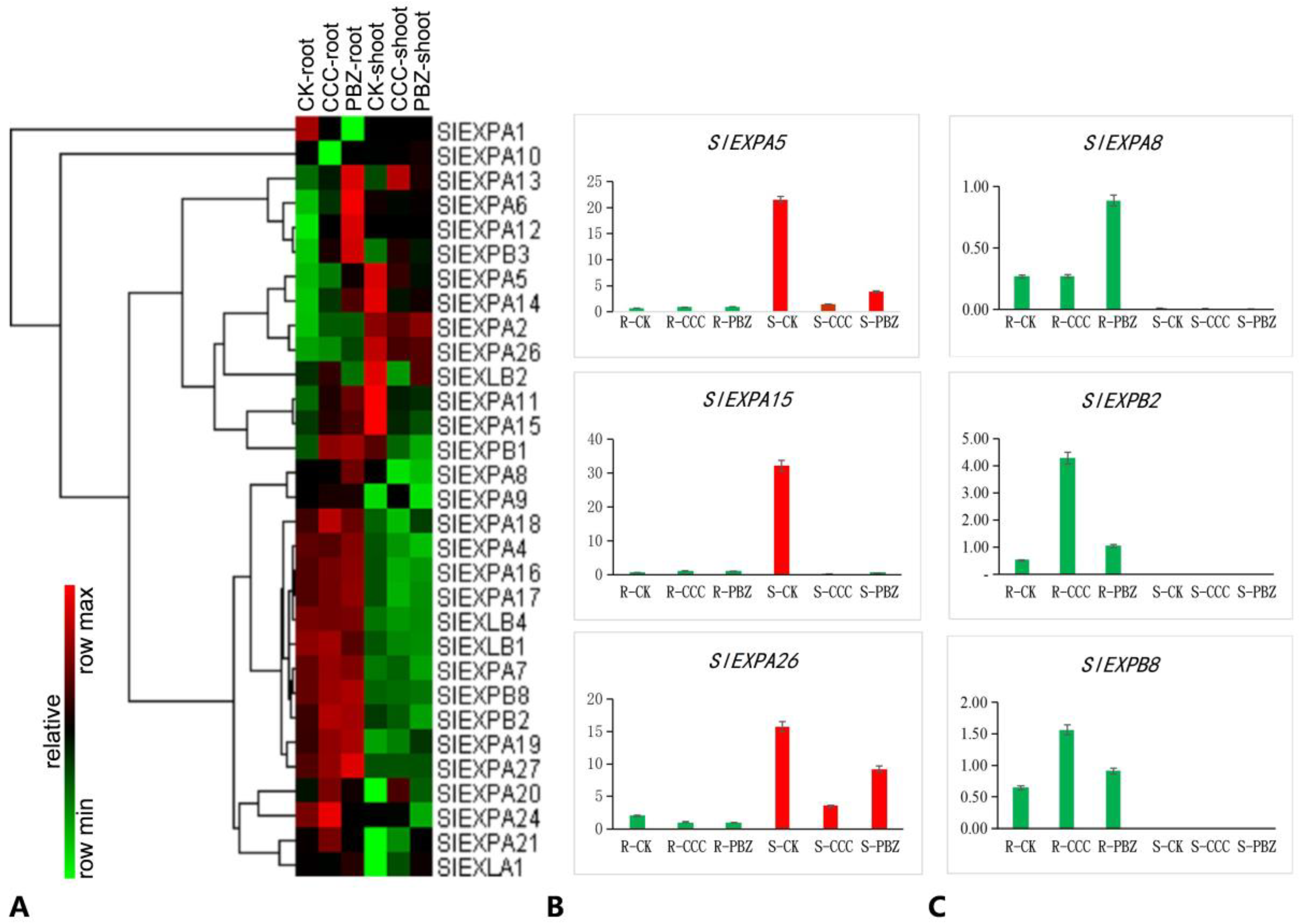
2.6. Plant Growth Retardants Caused an Opposite Regulation of Tissue-Specific SlEXPs Genes in Restraining Tomato Seedling Overgrowth
3. Discussion
3.1. Plant Growth Retardants Restrain GA Biosynthesis in Controlling Overgrowth
3.2. Plant Growth Retardants Regulate GA Signaling in an Opposite Way between Roots and Shoots
3.3. Plant Growth Retardants Control Overgrowth through the Tissue-Specific SlEXPs Genes
4. Materials and Methods
4.1. Plant Materials
4.2. Cytohistological Observations of Seedling Roots after PBZ and CCC Treatment
4.3. Total RNA Extraction and RNA-Seq Library Construction and Sequencing
4.4. Differentially Expressed Genes Affected by PBZ and CCC Treatment
4.5. Validation of Significantly Differentially Expressed Genes
4.6. Quantification of Endogenous GA Content by Enzyme-Linked Immunosorbent Assay (ELISA)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ilias, F.I.; Rajapakse, N. Prohexadione-calcium affects growth and flowering of petunia and impatiens grown under photoselective films. Sci. Hortic. 2005, 106, 190–202. [Google Scholar] [CrossRef]
- Berghage, R. Controlling height with temperature. HortTechnology 1998, 8, 535–539. [Google Scholar] [CrossRef]
- Maloupa-Ikonomou, E.; Jacques, M. On the flowering of weigela florida cultivar bristol ruby. Bull. De La Soc. Bot. De Fr. Lett. Bot. 1988, 135, 169–179. [Google Scholar]
- Roelofse, E.W.; Hand, D.W.; Hall, R.L. The effect of daylength on the development of glasshouse celery. J. Hortic. Sci. 1989, 64, 283–292. [Google Scholar] [CrossRef]
- Garner, L.C.; Bjorkman, T. Mechanical conditioning for controlling excessive elongation in tomato transplants: Sensitivity to dose, frequency and timing of brushing. Am. Soc. Hortic. Sci. 1996, 121, 894–900. [Google Scholar] [CrossRef]
- Samimy, C. Physical impedance retards top growth of tomato transplants. HortScience 1993, 28, 883–885. [Google Scholar] [CrossRef]
- Pasian, C.C.; Bennett, M.A. Paclobutrazol soaked marigold, geranium, and tomato seeds produce short seedlings. HortScience 2001, 36, 721–723. [Google Scholar] [CrossRef]
- Zayed, A.E.; El-Zawily, I.A.; Ibrahim, A.S. Growth, yield and chemical composition of okra plants as affected by some growth regulators. Angew. Bot. 1985, 59, 199–208. [Google Scholar]
- Syed, A.; Hussain, S.A.; Nawab, A.; Asghar, S.; Ali, N. Effect of exogenous growth regulators on growth, flowering and yield of okra (Abelmoschus esculentus L.). Sarhad J. Agric. 1997, 13, 449–453. [Google Scholar]
- Bhatt, R.M.; Srinivasa Rao, N.K. Physiology of Okra: Crop growth, development and yield. In Okra Handbook: Global Production, Processing and Crop Improvement; Dhankhar, B.S., Singh, R., Eds.; HNB Publishing: New York, NY, USA, 2009; pp. 37–60. [Google Scholar]
- Jiang, B.B.; Chen, S.M.; Miao, H.B.; Zhang, S.M.; Chen, F.D.; Fang, W.M. Changes of edogenous hormone levels during short-day inductive floral initiation and inflorescence differentiation of Chrysanthemum morifolium ‘Jingyun’. Int. J. Plant Prod. 2010, 4, 149–157. [Google Scholar]
- Rademacher, W. Growth retardants: Effects of gibberellin biosynthesis and other metabolic pathways. Annu. Rev. Plant Physiol. Mol. Biol. 2000, 51, 501–531. [Google Scholar] [CrossRef] [PubMed]
- Hedden, P.; Graebe, J.E. Inhibition of gibberellin biosynthesis by paclobutrozol in cell-free homogenates of Cucurbita maxima endosperm and Malus pumila embryos. J. Plant Growth Regul. 1985, 4, 111–122. [Google Scholar] [CrossRef]
- Hedden, P.; Thomas, S.G. Gibberellin biosynthesis and its regulation. Biochem. J. 2012, 444, 11–25. [Google Scholar] [CrossRef] [PubMed]
- Birnbaum, K.; Shasha, D.E.; Wang, J.Y.; Jung, J.W.; Lambert, G.M.; Galbraith, D.W.; Benfey, P.N. A gene expression map of the Arabidopsis root. Science 2003, 302, 1956–1960. [Google Scholar] [CrossRef] [PubMed]
- Desgagné-Penix, I.; Eakanunkul, S.; Coles, J.P.; Phillips, A.L.; Hedden, P.; Sponsel, V.M. The auxin transport inhibitor response3 (tir3) allele of BIG and auxin transport inhibitors affect the gibberellin status of Arabidopsis. Plant J. 2005, 41, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, M.; Itoh, H.; Inukai, Y.; Sakamoto, T.; Ueguchi-Tanaka, M.; Ashikari, M.; Matsuoka, M. Where do gibberellin biosynthesis and gibberellin signaling occur in rice plants? Plant J. 2003, 35, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Shani, E. Gibberellins accumulate in the elongating endodermal cells of Arabidopsis root. Proc. Natl. Acad. Sci. USA 2013, 110, 4834–4839. [Google Scholar] [CrossRef]
- Löfke, C.; Zwiewka, M.; Heilmann, I.; Van Montagu, M.C.; Teichmann, T.; Friml, J. Asymmetric gibberellin signaling regulates vacuolar trafficking of PIN auxin transporters during root gravitropism. Proc. Natl. Acad. Sci. USA 2013, 110, 3627–3632. [Google Scholar]
- Nelissen, H.; Rymen, B.; Jikumaru, Y.; Demuynck, K.; Van Lijsebettens, M.; Kamiya, Y.; Inzé, D. Beemster GT: A local maximum in gibberellin levels regulates maize leaf growth by spatial control of cell division. Curr. Biol. 2012, 22, 1183–1187. [Google Scholar] [CrossRef]
- Bai, M.Y.; Shang, J.X.; Oh, E.; Fan, M.; Bai, Y.; Zentella, R.; Sun, T.P.; Wang, Z.Y. Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 2012, 14, 810–817. [Google Scholar] [CrossRef]
- Zhang, L.Y.; Bai, M.Y.; Wu, J.; Zhu, J.Y.; Wang, H.; Zhang, Z.; Wang, W.; Sun, Y.; Zhao, J.; Sun, X.; et al. Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 2009, 21, 3767–3780. [Google Scholar] [CrossRef] [PubMed]
- Alabadí, D.; Gallego-Bartolomé, J.; Orlando, L.; García-Cárcel, L.; Rubio, V.; Martínez, C.; Frigerio, M.; Iglesias-Pedraz, J.M.; Espinosa, A.; Deng, X.W.; et al. Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness. Plant J. 2008, 53, 324–335. [Google Scholar] [CrossRef] [PubMed]
- Jing, Y.; Zhang, D.; Wang, X.; Tang, W.; Wang, W.; Huai, J.; Xu, G.; Chen, D.; Li, Y.; Lin, R. Arabidopsis chromatin remodeling factor PICKLE interacts with transcription factor HY5 to regulate hypocotyl cell elongation. Plant Cell 2013, 25, 242–256. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.L.; Yao, J.; Mei, C.S.; Tong, X.H.; Zeng, L.J.; Li, Q.; Xiao, L.T.; Sun, T.P.; Li, J.; Deng, X.W.; et al. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. USA 2012, 109, 1192–1200. [Google Scholar] [CrossRef] [PubMed]
- Murti, G.S.R.; Upreti, K.K. Plant hormones. In Advances in Plant Physiology; Hemantaranjan, A., Ed.; Scientific Publishers: Jodhpur, India, 2000; Volume 3, pp. 109–148. [Google Scholar]
- Wang, E.T.; Sandberg, R.; Luo, S.; Khrebtukova, I.; Zhang, L.; Mayr, C.; Kingsmore, S.F.; Schroth, G.P.; Burge, C.B. Alternative isoform regulation in human tissue transcriptomes. Nature 2008, 456, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Kende, H.; Bradford, K.; Brummell, D.; Cho, H.T.; Cosgrove, D.J.; Fleming, A.J.; Gehring, C.; Lee, Y.; McQueen-Mason, S.; Rose, J.K.C.; et al. Nomenclature for members of the expansin superfamily of genes and proteins. Plant Mol. Biol. 2004, 55, 311–314. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wei, T.; Wang, X.; Zhang, L.; Yang, M.; Chen, L.; Song, W.; Wang, C.; Chen, C. Transcriptome analyses from mutant Salvia miltiorrhiza reveals important roles for SmGASA4 during plant development. Int. J. Mol. Sci. 2018, 19, 2088. [Google Scholar] [CrossRef] [PubMed]
- Xiang, J.; Wu, H.; Zhang, Y.; Zhang, Y.; Wang, Y.; Li, Z.; Lin, H.; Chen, H.; Zhang, J.; Zhu, D. Transcriptomic analysis of gibberellin- and paclobutrazol-treated rice seedlings under submergence. Int. J. Mol. Sci. 2017, 18, 2225. [Google Scholar] [CrossRef]
- Guo, H.; Wang, Y.; Liu, H.; Hu, P.; Jia, Y.; Zhang, C.; Wang, Y.; Gu, S.; Yang, C.; Wang, C. Exogenous GA3 application enhances xylem development and induces the expression of secondary wall biosynthesis related genes in Betula Platyphylla. Int. J. Mol. Sci. 2015, 16, 22960–22975. [Google Scholar] [CrossRef]
- Claeys, H.; De Bodt, S.; Inze, D. Gibberellins and DELLAs: Central nodes in growth regulatory networks. Trends Plant Sci. 2014, 19, 231. [Google Scholar] [CrossRef]
- Ikeda, M.; Fujiwara, S.; Mitsuda, N.; Takagi, M.O. A Triantagonistic Basic Helix-Loop-Helix System Regulates Cell Elongation in Arabidopsis. Plant Cell 2012, 24, 4483–4497. [Google Scholar] [CrossRef] [PubMed]
- Huq, E.; Sady, B.A.; Hudson, M.; Kim, C.; Apel, K.; Peter, H. Quail Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 2004, 305, 24. [Google Scholar] [CrossRef] [PubMed]
- Bu, Q.; Castillon, A.; Chen, F.; Zhu, L.; Huq, E. Dimerization and blue light regulation of PIF1 interacting bHLH proteins in Arabidopsis. Plant Mol. Biol. 2011, 77, 501–511. [Google Scholar] [CrossRef] [PubMed]
- Soy, J.; Leivar, P.; Monte, E. PIF1 promotes phytochrome-regulated growth under photoperiodic conditions in Arabidopsis together with PIF3, PIF4, and PIF5. J. Exp. Bot. 2014, 65, 2925–2936. [Google Scholar] [CrossRef] [PubMed]
- Zhong, S.; Joung, J.G.; Zheng, Y.; Chen, Y.R.; Liu, B.; Shao, Y.; Xiang, J.Z.; Fei, Z.; Giovannoni, J.J. High-throughput illumina strand-specific RNA sequencing library preparation. Cold Spring Harb. Protoc. 2011, 2011, 940–949. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, J.; Wang, Z.; Zhu, Q.; Wang, W. Hormonal changes in the grains of rice subjected to water stress during grain filling. Plant Physiol. 2001, 127, 315–323. [Google Scholar] [CrossRef]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, Y.; Chen, G.; Zheng, X.; Zhong, Q.; Zhang, M.; Wu, Z.; Wen, C.; Liu, M. Comprehensive Transcriptome Reveals an Opposite Regulatory Effect of Plant Growth Retardants in Controlling Seedling Overgrowth between Roots and Shoots. Int. J. Mol. Sci. 2019, 20, 3307. https://doi.org/10.3390/ijms20133307
Ji Y, Chen G, Zheng X, Zhong Q, Zhang M, Wu Z, Wen C, Liu M. Comprehensive Transcriptome Reveals an Opposite Regulatory Effect of Plant Growth Retardants in Controlling Seedling Overgrowth between Roots and Shoots. International Journal of Molecular Sciences. 2019; 20(13):3307. https://doi.org/10.3390/ijms20133307
Chicago/Turabian StyleJi, Yanhai, Guanxing Chen, Xuyang Zheng, Qiwen Zhong, Mingyun Zhang, Zhanhui Wu, Changlong Wen, and Mingchi Liu. 2019. "Comprehensive Transcriptome Reveals an Opposite Regulatory Effect of Plant Growth Retardants in Controlling Seedling Overgrowth between Roots and Shoots" International Journal of Molecular Sciences 20, no. 13: 3307. https://doi.org/10.3390/ijms20133307
APA StyleJi, Y., Chen, G., Zheng, X., Zhong, Q., Zhang, M., Wu, Z., Wen, C., & Liu, M. (2019). Comprehensive Transcriptome Reveals an Opposite Regulatory Effect of Plant Growth Retardants in Controlling Seedling Overgrowth between Roots and Shoots. International Journal of Molecular Sciences, 20(13), 3307. https://doi.org/10.3390/ijms20133307




