The TAFs of TFIID Bind and Rearrange the Topology of the TATA-Less RPS5 Promoter
Abstract
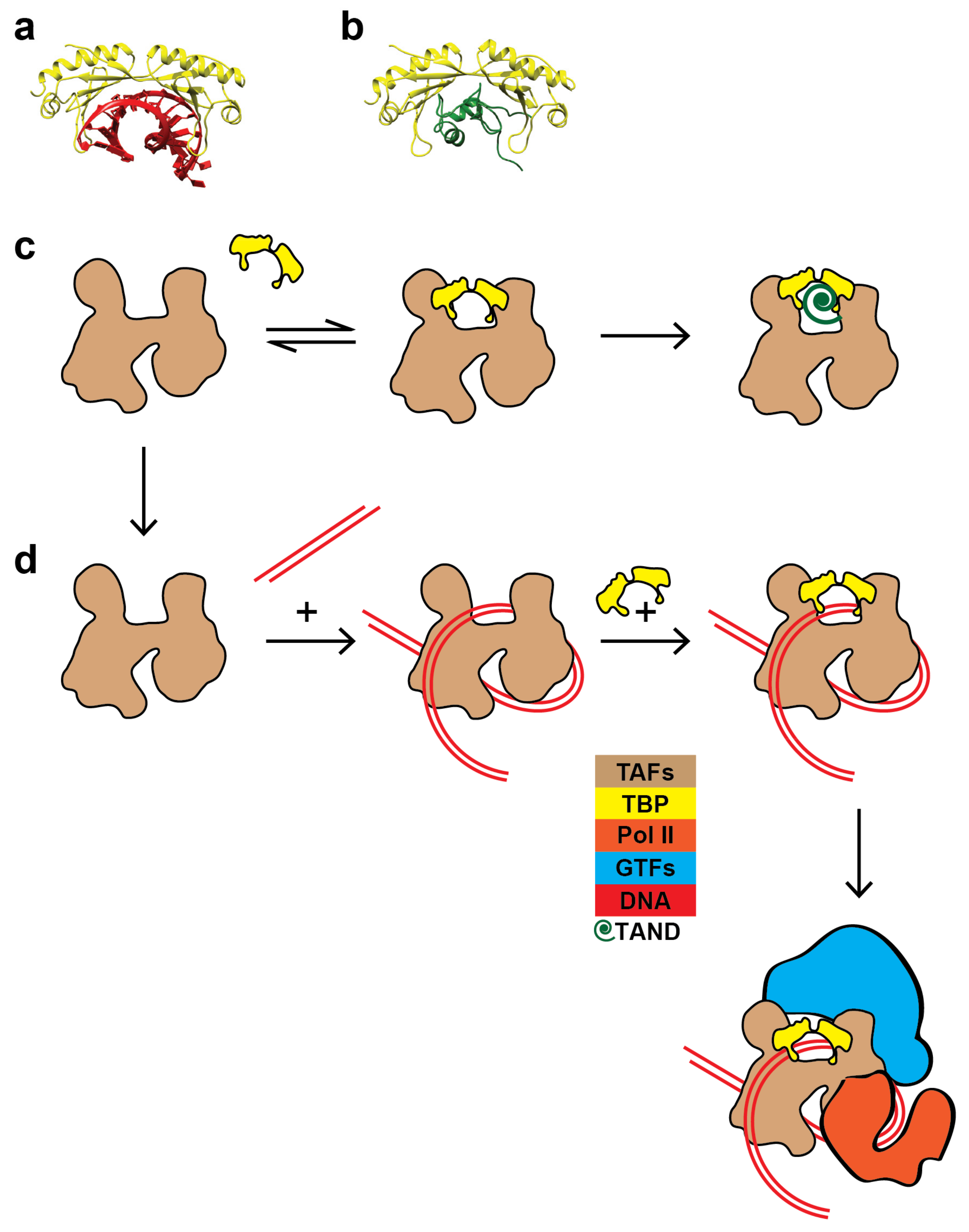
1. Introduction
2. Results
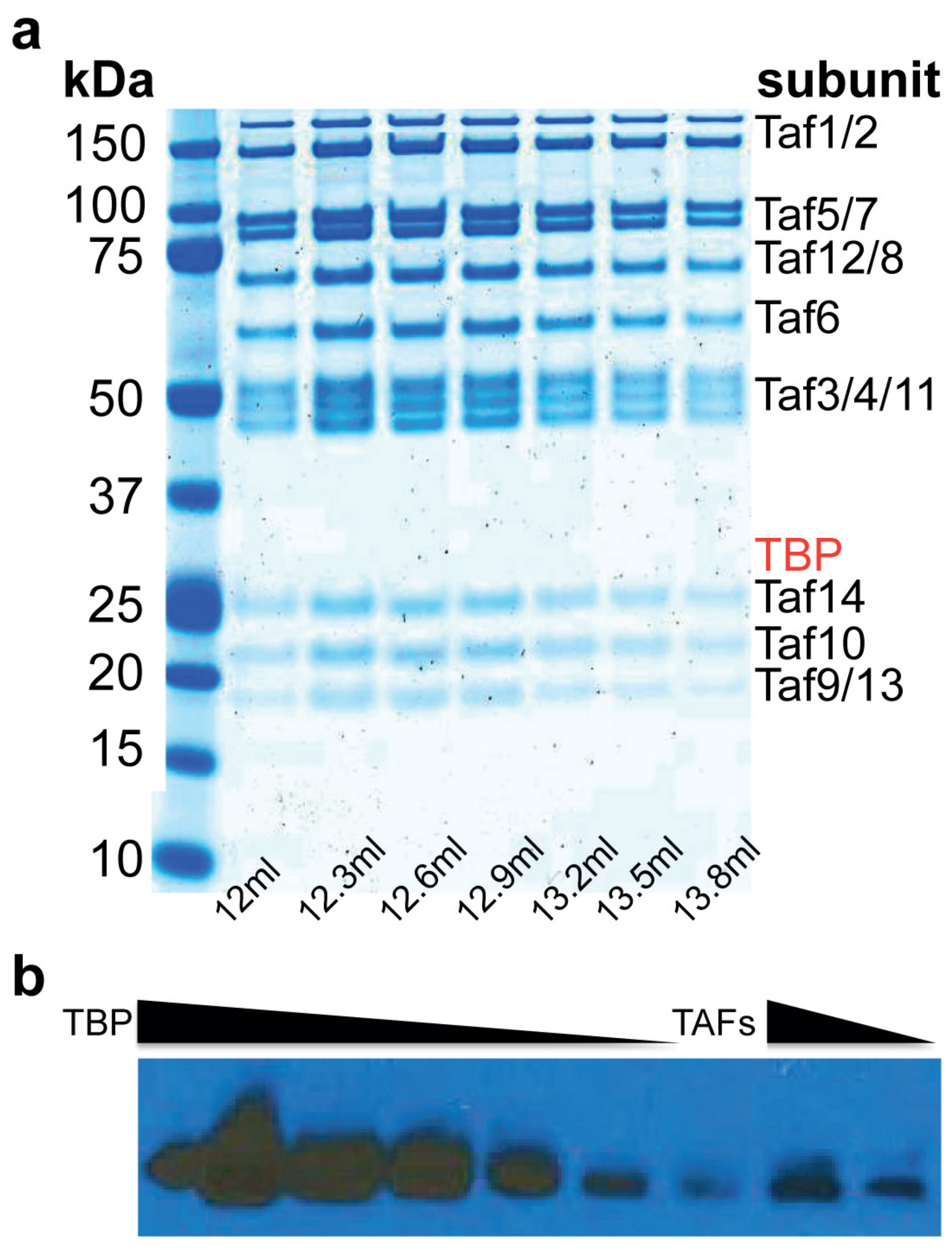
2.1. Purification of TFIID∆TBP (TAF Complex)
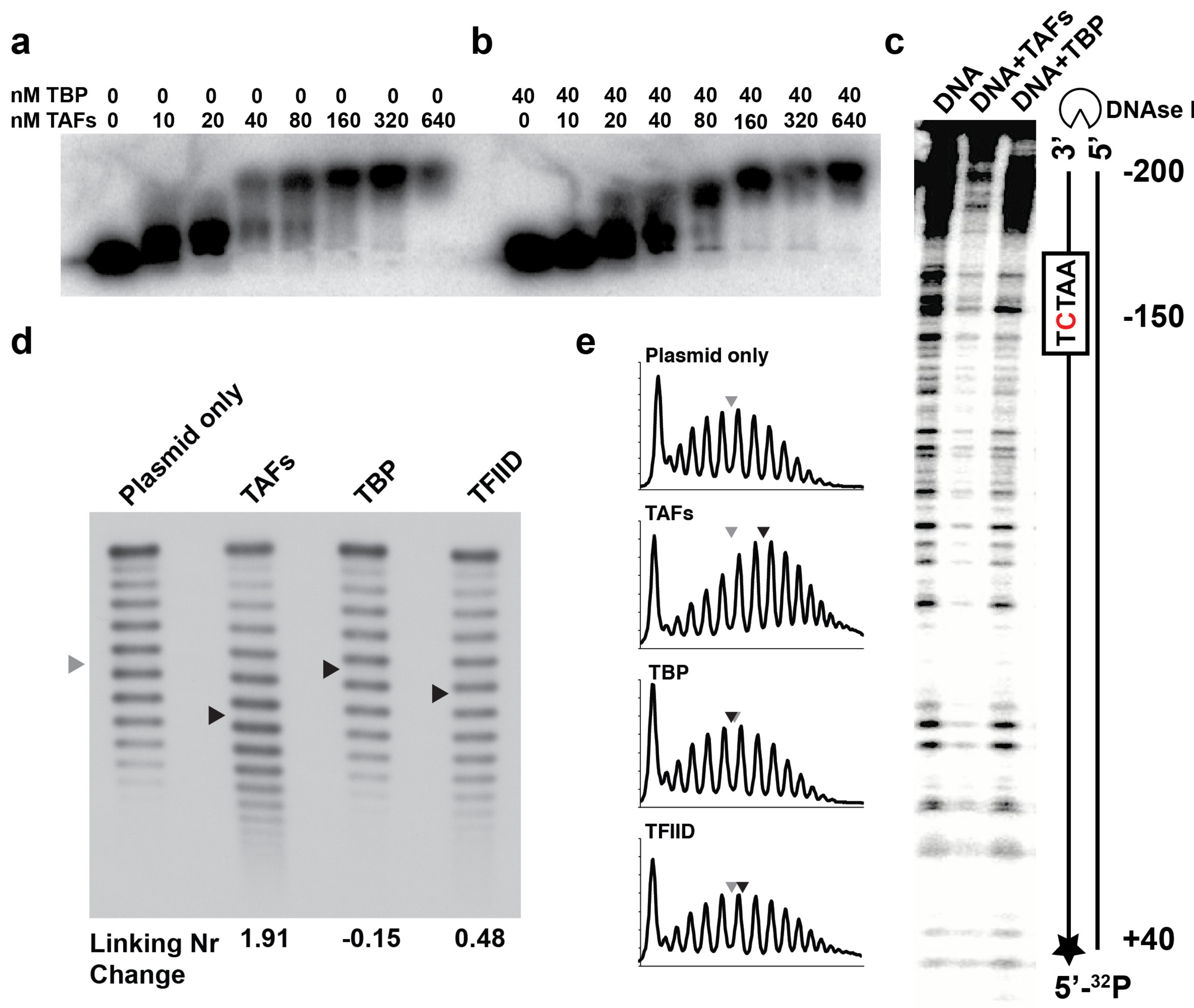
2.2. The TAFs Alone Bind and Introduce Significant Topological Changes to the TATA-Less RPS5 Promoter
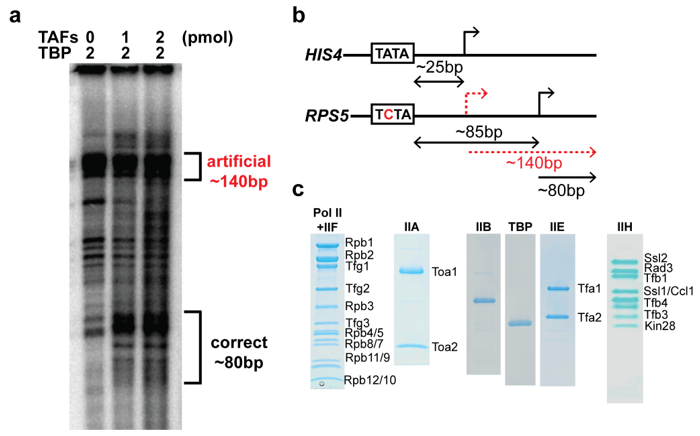
2.3. The TAF Complex Provides the Nucleation Point for Correct Housekeeping Gene Transcription In Vitro
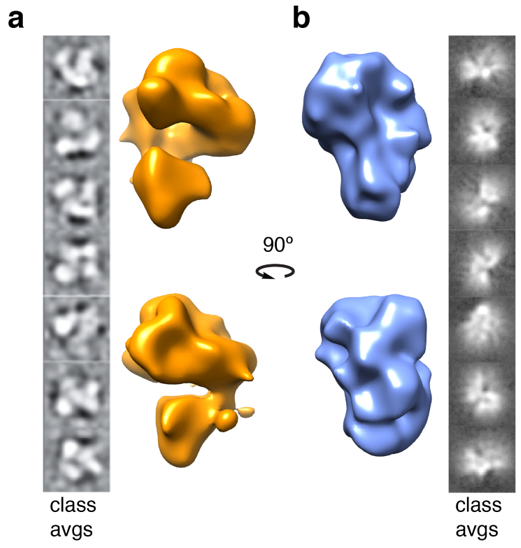
2.4. Structural Rearrangements of DNA Bound and Unbound TAF Complexes
3. Discussion
4. Materials and Methods
4.1. Oligonucleotides
4.2. Protein Purification
4.3. Quantitation of the Amount of TBP in the TAF Complex Preparation
4.4. Electrophoretic Mobility Shift Assay
4.5. DNase I Footprinting
4.6. Topological Analysis
4.7. Run-Off Transcription Assay
4.8. 3D Reconstructions of Apo-TAFs and TAFs–DNA Complexes
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| bp | Base pairs |
| EMSA | Electrophoretic mobility gel-shift assay |
| GTFs | General transcription factors |
| ΔLk | Linking number change |
| PIC | Pre-initiation complex |
| Pol II | RNA polymerase II |
| RPS5 | Ribosomal protein 5 |
| TAFs | TBP-associated factors |
| TAND | N-terminal domain of TAF1 |
| TBP | TATA-box binding protein |
| TSS | Transcription start site |
References
- Conaway, R.C.; Conaway, J.W. General initiation factors for RNA polymerase II. Annu. Rev. Biochem. 1993, 62, 161–190. [Google Scholar] [CrossRef] [PubMed]
- Kornberg, R.D. The molecular basis of eukaryotic transcription. Proc. Natl. Acad. Sci. USA 2007, 104, 12955–12961. [Google Scholar] [CrossRef] [PubMed]
- Basehoar, A.D.; Zanton, S.J.; Pugh, B.F. Identification and distinct regulation of yeast TATA box-containing genes. Cell 2004, 116, 699–709. [Google Scholar] [CrossRef]
- Yang, C.; Bolotin, E.; Jiang, T.; Sladek, F.M.; Martinez, E. Prevalence of the initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters. Gene 2007, 389, 52–65. [Google Scholar] [CrossRef]
- Lee, D.H.; Gershenzon, N.; Gupta, M.; Ioshikhes, I.P.; Reinberg, D.; Lewis, B.A. Functional characterization of core promoter elements: The downstream core element is recognized by TAF1. Mol. Cell. Biol. 2005, 25, 9674–9686. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Y.; Bhaumik, S.R.; Green, M.R. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science 2000, 288, 1242–1244. [Google Scholar] [CrossRef]
- Kim, Y.; Geiger, J.H.; Hahn, S.; Sigler, P.B. Crystal structure of a yeast TBP/TATA-box complex. Nature 1993, 365, 512–520. [Google Scholar] [CrossRef]
- Kamenova, I.; Warfield, L.; Hahn, S. Mutations on the DNA binding surface of TBP discriminate between yeast TATA and TATA-less gene transcription. Mol. Cell. Biol. 2014, 34, 2929–2943. [Google Scholar] [CrossRef]
- Kuras, L.; Kosa, P.; Mencia, M.; Struhl, K. TAF-Containing and TAF-independent forms of transcriptionally active TBP in vivo. Science 2000, 288, 1244–1248. [Google Scholar] [CrossRef]
- Rhee, H.S.; Pugh, B.F. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature 2012, 483, 295–301. [Google Scholar] [CrossRef]
- Warfield, L.; Ramachandran, S.; Baptista, T.; Devys, D.; Tora, L.; Hahn, S. Transcription of nearly all yeast RNA polymerase II-transcribed genes is dependent of transcription factor TFIID. Mol. Cell 2017, 68, 118–129. [Google Scholar] [CrossRef]
- Vinayachandran, V.; Reja, R.; Rossi, M.J.; Park, B.; Rieber, L.; Mittal, C.; Mahony, S.; Pugh, B.F. Widespread and precise reprogramming of yeast protein-genome interactions in response to heat shock. Genome Res. 2018, 28, 357–366. [Google Scholar] [CrossRef]
- Sugihara, F.; Kasahara, K.; Kokubo, T. Highly redundant function of multiple AT-rich sequences as core promoter elements in the TATA-less RPS5 promoter of Saccharomyces cerevisiae. Nucleic Acids Res. 2011, 39, 59–75. [Google Scholar] [CrossRef]
- Pugh, B.F.; Tjian, R. Transcription from a TATA-less promoter requires a multisubunit TFIID complex. Genes Dev. 1991, 5, 1935–1945. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Lieberman, P.M.; Boyer, T.G.; Berk, A.J. Holo-TFIID supports transcriptional stimulation by diverse activators and from a TATA-less promoter. Genes Dev. 1992, 6, 1964–1974. [Google Scholar] [CrossRef] [PubMed]
- Guermah, M.; Tao, Y.; Roeder, R.G. Positive and negative TAF(II) functions that suggest a dynamic TFIID structure and elicit synergy with TRAPs in activator-induced transcription. Mol. Cell. Biol. 2001, 21, 6882–6894. [Google Scholar] [CrossRef] [PubMed]
- Sanders, S.L.; Garbett, K.A.; Weil, P.A. Molecular characterization of Saccharomyces cerevisiae TFIID. Mol. Cell. Biol. 2002, 22, 6000–6013. [Google Scholar] [CrossRef]
- Liu, W.L.; Coleman, R.A.; Grob, P.; King, D.S.; Florens, L.; Washburn, M.P.; Geles, K.G.; Yang, J.L.; Ramey, V.; Nogales, E.; et al. Structural changes in TAF4b-TFIID correlate with promoter selectivity. Mol. Cell 2008, 29, 81–91. [Google Scholar] [CrossRef]
- Layer, J.H.; Miller, S.G.; Weil, P.A. Direct transactivator-transcription factor IID (TFIID) contacts drive yeast ribosomal protein gene transcription. J. Biol. Chem. 2010, 285, 15489–15499. [Google Scholar] [CrossRef]
- Elmlund, H.; Baraznenok, V.; Linder, T.; Szilagyi, Z.; Rofougaran, R.; Hofer, A.; Hebert, H.; Lindahl, M.; Gustafsson, C.M. Cryo-EM reveals promoter DNA binding and conformational flexibility of the general transcription factor TFIID. Structure 2009, 17, 1442–1452. [Google Scholar] [CrossRef]
- Yudkovsky, N.; Ranish, J.A.; Hahn, S. A transcription reinitiation intermediate that is stabilized by activator. Nature 2000, 408, 225–229. [Google Scholar] [CrossRef] [PubMed]
- Crick, F.H. Linking numbers and nucleosomes. Proc. Natl. Acad. Sci. USA 1976, 73, 2639–2643. [Google Scholar] [CrossRef] [PubMed]
- Murakami, K.; Calero, G.; Brown, C.R.; Liu, X.; Davis, R.E.; Boeger, H.; Kornberg, R.D. Formation and fate of a complete 31-protein RNA polymerase II transcription preinitiation complex. J. Biol. Chem. 2013, 288, 6325–6332. [Google Scholar] [CrossRef] [PubMed]
- Murakami, K.; Elmlund, H.; Kalisman, N.; Bushnell, D.A.; Adams, C.M.; Azubel, M.; Elmlund, D.; Levi-Kalisman, Y.; Liu, X.; Gibbons, B.J.; et al. Architecture of an RNA polymerase II transcription pre-initiation complex. Science 2013, 342, 1238724. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.B.; Louder, R.K.; Greber, B.J.; Grünberg, S.; Luo, J.; Fang, J.; Liu, Y.; Ranish, J.; Hahn, S.; Nogales, E. Structure of human TFIID and mechanism of TBP loading onto promoter DNA. Science 2018, 362, 1376. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.J.; Ishima, R.; Tong, K.I.; Bagby, S.; Kokubo, T.; Muhandiram, D.R.; Kay, L.E.; Nakatani, Y.; Ikura, M. Solution structure of a TBP-TAF(II)230 complex: Protein mimicry of the minor groove surface of the TATA box unwound by TBP. Cell 1998, 94, 573–583. [Google Scholar] [CrossRef]
- Cheng, J.X.; Nevado, J.; Lu, Z.; Ptashne, M. The TBP-inhibitory domain of TAF145 limits the effects of nonclassical transcriptional activators. Curr. Biol. 2002, 12, 934–937. [Google Scholar] [CrossRef]
- Boeger, H.; Griesenbeck, J.; Strattan, J.S.; Kornberg, R.D. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell 2003, 11, 1587–1598. [Google Scholar] [CrossRef]
- Stark, H. GraFix: Stablization of fragile macromolecular complexes for single particle cryo-EM. Methods Enzymol. 2010, 481, 109–126. [Google Scholar] [CrossRef]
- Kastner, B.; Fischer, N.; Golas, M.M.; Sander, B.; Dube, P.; Boehringer, D.; Hartmuth, K.; Deckert, J.; Hauer, F.; Wolf, E.; et al. GraFix: Sample preparation for single-particle electron cryomicroscopy. Nat. Methods 2008, 5, 53–55. [Google Scholar] [CrossRef]
- Elmlund, D.; Elmlund, H. SIMPLE: Software for ab initio reconstruction of heterogeneous single-particles. J. Struct. Biol. 2012, 180, 420–427. [Google Scholar] [CrossRef] [PubMed]
- Elmlund, D.; Davis, R.; Elmlund, H. Ab initio structure determination from electron microscopic images of single molecules coexisting in different functional states. Structure 2010, 18, 777–786. [Google Scholar] [CrossRef] [PubMed]
- Tang, G.; Peng, L.; Baldwin, P.R.; Mann, D.S.; Jiang, W.; Rees, I.; Ludtke, S.J. EMAN2: An extensible image processing suite for electron microscopy. J. Struct. Biol. 2007, 157, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Petterson, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Le, S.N.; Brown, C.R.; Harvey, S.; Boeger, H.; Elmlund, H.; Elmlund, D. The TAFs of TFIID Bind and Rearrange the Topology of the TATA-Less RPS5 Promoter. Int. J. Mol. Sci. 2019, 20, 3290. https://doi.org/10.3390/ijms20133290
Le SN, Brown CR, Harvey S, Boeger H, Elmlund H, Elmlund D. The TAFs of TFIID Bind and Rearrange the Topology of the TATA-Less RPS5 Promoter. International Journal of Molecular Sciences. 2019; 20(13):3290. https://doi.org/10.3390/ijms20133290
Chicago/Turabian StyleLe, Sarah N., Christopher R. Brown, Stacy Harvey, Hinrich Boeger, Hans Elmlund, and Dominika Elmlund. 2019. "The TAFs of TFIID Bind and Rearrange the Topology of the TATA-Less RPS5 Promoter" International Journal of Molecular Sciences 20, no. 13: 3290. https://doi.org/10.3390/ijms20133290
APA StyleLe, S. N., Brown, C. R., Harvey, S., Boeger, H., Elmlund, H., & Elmlund, D. (2019). The TAFs of TFIID Bind and Rearrange the Topology of the TATA-Less RPS5 Promoter. International Journal of Molecular Sciences, 20(13), 3290. https://doi.org/10.3390/ijms20133290





