Diversifying Evolution of the Ubiquitin-26S Proteasome System in Brassicaceae and Poaceae
Abstract
1. Introduction
2. Results
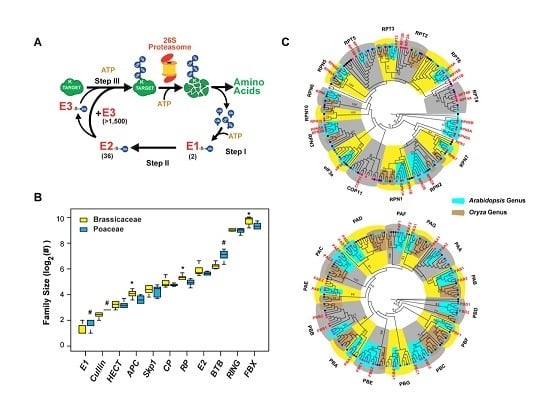
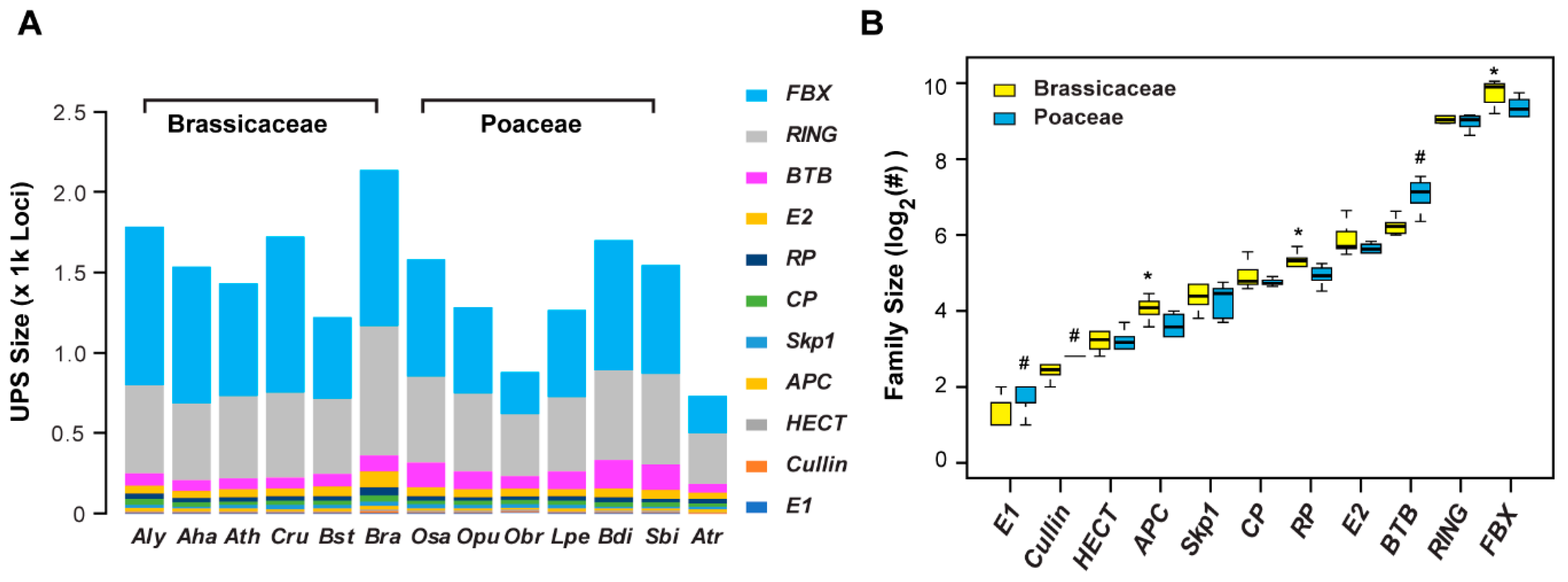
2.1. Re-Annotation of UPS Gene Families
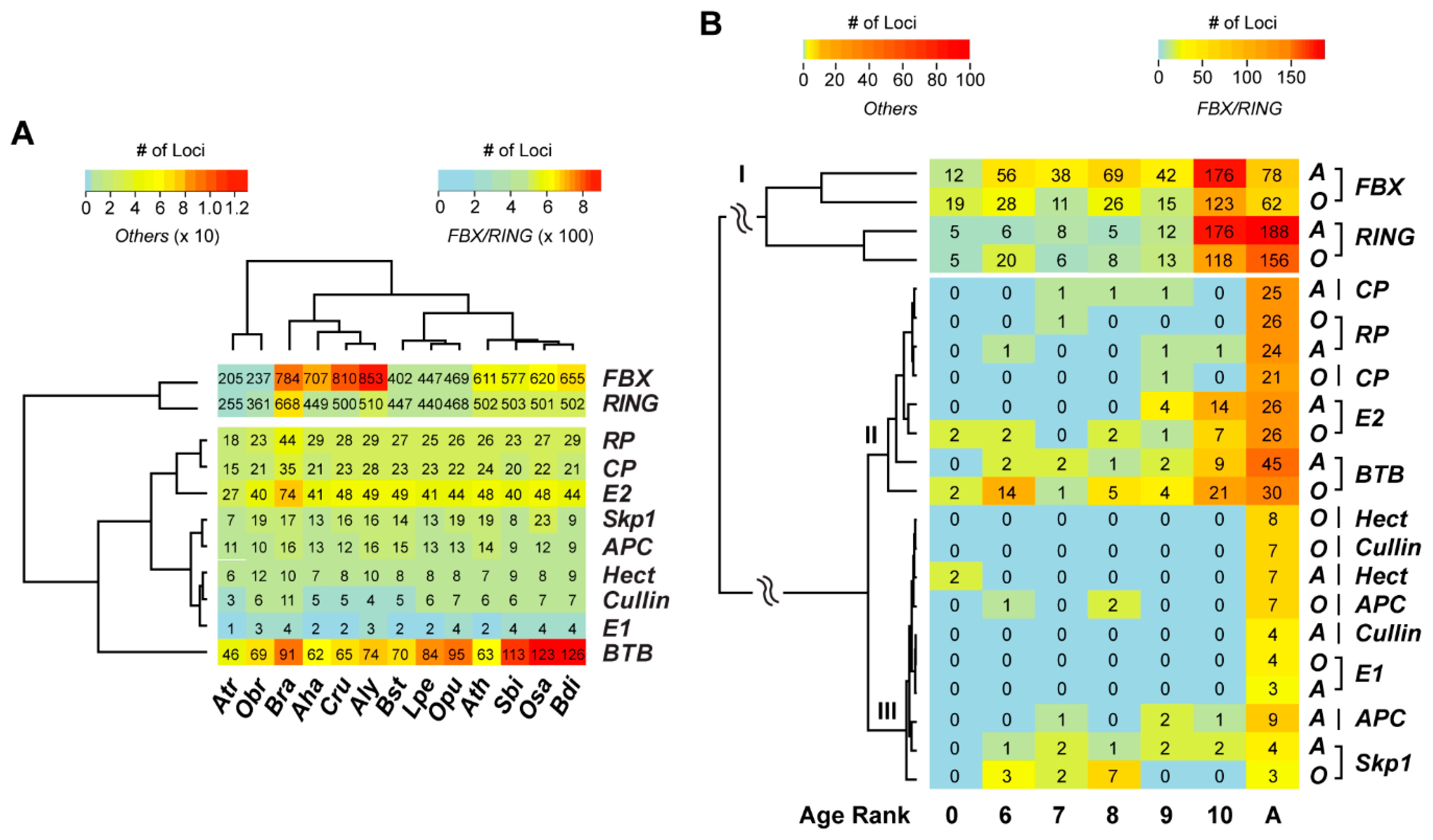
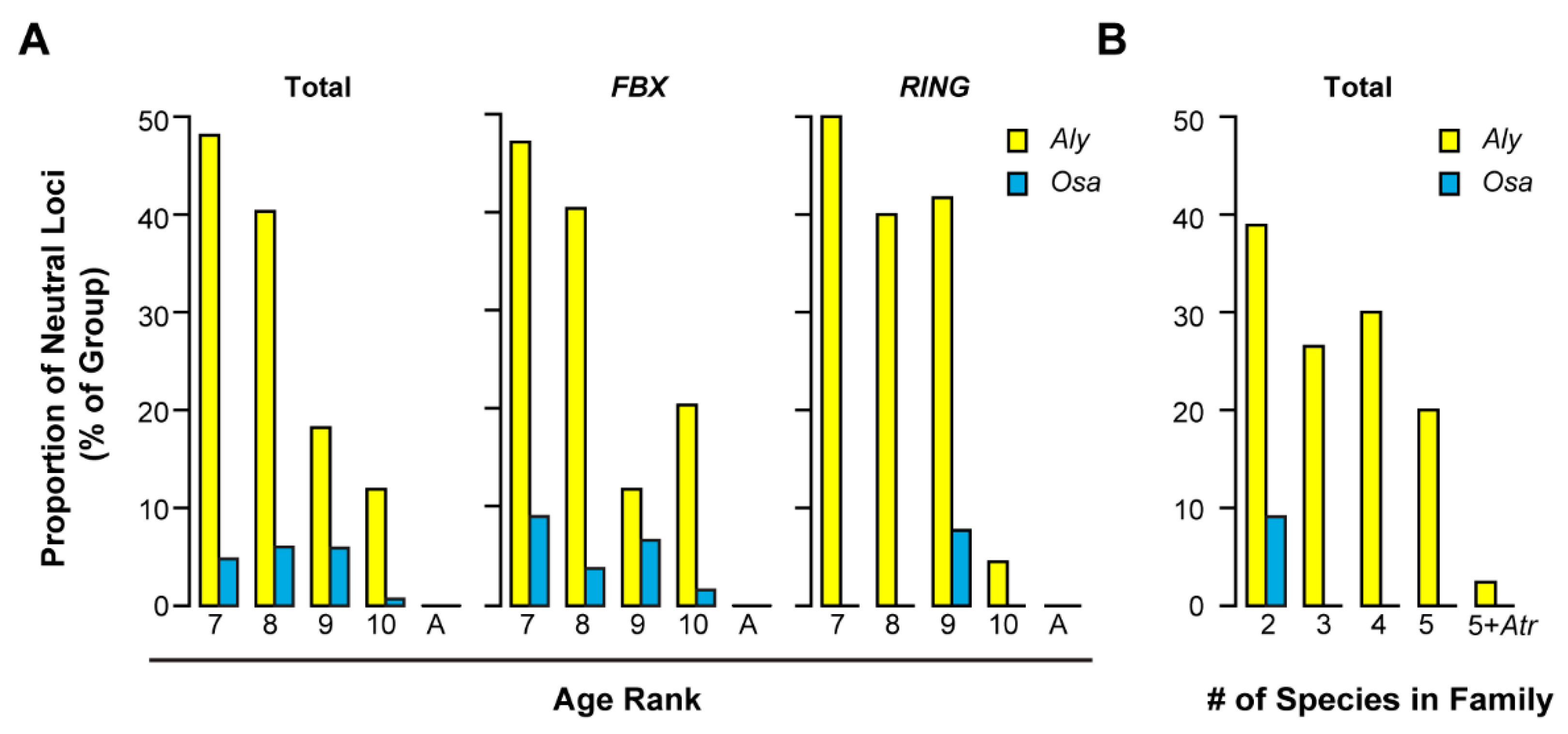
2.2. Variance of Expansion History of the UPS Families
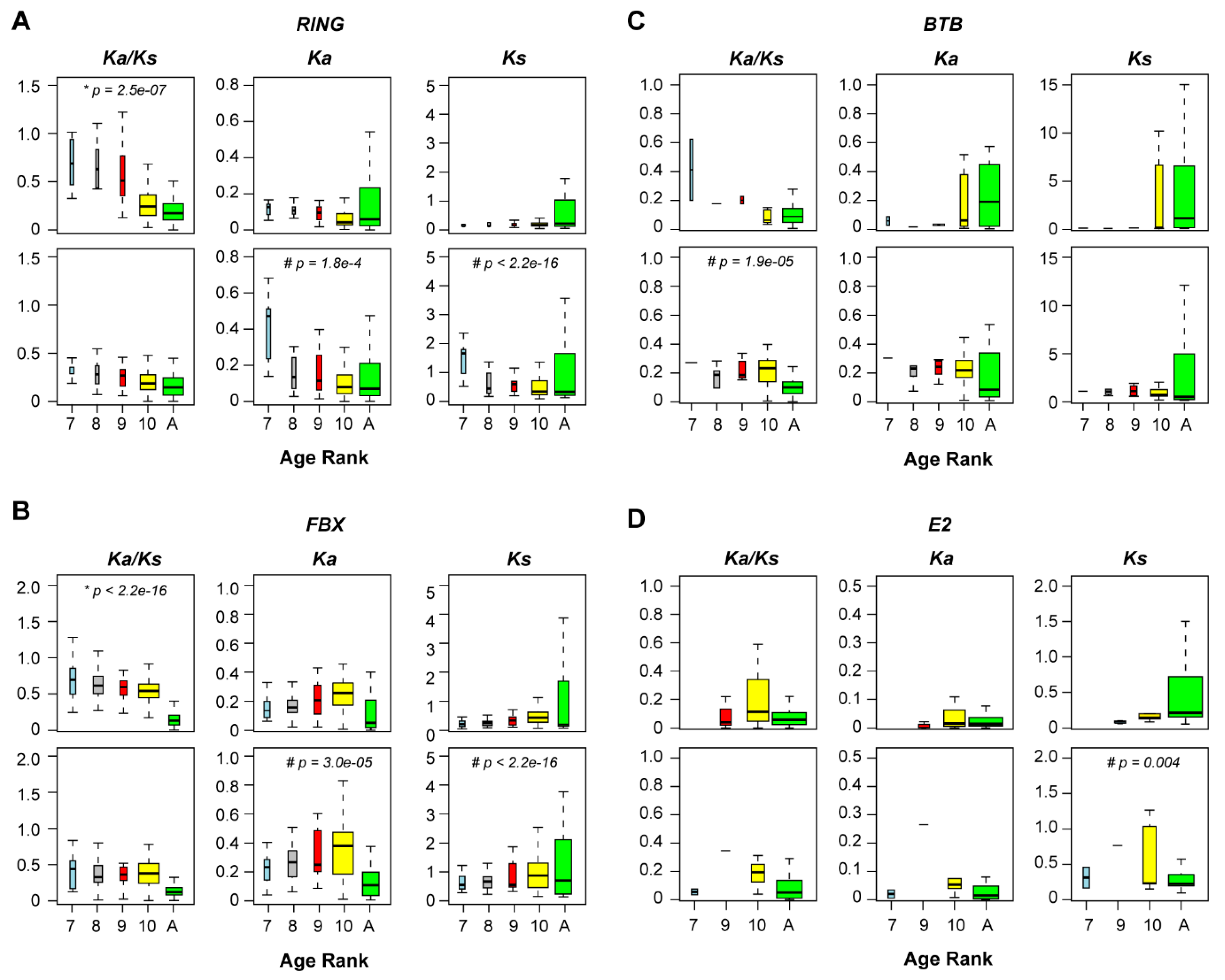
2.3. Differential Evolutionary Selection
2.4. Relaxed Negative Selection Resulted in Neutralization
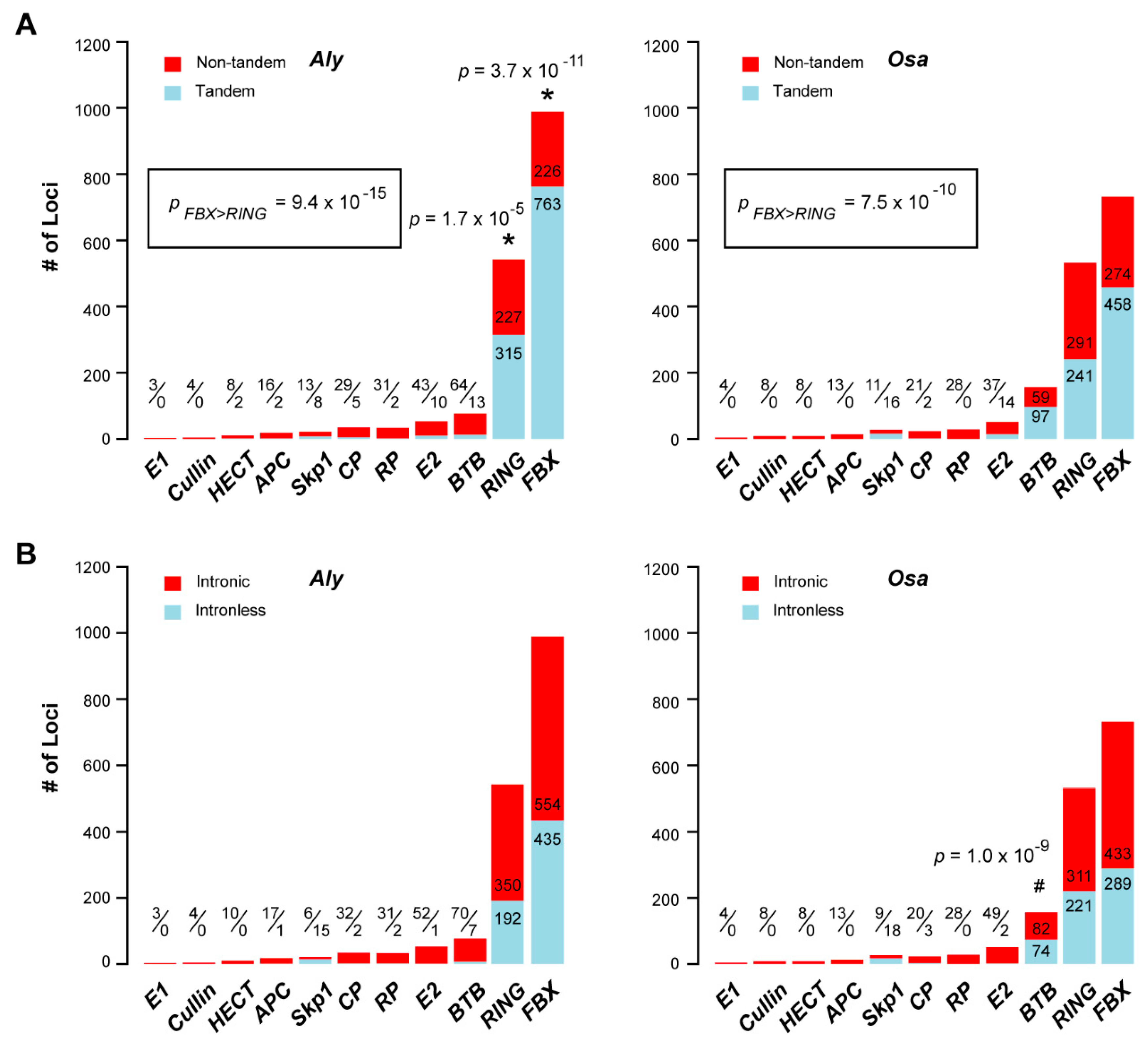
2.5. Contribution of Recent Duplications to the Size Expansion of UPS Families
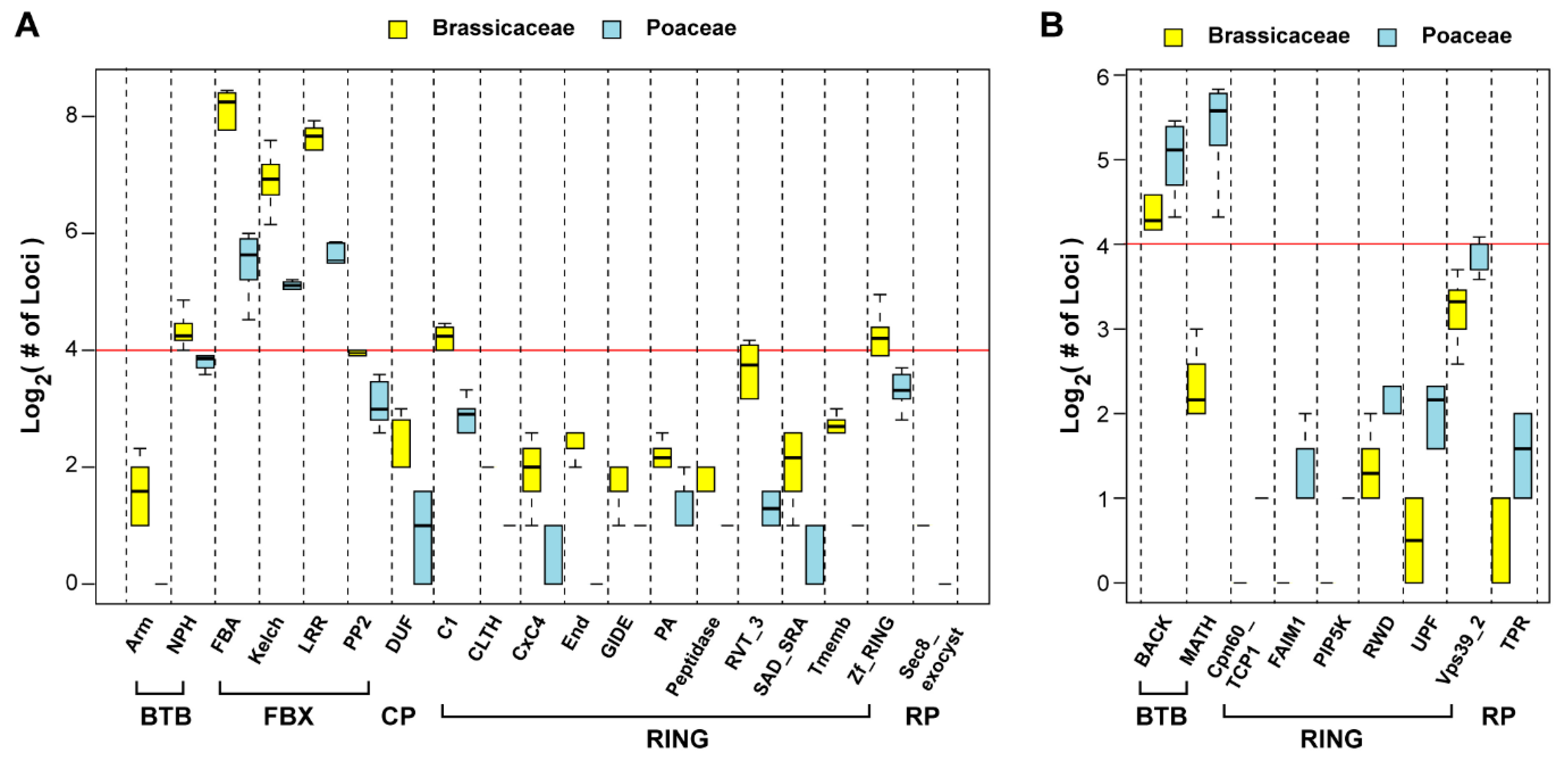
2.6. Functional Variance of the UPS Gene Families between Aly and Osa
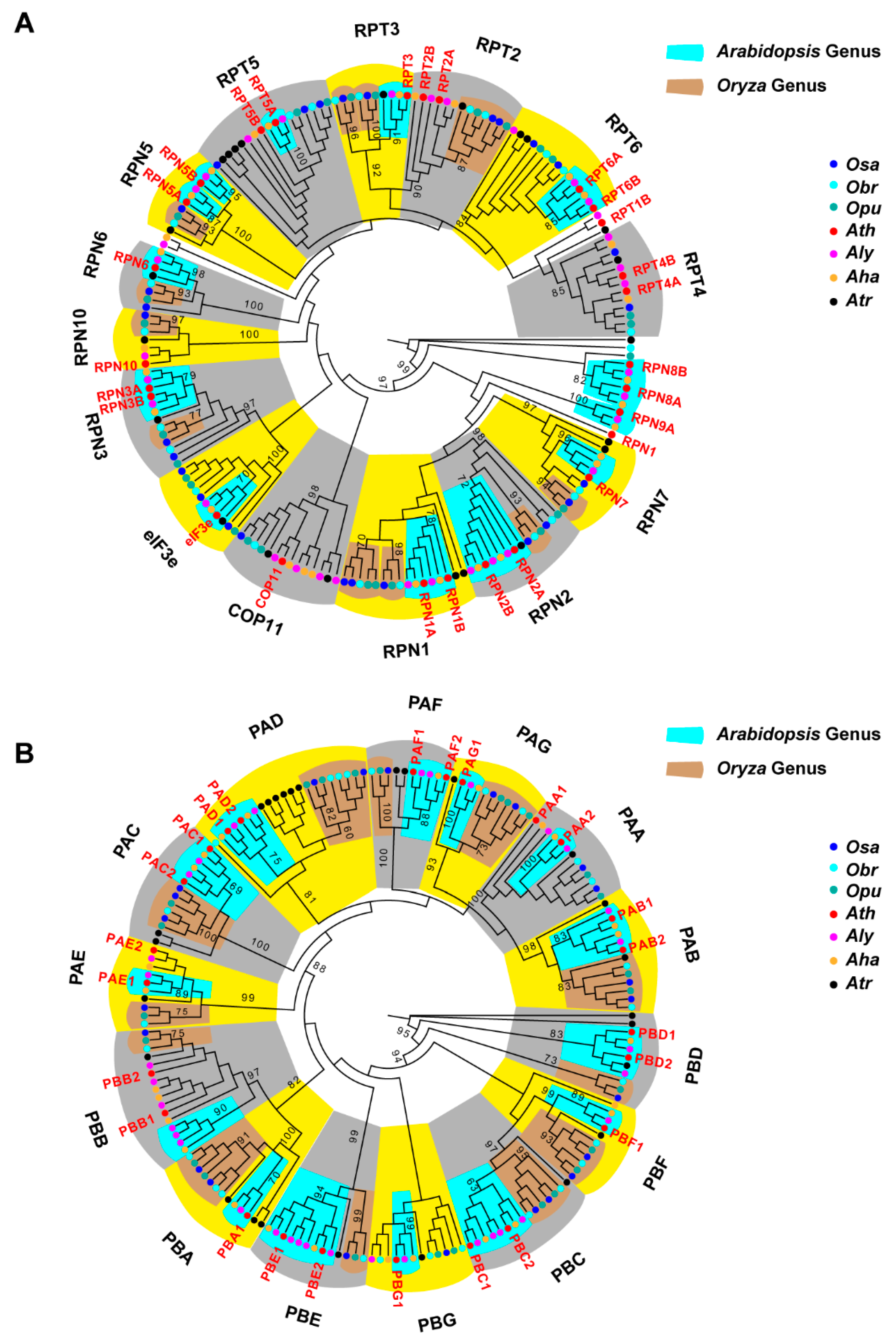
2.7. Diversifying Evolution of the 26S Proteasome in the Genera of Arabidopsis and Oryza
3. Discussion
4. Materials and Methods
4.1. Data Resources and UPS Family Annotation
4.2. Age Ranked Orthologous Group Analysis
4.3. Clustering Analysis
4.4. Detection of Recent Gene Duplications
4.5. Mutation Rate and Neutral Evolution Analysis
4.6. Proteasome Sequence Alignment and Phylogenetic Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Morris, J.L.; Puttick, M.N.; Clark, J.W.; Edwards, D.; Kenrick, P.; Pressel, S.; Wellman, C.H.; Yang, Z.; Schneider, H.; Donoghue, P.C.J. The timescale of early land plant evolution. Proc. Natl. Acad. Sci. USA 2018, 115, E2274–E2283. [Google Scholar] [CrossRef] [PubMed]
- Hua, Z.; Vierstra, R.D. The cullin-ring ubiquitin-protein ligases. Annu Rev. Plant. Biol 2011, 62, 299–334. [Google Scholar] [CrossRef] [PubMed]
- Haak, D.C.; Fukao, T.; Grene, R.; Hua, Z.; Ivanov, R.; Perrella, G.; Li, S. Multilevel regulation of abiotic stress responses in plants. Front. Plant Sci. 2017, 8, 1564. [Google Scholar] [CrossRef] [PubMed]
- Callis, J. The ubiquitination machinery of the ubiquitin system. Arabidopsis Book 2014, 12, e0174. [Google Scholar] [CrossRef] [PubMed]
- Zheng, N.; Shabek, N. Ubiquitin ligases: Structure, function, and regulation. Annu. Rev. Biochem. 2017, 86, 129–157. [Google Scholar] [CrossRef] [PubMed]
- Serrano, I.; Campos, L.; Rivas, S. Roles of e3 ubiquitin-ligases in nuclear protein homeostasis during plant stress responses. Front. Plant Sci. 2018, 9, 139. [Google Scholar] [CrossRef] [PubMed]
- Hua, Z.; Vierstra, R.D. Ubiquitin goes green. Trends Cell Biol. 2016, 26, 3–5. [Google Scholar] [CrossRef]
- Santner, A.; Estelle, M. The ubiquitin-proteasome system regulates plant hormone signaling. Plant J. 2010, 61, 1029–1040. [Google Scholar] [CrossRef]
- Goritschnig, S.; Zhang, Y.; Li, X. The ubiquitin pathway is required for innate immunity in arabidopsis. Plant J. 2007, 49, 540–551. [Google Scholar] [CrossRef]
- Yau, R.; Rape, M. The increasing complexity of the ubiquitin code. Nat. Cell Biol. 2016, 18, 579–586. [Google Scholar] [CrossRef]
- Hua, Z.; Zou, C.; Shiu, S.H.; Vierstra, R.D. Phylogenetic comparison of f-box (fbx) gene superfamily within the plant kingdom reveals divergent evolutionary histories indicative of genomic drift. PLoS ONE 2011, 6, e16219. [Google Scholar] [CrossRef] [PubMed]
- Gagne, J.M.; Downes, B.P.; Shiu, S.H.; Durski, A.M.; Vierstra, R.D. The f-box subunit of the scf e3 complex is encoded by a diverse superfamily of genes in arabidopsis. Proc. Natl. Acad. Sci. USA 2002, 99, 11519–11524. [Google Scholar] [CrossRef] [PubMed]
- Gingerich, D.J.; Hanada, K.; Shiu, S.H.; Vierstra, R.D. Large-scale, lineage-specific expansion of a bric-a-brac/tramtrack/broad complex ubiquitin-ligase gene family in rice. Plant Cell 2007, 19, 2329–2348. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.; Landherr, L.L.; Frohlich, M.W.; Leebens-Mack, J.; Ma, H.; dePamphilis, C.W. Patterns of gene duplication in the plant skp1 gene family in angiosperms: Evidence for multiple mechanisms of rapid gene birth. Plant J. 2007, 50, 873–885. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Terzaghi, W.; Gusmaroli, G.; Charron, J.B.; Yoon, H.J.; Chen, H.; He, Y.J.; Xiong, Y.; Deng, X.W. Characterization of arabidopsis and rice dwd proteins and their roles as substrate receptors for cul4-ring e3 ubiquitin ligases. Plant Cell 2008, 20, 152–167. [Google Scholar] [CrossRef] [PubMed]
- Risseeuw, E.P.; Daskalchuk, T.E.; Banks, T.W.; Liu, E.; Cotelesage, J.; Hellmann, H.; Estelle, M.; Somers, D.E.; Crosby, W.L. Protein interaction analysis of scf ubiquitin e3 ligase subunits from arabidopsis. Plant J. 2003, 34, 753–767. [Google Scholar] [CrossRef] [PubMed]
- Stone, S.L.; Hauksdottir, H.; Troy, A.; Herschleb, J.; Kraft, E.; Callis, J. Functional analysis of the ring-type ubiquitin ligase family of arabidopsis. Plant Physiol. 2005, 137, 13–30. [Google Scholar] [CrossRef] [PubMed]
- Hua, Z.; Doroodian, P.; Vu, W. Contrasting duplication patterns reflect functional diversities of ubiquitin and ubiquitin-like protein modifiers in plants. Plant J. 2018, 95, 296–311. [Google Scholar] [CrossRef] [PubMed]
- Vierstra, R.D. The ubiquitin-26s proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 2009, 10, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Kang, H.; Liu, W.; Wang, G.L. Comprehensive profiling of the rice ubiquitome reveals the significance of lysine ubiquitination in young leaves. J. Proteome Res. 2015, 14, 2017–2025. [Google Scholar] [CrossRef]
- Kim, D.Y.; Scalf, M.; Smith, L.M.; Vierstra, R.D. Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in arabidopsis. Plant Cell 2013, 25, 1523–1540. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Hernandez, V.; Kim, D.Y.; Stankey, R.J.; Scalf, M.; Smith, L.M.; Vierstra, R.D. Mass spectrometric analyses reveal a central role for ubiquitylation in remodeling the arabidopsis proteome during photomorphogenesis. Mol. Plant 2017, 10, 846–865. [Google Scholar] [CrossRef] [PubMed]
- Marshall, R.S.; Hua, Z.; Mali, S.; McLoughlin, F.; Vierstra, R.D. Atg8-binding uim proteins define a new class of autophagy adaptors and receptors. Cell 2019, 177, 766–781.e24. [Google Scholar] [CrossRef] [PubMed]
- Marshall, R.S.; McLoughlin, F.; Vierstra, R.D. Autophagic turnover of inactive 26s proteasomes in yeast is directed by the ubiquitin receptor cue5 and the hsp42 chaperone. Cell Rep. 2016, 16, 1717–1732. [Google Scholar] [CrossRef] [PubMed]
- Marshall, R.S.; Vierstra, R.D. To save or degrade: Balancing proteasome homeostasis to maximize cell survival. Autophagy 2018, 14, 2029–2031. [Google Scholar] [CrossRef] [PubMed]
- Marshall, R.S.; Vierstra, R.D. Proteasome storage granules protect proteasomes from autophagic degradation upon carbon starvation. Elife 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Marshall, R.S.; Vierstra, R.D. Autophagy: The master of bulk and selective recycling. Annu Rev. Plant Biol. 2018, 69, 173–208. [Google Scholar] [CrossRef]
- Marshall, R.S.; Li, F.; Gemperline, D.C.; Book, A.J.; Vierstra, R.D. Autophagic degradation of the 26s proteasome is mediated by the dual atg8/ubiquitin receptor rpn10 in arabidopsis. Mol. Cell 2015, 58, 1053–1066. [Google Scholar] [CrossRef]
- Stein, J.C.; Yu, Y.; Copetti, D.; Zwickl, D.J.; Zhang, L.; Zhang, C.; Chougule, K.; Gao, D.; Iwata, A.; Goicoechea, J.L.; et al. Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus oryza. Nat. Genet. 2018, 50, 285–296. [Google Scholar] [CrossRef]
- Hua, Z.; Early, M.J. Closing target trimming: A perl package for discovering hidden superfamily loci in genomes. PLoS One 2019, (in press).
- Ichihara, A. Reminiscence of 40-year research on nitrogen metabolism. Proc. Jpn Acad Ser. B Phys. Biol. Sci. 2010, 86, 707–716. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sivakumar, S.; Gorbsky, G.J. Spatiotemporal regulation of the anaphase-promoting complex in mitosis. Nat. Rev. Mol. Cell Biol. 2015, 16, 82–94. [Google Scholar] [CrossRef] [PubMed]
- Winklhofer, K.F. Parkin and mitochondrial quality control: Toward assembling the puzzle. Trends Cell Biol. 2014, 24, 332–341. [Google Scholar] [CrossRef] [PubMed]
- Beilstein, M.A.; Nagalingum, N.S.; Clements, M.D.; Manchester, S.R.; Mathews, S. Dated molecular phylogenies indicate a miocene origin for Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2010, 107, 18724–18728. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Defoort, J.; Tasdighian, S.; Maere, S.; Van de Peer, Y.; De Smet, R. Gene duplicability of core genes is highly consistent across all angiosperms. Plant Cell 2016, 28, 326–344. [Google Scholar] [CrossRef]
- Roux, C.; Castric, V.; Pauwels, M.; Wright, S.I.; Saumitou-Laprade, P.; Vekemans, X. Does speciation between arabidopsis halleri and arabidopsis lyrata coincide with major changes in a molecular target of adaptation? PLoS ONE 2011, 6, e26872. [Google Scholar] [CrossRef] [PubMed]
- Amborella Genome, P. The amborella genome and the evolution of flowering plants. Science 2013, 342, 1241089. [Google Scholar] [CrossRef]
- Heslop-Harrison, J.S.; Schwarzacher, T. Organisation of the plant genome in chromosomes. Plant J. 2011, 66, 18–33. [Google Scholar] [CrossRef]
- Schlotterer, C. Genes from scratch--the evolutionary fate of de novo genes. Trends Genet. 2015, 31, 215–219. [Google Scholar] [CrossRef]
- Yang, Z. Paml 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Hua, Z.; Gao, Z. Adaptive and degenerative evolution of the s-phase kinase-associated protein 1-like family in Arabidopsis thaliana. PeerJ 2019, 7, e6740. [Google Scholar] [CrossRef] [PubMed]
- Book, A.J.; Gladman, N.P.; Lee, S.S.; Scalf, M.; Smith, L.M.; Vierstra, R.D. Affinity purification of the arabidopsis 26 s proteasome reveals a diverse array of plant proteolytic complexes. J. Biol. Chem. 2010, 285, 25554–25569. [Google Scholar] [CrossRef] [PubMed]
- Finley, D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 2009, 78, 477–513. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutierrez, S.; Silla-Martinez, J.M.; Gabaldon, T. Trimal: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. Raxml version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Burks, E.A.; Bezerra, P.P.; Le, H.; Gallie, D.R.; Browning, K.S. Plant initiation factor 3 subunit composition resembles mammalian initiation factor 3 and has a novel subunit. J. Biol. Chem. 2001, 276, 2122–2131. [Google Scholar] [CrossRef]
- Dessau, M.; Halimi, Y.; Erez, T.; Chomsky-Hecht, O.; Chamovitz, D.A.; Hirsch, J.A. The arabidopsis cop9 signalosome subunit 7 is a model pci domain protein with subdomains involved in cop9 signalosome assembly. Plant Cell 2008, 20, 2815–2834. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.J.; Zhu, T.; Xia, E.H.; Shi, C.; Liu, Y.L.; Zhang, Y.; Liu, Y.; Jiang, W.K.; Zhao, Y.J.; Mao, S.Y.; et al. Rapid diversification of five oryza aa genomes associated with rice adaptation. Proc. Natl. Acad. Sci. USA 2014, 111, E4954–E4962. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: Oxford, UK, 2000. [Google Scholar]
- Hua, Z. Using ctt for comprehensive superfamily gene annotations. Protocols.io. 2019. Available online: https://www.protocols.io/view/using-ctt-for-comprehensive-superfamily-gene-annot-zf4f3qw (accessed on 23 March 2019).
- Bateman, A.; Finn, R.D. Scoop: A simple method for identification of novel protein superfamily relationships. Bioinformatics 2007, 23, 809–814. [Google Scholar] [CrossRef] [PubMed]
- Burroughs, A.M.; Iyer, L.M.; Aravind, L. Functional diversification of the ring finger and other binuclear treble clef domains in prokaryotes and the early evolution of the ubiquitin system. Mol. Biosyst. 2011, 7, 2261–2277. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Stoeckert, C.J., Jr.; Roos, D.S. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003, 13, 2178–2189. [Google Scholar] [CrossRef] [PubMed]
- Taly, J.F.; Magis, C.; Bussotti, G.; Chang, J.M.; Di Tommaso, P.; Erb, I.; Espinosa-Carrasco, J.; Kemena, C.; Notredame, C. Using the t-coffee package to build multiple sequence alignments of protein, rna, DNA sequences and 3d structures. Nat. Protoc. 2011, 6, 1669–1682. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Muscle: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hua, Z.; Yu, P. Diversifying Evolution of the Ubiquitin-26S Proteasome System in Brassicaceae and Poaceae. Int. J. Mol. Sci. 2019, 20, 3226. https://doi.org/10.3390/ijms20133226
Hua Z, Yu P. Diversifying Evolution of the Ubiquitin-26S Proteasome System in Brassicaceae and Poaceae. International Journal of Molecular Sciences. 2019; 20(13):3226. https://doi.org/10.3390/ijms20133226
Chicago/Turabian StyleHua, Zhihua, and Peifeng Yu. 2019. "Diversifying Evolution of the Ubiquitin-26S Proteasome System in Brassicaceae and Poaceae" International Journal of Molecular Sciences 20, no. 13: 3226. https://doi.org/10.3390/ijms20133226
APA StyleHua, Z., & Yu, P. (2019). Diversifying Evolution of the Ubiquitin-26S Proteasome System in Brassicaceae and Poaceae. International Journal of Molecular Sciences, 20(13), 3226. https://doi.org/10.3390/ijms20133226