Small RNA Functions as a Trafficking Effector in Plant Immunity
Abstract
1. Introduction
2. Biosynthesis of sRNA Induced by Pathogen Invasion
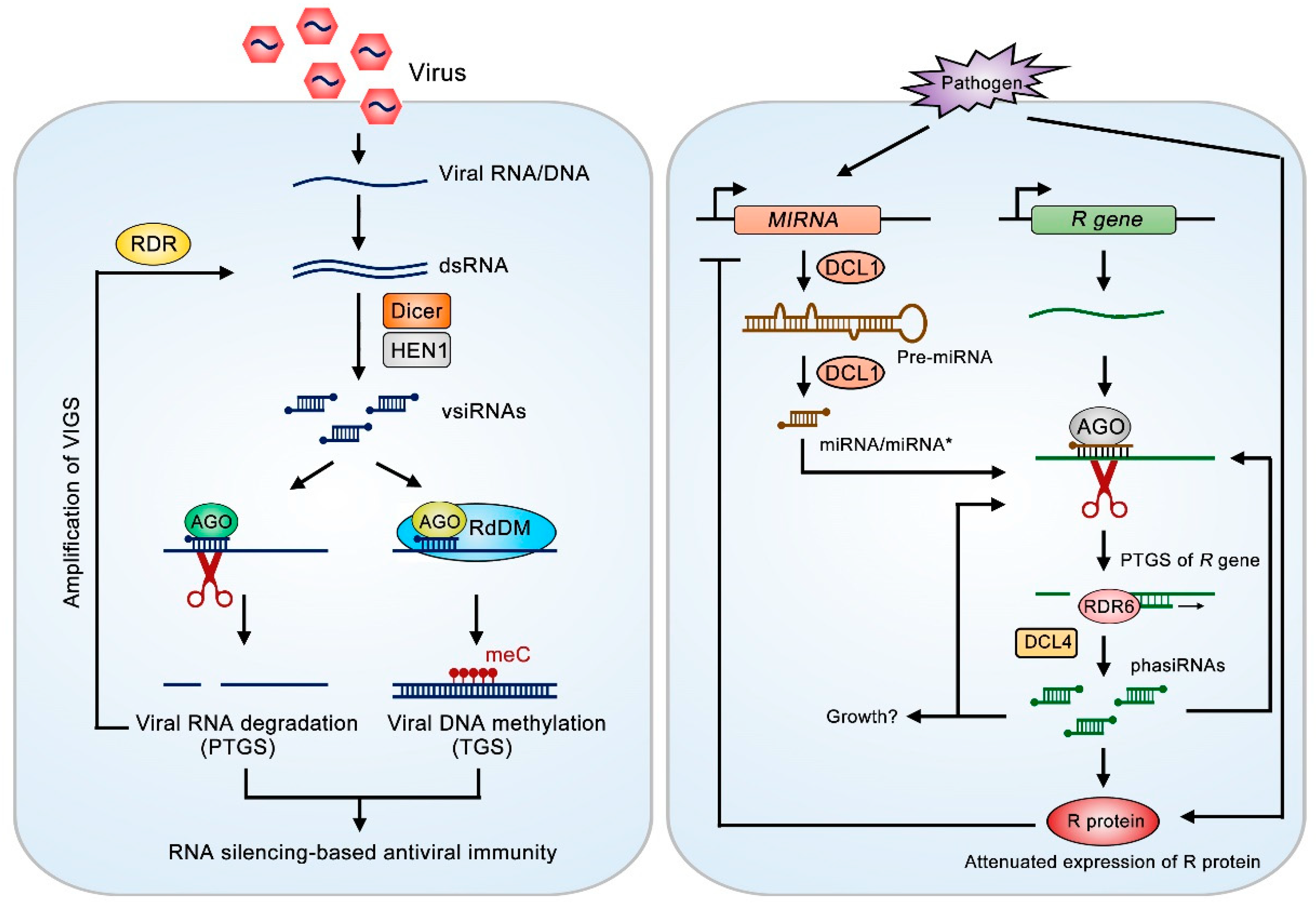
2.1. sRNAs in Plant-Virus Interaction
2.2. sRNAs in Plant-Bacteria Interaction
2.3. sRNAs in Plant-Fungi and Plant-Oomycetes Interaction
3. The Transportation of Trafficking sRNAs in Plants
3.1. sRNA Transportation within Plants
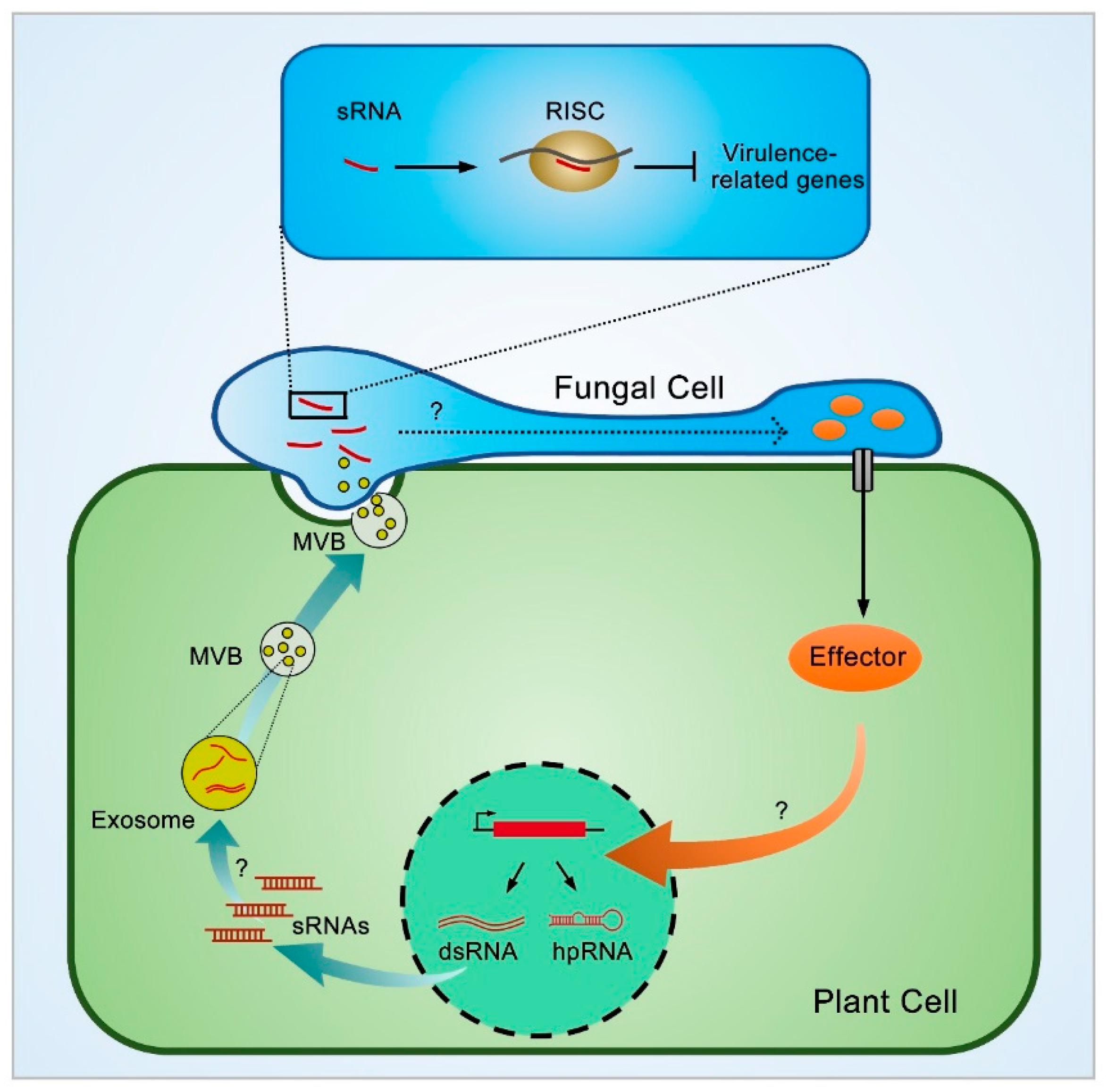
3.2. Cross-Kingdom Transportation of Plant sRNAs through Extracellular Vesicles (EVs)
4. Plant Mobile sRNA-Mediated Gene Silencing in Pathogens
4.1. The Working Mechanism of Plant sRNA in Fungi
4.2. Application of Trafficking sRNAs in Improving Plant Resistance
5. Concluding Remarks and Future Perspectives
- (1)
- What are the common molecular components (effectors or small molecular compounds) involved in pathogen-induced sRNA biogenesis in plants?
- (2)
- Is any RBP involved in transporting plant sRNAs into pathogen cells?
- (3)
- How are trafficking sRNAs selected? In other words, what is the selective criterion for sRNA translocation in EVs?
- (4)
- How do EVs establish connections with fungus or parasite cells? How are sRNAs released from plant cells and absorbed by pathogen cells?
- (5)
- What is the molecular form of plant mobile sRNAs in pathogen cells?
- (6)
- Which components are required for plant mobile sRNA-mediated gene silencing in pathogen cells?
Acknowledgments
Conflicts of Interest
References
- Baulcombe, D. RNA silencing in plants. Nature 2004, 431, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Qi, Y. RNAi in Plants: An Argonaute-Centered View. Plant Cell 2016, 28, 272–285. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Li, Y.; Cao, X.; Qi, Y. MicroRNAs and Their Regulatory Roles in Plant–Environment Interactions. Annu. Rev. Plant Biol. 2019, 70, 489–525. [Google Scholar] [CrossRef] [PubMed]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Duan, C.G.; Wang, C.H.; Guo, H.S. Application of RNA silencing to plant disease resistance. Silence 2012, 3, 5. [Google Scholar] [CrossRef]
- Chaloner, T.; van Kan, J.A.L.; Grant-Downton, R.T. RNA ‘Information Warfare’ in Pathogenic and Mutualistic Interactions. Trends Plant Sci. 2016, 21, 738–748. [Google Scholar] [CrossRef] [PubMed]
- D’Ario, M.; Griffiths-Jones, S.; Kim, M. Small RNAs: Big Impact on Plant Development. Trends Plant Sci. 2017, 22, 1056–1068. [Google Scholar] [CrossRef]
- Hamilton, A.J.; Baulcombe, D.C. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 1999, 286, 950–952. [Google Scholar] [CrossRef]
- Zamore, P.D.; Tuschl, T.; Sharp, P.A.; Bartel, D.P. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 2000, 101, 25–33. [Google Scholar] [CrossRef]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366. [Google Scholar] [CrossRef]
- Elbashir, S.M.; Lendeckel, W.; Tuschl, T. RNA interference is mediated by 21-and 22-nucleotide RNAs. Genes Dev. 2001, 15, 188–200. [Google Scholar] [CrossRef] [PubMed]
- Bigeard, J.; Colcombet, J.; Hirt, H. Signaling Mechanisms in Pattern-Triggered Immunity (PTI). Mol. Plant 2015, 8, 521–539. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Tsuda, K.; Parker, J.E. Effector-triggered immunity: From pathogen perception to robust defense. Annu. Rev. Plant Biol. 2015, 66, 487–511. [Google Scholar] [CrossRef] [PubMed]
- De Wit, P.J. Cladosporium fulvum Effectors: Weapons in the Arms Race with Tomato. Annu. Rev. Phytopathol. 2016, 54, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.X.; Dodds, P.N.; Bernoux, M. What Do We Know About NOD-Like Receptors in Plant Immunity? Annu. Rev. Phytopathol. 2017, 55, 205–229. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, F.; Nishimura, M.T. Structural, Functional, and Genomic Diversity of Plant NLR Proteins: An Evolved Resource for Rational Engineering of Plant Immunity. Annu. Rev. Phytopathol. 2018, 56, 243–267. [Google Scholar] [CrossRef] [PubMed]
- Klosterman, S.J.; Subbarao, K.V.; Kang, S.C.; Veronese, P.; Gold, S.E.; Thomma, B.P.H.J.; Chen, Z.H.; Henrissat, B.; Lee, Y.H.; Park, J.; et al. Comparative Genomics Yields Insights into Niche Adaptation of Plant Vascular Wilt Pathogens. PLoS Pathog. 2011, 7, e1002137. [Google Scholar] [CrossRef]
- Flor, H.H. Current Status of the Gene-For-Gene Concept. Annu. Rev. Phytopathol. 1971, 9, 275–296. [Google Scholar] [CrossRef]
- Guo, Z.X.; Li, Y.; Ding, S.W. Small RNA-based antimicrobial immunity. Nat. Rev. Immunol. 2019, 19, 31–44. [Google Scholar] [CrossRef]
- Fei, Q.; Xia, R.; Meyers, B.C. Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell 2013, 25, 2400–2415. [Google Scholar] [CrossRef]
- Li, S.; Castillo-Gonzalez, C.; Yu, B.; Zhang, X. The functions of plant small RNAs in development and in stress responses. Plant J. Cell Mol. Biol. 2017, 90, 654–670. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Chen, Y.; Tian, X.; Zhu, X.; Jin, W. Genome-wide identification and characterization of phased small interfering RNA genes in response to Botrytis cinerea infection in Solanum lycopersicum. Sci. Rep. 2017, 7, 3019. [Google Scholar] [CrossRef]
- Jin, Y.; Zhao, P.; Fang, Y.Y.; Gao, F.; Guo, H.S.; Zhao, J.H. Genome-wide profiling of sRNAs in the Verticillium dahliae-infected Arabidopsis roots. Mycology 2018, 9, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Weiberg, A.; Wang, M.; Lin, F.M.; Zhao, H.; Zhang, Z.; Kaloshian, I.; Huang, H.D.; Jin, H. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 2013, 342, 118–123. [Google Scholar] [CrossRef]
- Wang, M.; Weiberg, A.; Lin, F.M.; Thomma, B.P.; Huang, H.D.; Jin, H. Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants 2016, 2, 16151. [Google Scholar] [CrossRef] [PubMed]
- Weiberg, A.; Wang, M.; Bellinger, M.; Jin, H. Small RNAs: A new paradigm in plant-microbe interactions. Annu. Rev. Phytopathol. 2014, 52, 495–516. [Google Scholar] [CrossRef]
- Weiberg, A.; Jin, H.L. Small RNAs—The secret agents in the plant-pathogen interactions. Curr. Opin. Plant Biol. 2015, 26, 87–94. [Google Scholar] [CrossRef]
- Baulcombe, D. Viruses and gene silencing in plants. Arch. Virol. Suppl. 1999, 15, 189–201. [Google Scholar] [PubMed]
- Ding, S.W.; Voinnet, O. Antiviral immunity directed by small RNAs. Cell 2007, 130, 413–426. [Google Scholar] [CrossRef]
- Waterhouse, P.M.; Fusaro, A.F. Plant science. Viruses face a double defense by plant small RNAs. Science 2006, 313, 54–55. [Google Scholar] [CrossRef] [PubMed]
- Bouche, N.; Lauressergues, D.; Gasciolli, V.; Vaucheret, H. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 2006, 25, 3347–3356. [Google Scholar] [CrossRef] [PubMed]
- Deleris, A.; Gallego-Bartolome, J.; Bao, J.; Kasschau, K.D.; Carrington, J.C.; Voinnet, O. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 2006, 313, 68–71. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Pendon, J.A.; Li, F.; Li, W.X.; Ding, S.W. Suppression of antiviral silencing by cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell 2007, 19, 2053–2063. [Google Scholar] [CrossRef] [PubMed]
- Blevins, T.; Rajeswaran, R.; Aregger, M.; Borah, B.K.; Schepetilnikov, M.; Baerlocher, L.; Farinelli, L.; Meins, F., Jr.; Hohn, T.; Pooggin, M.M. Massive production of small RNAs from a non-coding region of Cauliflower mosaic virus in plant defense and viral counter-defense. Nucleic Acids Res. 2011, 39, 5003–5014. [Google Scholar] [CrossRef] [PubMed]
- Aregger, M.; Borah, B.K.; Seguin, J.; Rajeswaran, R.; Gubaeva, E.G.; Zvereva, A.S.; Windels, D.; Vazquez, F.; Blevins, T.; Farinelli, L.; et al. Primary and secondary siRNAs in geminivirus-induced gene silencing. PLoS Pathog. 2012, 8, e1002941. [Google Scholar] [CrossRef] [PubMed]
- De Jonge, R.; van Esse, H.P.; Maruthachalam, K.; Bolton, M.D.; Santhanam, P.; Saber, M.K.; Zhang, Z.; Usami, T.; Lievens, B.; Subbarao, K.V.; et al. Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc. Natl. Acad. Sci. USA 2012, 109, 5110–5115. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.B.; Wu, Q.F.; Ito, T.; Cillo, F.; Li, W.X.; Chen, X.M.; Yu, J.L.; Ding, S.W. RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2010, 107, 484–489. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.X.; Allen, E.; Wilken, A.; Carrington, J.C. DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2005, 102, 12984–12989. [Google Scholar] [CrossRef] [PubMed]
- Duan, C.G.; Fang, Y.Y.; Zhou, B.J.; Zhao, J.H.; Hou, W.N.; Zhu, H.; Ding, S.W.; Guo, H.S. Suppression of Arabidopsis ARGONAUTE1-Mediated Slicing, Transgene-Induced RNA Silencing, and DNA Methylation by Distinct Domains of the Cucumber mosaic virus 2b Protein. Plant Cell 2012, 24, 259–274. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Lu, J.; Wang, X.; Zhan, B.; Li, W.; Ding, S.W. Lipid flippases promote antiviral silencing and the biogenesis of viral and host siRNAs in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, 1377–1382. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.X.; Fan, B.F.; Chen, C.H.; Chen, Z.X. An important role of an inducible RNA-dependent RNA polymerase in plant antiviral defense. Proc. Natl. Acad. Sci. USA 2001, 98, 6516–6521. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.C.; Jiao, X.M.; Kong, X.Y.; Hamera, S.; Wu, Y.; Chen, X.Y.; Fang, R.X.; Yan, Y.S. A Signaling Cascade from miR444 to RDR1 in Rice Antiviral RNA Silencing Pathway. Plant Physiol. 2016, 170, 2365–2377. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Pignatta, D.; Bendix, C.; Brunkard, J.O.; Cohn, M.M.; Tung, J.; Sun, H.; Kumar, P.; Baker, B. MicroRNA regulation of plant innate immune receptors. Proc. Natl. Acad. Sci. USA 2012, 109, 1790–1795. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Liang, C.; Wang, S.; Hou, Y.; Gao, L.; Liu, L.; He, W.; Ma, W.; Mo, B.; Chen, X. The disease resistance protein SNC1 represses the biogenesis of microRNAs and phased siRNAs. Nat. Commun. 2018, 9, 5080. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Wang, J.; Tung, J.; Liu, D.; Zhou, Y.; He, S.; Du, Y.; Baker, B.; Li, F. A role for small RNA in regulating innate immunity during plant growth. PLoS Pathog. 2018, 14, e1006756. [Google Scholar] [CrossRef]
- Navarro, L.; Jay, F.; Nomura, K.; He, S.Y.; Voinnet, O. Suppression of the microRNA pathway by bacterial effector proteins. Science 2008, 321, 964–967. [Google Scholar] [CrossRef]
- Ruiz-Ferrer, V.; Voinnet, O. Roles of plant small RNAs in biotic stress responses. Annu. Rev. Plant Biol. 2009, 60, 485–510. [Google Scholar] [CrossRef]
- Navarro, L.; Dunoyer, P.; Jay, F.; Arnold, B.; Dharmasiri, N.; Estelle, M.; Voinnet, O.; Jones, J.D. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 2006, 312, 436–439. [Google Scholar] [CrossRef]
- Zhang, X.M.; Zhao, H.W.; Gao, S.; Wang, W.C.; Katiyar-Agarwal, S.; Huang, H.D.; Raikhel, N.; Jin, H.L. Arabidopsis Argonaute 2 Regulates Innate Immunity via miRNA393*-Mediated Silencing of a Golgi-Localized SNARE Gene, MEMB12. Mol. Cell 2011, 42, 356–366. [Google Scholar] [CrossRef]
- Katiyar-Agarwal, S.; Morgan, R.; Dahlbeck, D.; Borsani, O.; Villegas, A., Jr.; Zhu, J.K.; Staskawicz, B.J.; Jin, H. A pathogen-inducible endogenous siRNA in plant immunity. Proc. Natl. Acad. Sci. USA 2006, 103, 18002–18007. [Google Scholar] [CrossRef]
- Katiyar-Agarwal, S.; Gao, S.; Vivian-Smith, A.; Jin, H. A novel class of bacteria-induced small RNAs in Arabidopsis. Genes Dev. 2007, 21, 3123–3134. [Google Scholar] [CrossRef] [PubMed]
- Oh, C.S.; Martin, G.B. Effector-triggered immunity mediated by the Pto kinase. Trends Plant Sci. 2011, 16, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.L.; Liu, L.; Xiong, Q.; Flores, C.; Wong, J.; Shi, J.X.; Wang, X.B.; Liu, X.G.; Xiang, Q.J.; Jiang, S.S.; et al. Oomycete pathogens encode RNA silencing suppressors. Nat. Genet. 2013, 45, 330–333. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.L.; Shi, J.X.; Zhai, Y.; Hou, Y.N.; Ma, W.B. Phytophthora effector targets a novel component of small RNA pathway in plants to promote infection. Proc. Natl. Acad. Sci. USA 2015, 112, 5850–5855. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Zhai, Y.; Feng, L.; Karimi, H.Z.; Rutter, B.D.; Zeng, L.; Choi, D.S.; Zhang, B.; Gu, W.; Chen, X.; et al. A Phytophthora Effector Suppresses Trans-Kingdom RNAi to Promote Disease Susceptibility. Cell Host Microbe 2019, 25, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Ye, W.; Ma, W. Filamentous pathogen effectors interfering with small RNA silencing in plant hosts. Curr. Opin. Microbiol. 2016, 32, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Fradin, E.F.; Thomma, B.P. Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol. Plant Pathol. 2006, 7, 71–86. [Google Scholar] [CrossRef]
- Jian, H.; Ma, J.; Wei, L.; Liu, P.; Zhang, A.; Yang, B.; Li, J.; Xu, X.; Liu, L. Integrated mRNA, sRNA, and degradome sequencing reveal oilseed rape complex responses to Sclerotinia sclerotiorum (Lib.) infection. Sci. Rep. 2018, 8, 10987. [Google Scholar] [CrossRef]
- Huang, G.Z.; Allen, R.; Davis, E.L.; Baum, T.J.; Hussey, R.S. Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Proc. Natl. Acad. Sci. USA 2006, 103, 14302–14306. [Google Scholar] [CrossRef]
- Nowara, D.; Gay, A.; Lacomme, C.; Shaw, J.; Ridout, C.; Douchkov, D.; Hensel, G.; Kumlehn, J.; Schweizer, P. HIGS: Host-Induced Gene Silencing in the Obligate Biotrophic Fungal Pathogen Blumeria graminis. Plant Cell 2010, 22, 3130–3141. [Google Scholar] [CrossRef]
- Alakonya, A.; Kumar, R.; Koenig, D.; Kimura, S.; Townsley, B.; Runo, S.; Garces, H.M.; Kang, J.; Yanez, A.; David-Schwartz, R.; et al. Interspecific RNA Interference of SHOOT MERISTEMLESS-Like Disrupts Cuscuta pentagona Plant Parasitism. Plant Cell 2012, 24, 3153–3166. [Google Scholar] [CrossRef] [PubMed]
- Nunes, C.C.; Dean, R.A. Host-induced gene silencing: A tool for understanding fungal host interaction and for developing novel disease control strategies. Mol. Plant Pathol. 2012, 13, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Koch, A.; Kumar, N.; Weber, L.; Keller, H.; Imani, J.; Kogel, K.H. Host-induced gene silencing of cytochrome P450 lanosterol C14 alpha-demethylase-encoding genes confers strong resistance to Fusarium species. Proc. Natl. Acad. Sci. USA 2013, 110, 19324–19329. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Jin, Y.; Zhao, J.H.; Gao, F.; Zhou, B.J.; Fang, Y.Y.; Guo, H.S. Host-Induced Gene Silencing of the Target Gene in Fungal Cells Confers Effective Resistance to the Cotton Wilt Disease Pathogen Verticillium dahliae. Mol. Plant 2016, 9, 939–942. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhao, Y.L.; Zhao, J.H.; Wang, S.; Jin, Y.; Chen, Z.Q.; Fang, Y.Y.; Hua, C.L.; Ding, S.W.; Guo, H.S. Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nat. Plants 2016, 2, 16153. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Qiao, L.; Wang, M.; He, B.; Lin, F.M.; Palmquist, J.; Huang, S.D.; Jin, H. Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 2018, 360, 1126–1129. [Google Scholar] [CrossRef] [PubMed]
- Shahid, S.; Kim, G.; Johnson, N.R.; Wafula, E.; Wang, F.; Coruh, C.; Bernal-Galeano, V.; Phifer, T.; dePamphilis, C.W.; Westwood, J.H.; et al. MicroRNAs from the parasitic plant Cuscuta campestris target host messenger RNAs. Nature 2018, 553, 82–85. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Weiberg, A.; Dellota, E.; Yamane, D.; Jin, H.L. Botrytis small RNA Bc-siR37 suppresses plant defense genes by cross-kingdom RNAi. RNA Biol. 2017, 14, 421–428. [Google Scholar] [CrossRef]
- Wang, B.; Sun, Y.F.; Song, N.; Zhao, M.X.; Liu, R.; Feng, H.; Wang, X.J.; Kang, Z.S. Puccinia striiformis f. sp tritici microRNA-like RNA 1 (Pst-milR1), an important pathogenicity factor of Pst, impairs wheat resistance to Pst by suppressing the wheat pathogenesis-related 2 gene. New Phytol. 2017, 215, 338–350. [Google Scholar] [CrossRef]
- Lalonde, S.; Wipf, D.; Frommer, W.B. Transport mechanisms for organic forms of carbon and nitrogen between source and sink. Annu. Rev. Plant Biol. 2004, 55, 341–372. [Google Scholar] [CrossRef]
- Otero, S.; Helariutta, Y.; Benitez-Alfonso, Y. Symplastic communication in organ formation and tissue patterning. Curr. Opin. Plant Biol. 2016, 29, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Kragler, F.; Lucas, W.J.; Monzer, J. Plasmodesmata: Dynamics, domains and patterning. Ann. Bot. 1998, 81, 1–10. [Google Scholar] [CrossRef]
- Lucas, W.J.; Bouchepillon, S.; Jackson, D.P.; Nguyen, L.; Baker, L.; Ding, B.; Hake, S. Selective Trafficking of Knotted1 Homeodomain Protein and Its Messenger-Rna Through Plasmodesmata. Science 1995, 270, 1980–1983. [Google Scholar] [CrossRef] [PubMed]
- Oparka, K.J.; Roberts, A.G.; Boevink, P.; Santa Cruz, S.; Roberts, I.; Pradel, K.S.; Imlau, A.; Kotlizky, G.; Sauer, N.; Epell, B. Simple, but Not Branched, Plasmodesmata Allow the Nonspecific Trafficking of Proteins in Developing Tobacco Leaves. Cell 1999, 97, 743–754. [Google Scholar] [CrossRef]
- Kragler, F.; Monzer, J.; Xoconostle-Cazares, B.; Lucas, W.J. Peptide antagonists of the plasmodesmal macromolecular trafficking pathway. EMBO J. 2000, 19, 2856–2868. [Google Scholar] [CrossRef] [PubMed]
- Corbesier, L.; Vincent, C.; Jang, S.; Fornara, F.; Fan, Q.; Searle, I.; Giakountis, A.; Farrona, S.; Gissot, L.; Turnbull, C.; et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 2007, 316, 1030–1033. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.M.; Wang, J.; Xuan, Z.Y.; Goldshmidt, A.; Borrill, P.G.M.; Hariharan, N.; Kim, J.Y.; Jackson, D. Chaperonins Facilitate KNOTTED1 Cell-to-Cell Trafficking and Stem Cell Function. Science 2011, 333, 1141–1144. [Google Scholar] [CrossRef] [PubMed]
- Voinnet, O.; Baulcombe, D.C. Systemic signalling in gene silencing. Nature 1997, 389, 553. [Google Scholar] [CrossRef] [PubMed]
- Dalmay, T.; Hamilton, A.; Rudd, S.; Angell, S.; Baulcombe, D.C. An RNA-Dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 2000, 101, 543–553. [Google Scholar] [CrossRef]
- Dunoyer, P.; Himber, C.; Ruiz-Ferrer, V.; Alioua, A.; Voinnet, O. Intra- and intercellular RNA interference in Arabidopsis thaliana requires components of the microRNA and heterochromatic silencing pathways. Nat. Genet. 2007, 39, 848–856. [Google Scholar] [CrossRef]
- Himber, C.; Dunoyer, P.; Moissiard, G.; Ritzenthaler, C.; Voinnet, O. Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J. 2003, 22, 4523–4533. [Google Scholar] [CrossRef] [PubMed]
- Dunoyer, P.; Himber, C.; Voinnet, O. Induction, suppression and requirement of RNA silencing pathways in virulent Agrobacterium tumefaciens infections. Nat. Genet. 2015, 47, 847. [Google Scholar] [CrossRef] [PubMed]
- Pant, B.D.; Buhtz, A.; Kehr, J.; Scheible, W.R. MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. 2008, 53, 731–738. [Google Scholar] [CrossRef] [PubMed]
- Tsikou, D.; Yan, Z.; Holt, D.B.; Abel, N.B.; Reid, D.E.; Madsen, L.H.; Bhasin, H.; Sexauer, M.; Stougaard, J.; Markmann, K. Systemic control of legume susceptibility to rhizobial infection by a mobile microRNA. Science 2018, 362, 233–235. [Google Scholar] [CrossRef] [PubMed]
- Buhtz, A.; Pieritz, J.; Springer, F.; Kehr, J. Phloem small RNAs, nutrient stress responses, and systemic mobility. BMC Plant Biol. 2010, 10, 64. [Google Scholar] [CrossRef]
- Paultre, D.S.G.; Gustin, M.P.; Molnar, A.; Oparka, K.J. Lost in Transit: Long-Distance Trafficking and Phloem Unloading of Protein Signals in Arabidopsis Homografts. Plant Cell 2016, 28, 2016–2025. [Google Scholar] [CrossRef]
- Kehr, J.; Kragler, F. Long distance RNA movement. New Phytol. 2018, 218, 29–40. [Google Scholar] [CrossRef]
- Yoo, B.C.; Kragler, F.; Varkonyi-Gasic, E.; Haywood, V.; Archer-Evans, S.; Lee, Y.M.; Lough, T.J.; Lucas, W.J. A systemic small RNA signaling system in plants. Plant Cell 2004, 16, 1979–2000. [Google Scholar] [CrossRef]
- Ham, B.K.; Li, G.; Jia, W.; Leary, J.A.; Lucas, W.J. Systemic delivery of siRNA in pumpkin by a plant PHLOEM SMALL RNA-BINDING PROTEIN 1-ribonucleoprotein complex. Plant J. Cell Mol. Biol. 2014, 80, 683–694. [Google Scholar] [CrossRef]
- Mittelbrunn, M.; Sanchez-Madrid, F. Intercellular communication: Diverse structures for exchange of genetic information. Nat. Rev. Mol. Cell Biol. 2012, 13, 328–335. [Google Scholar] [CrossRef]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654. [Google Scholar] [CrossRef] [PubMed]
- Bobrie, A.; Colombo, M.; Raposo, G.; Thery, C. Exosome Secretion: Molecular Mechanisms and Roles in Immune Responses. Traffic 2011, 12, 1659–1668. [Google Scholar] [CrossRef] [PubMed]
- Mittelbrunn, M.; Gutierrez-Vazquez, C.; Villarroya-Beltri, C.; Gonzalez, S.; Sanchez-Cabo, F.; Gonzalez, M.A.; Bernad, A.; Sanchez-Madrid, F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2011, 2, 282. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed]
- An, Q.L.; Huckelhoven, R.; Kogel, K.H.; Van Bel, A.J.E. Multivesicular bodies participate in a cell wall-associated defence response in barley leaves attacked by the pathogenic powdery mildew fungus. Cell Microbiol. 2006, 8, 1009–1019. [Google Scholar] [CrossRef] [PubMed]
- Bohlenius, H.; Morch, S.M.; Godfrey, D.; Nielsen, M.E.; Thordal-Christensen, H. The Multivesicular Body-Localized GTPase ARFA1b/1c Is Important for Callose Deposition and ROR2 Syntaxin-Dependent Preinvasive Basal Defense in Barley. Plant Cell 2010, 22, 3831–3844. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, M.E.; Thordal-Christensen, H. Transcytosis shuts the door for an unwanted guest. Trends Plant Sci. 2013, 18, 611–616. [Google Scholar] [CrossRef] [PubMed]
- Hua, C.; Zhao, J.H.; Guo, H.S. Trans-Kingdom RNA Silencing in Plant-Fungal Pathogen Interactions. Mol. Plant 2018, 11, 235–244. [Google Scholar] [CrossRef]
- Thomma, B.; Cook, D.E. Targeting microbial pathogens. Science 2018, 360, 1070–1071. [Google Scholar] [CrossRef]
- Roth, R.; Hillmer, S.; Funaya, C.; Chiapello, M.; Schumacher, K.; Lo Presti, L.; Kahmann, R.; Paszkowski, U. Arbuscular cell invasion coincides with extracellular vesicles and membrane tubules. Nat. Plants 2019, 5, 204–211. [Google Scholar] [CrossRef]
- Ivanov, S.; Austin, J.; Berg, R.H.; Harrison, M.J. Extensive membrane systems at the host-arbuscular mycorrhizal fungus interface. Nat. Plants 2019, 5, 194. [Google Scholar] [CrossRef] [PubMed]
- Collins, N.C.; Thordal-Christensen, H.; Lipka, V.; Bau, S.; Kombrink, E.; Qiu, J.L.; Huckelhoven, R.; Stein, M.; Freialdenhoven, A.; Somerville, S.C.; et al. SNARE-protein-mediated disease resistance at the plant cell wall. Nature 2003, 425, 973–977. [Google Scholar] [CrossRef] [PubMed]
- Lipka, V.; Dittgen, J.; Bednarek, P.; Bhat, R.; Wiermer, M.; Stein, M.; Landtag, J.; Brandt, W.; Rosahl, S.; Scheel, D.; et al. Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 2005, 310, 1180–1183. [Google Scholar] [CrossRef] [PubMed]
- Clay, N.K.; Adio, A.M.; Denoux, C.; Jander, G.; Ausubel, F.M. Glucosinolate Metabolites Required for an Arabidopsis Innate Immune Response. Science 2009, 323, 95–101. [Google Scholar] [CrossRef]
- Regente, M.; Monzon, G.C.; de la Canal, L. Phospholipids are present in extracellular fluids of imbibing sunflower seeds and are modulated by hormonal treatments. J. Exp. Bot. 2008, 59, 553–562. [Google Scholar] [CrossRef] [PubMed]
- Regente, M.; Pinedo, M.; San Clemente, H.; Balliau, T.; Jamet, E.; de la Canal, L. Plant extracellular vesicles are incorporated by a fungal pathogen and inhibit its growth. J. Exp. Bot. 2017, 68, 5485–5495. [Google Scholar] [CrossRef]
- Gopinath, K.; Kao, C.C. Replication-independent long-distance trafficking by viral RNAs in Nicotiana benthamiana. Plant Cell 2007, 19, 1179–1191. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.N.; Thieme, C.J.; Kollwig, G.; Apelt, F.; Yang, L.; Winter, N.; Andresen, N.; Walther, D.; Kragler, F. tRNA-Related Sequences Trigger Systemic mRNA Transport in Plants. Plant Cell 2016, 28, 1237–1249. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Raposo, G.; Thery, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Ann. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.S.; Zhang, Z.Y.; Liu, Y. RNA Interference Pathways in Fungi: Mechanisms and Functions. Ann. Rev. Microbiol. 2012, 66, 305–323. [Google Scholar] [CrossRef] [PubMed]
- Brosnan, C.A.; Mitter, N.; Christie, M.; Smith, N.A.; Waterhouse, P.M.; Carroll, B.J. Nuclear gene silencing directs reception of long-distance mRNA silencing in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 14741–14746. [Google Scholar] [CrossRef] [PubMed]
- Koch, A.; Biedenkopf, D.; Furch, A.; Weber, L.; Rossbach, O.; Abdellatef, E.; Linicus, L.; Johannsmeier, J.; Jelonek, L.; Goesmann, A.; et al. An RNAi-Based Control of Fusarium graminearum Infections Through Spraying of Long dsRNAs Involves a Plant Passage and Is Controlled by the Fungal Silencing Machinery. PLoS Pathog. 2016, 12, e1005910. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, Q.; Iritani, A.; Ohkita, S.; Vu, B.V.; Yokoya, K.; Matsubara, A.; Ikeda, K.; Suzuki, N.; Nakayashiki, H. A fungal Argonaute interferes with RNA interference. Nucleic Acids Res. 2018, 46, 2495–2508. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Jin, H.L. Spray-Induced Gene Silencing: A Powerful Innovative Strategy for Crop Protection. Trends Microbiol. 2017, 25, 4–6. [Google Scholar] [CrossRef] [PubMed]
- Mitter, N.; Worrall, E.A.; Robinson, K.E.; Li, P.; Jain, R.G.; Taochy, C.; Fletcher, S.J.; Carroll, B.J.; Lu, G.Q.; Xu, Z.P.; et al. Clay nanosheets for topical delivery of RNAi for sustained protection against plant viruses. Nat. Plants 2017, 3, 16207. [Google Scholar] [CrossRef] [PubMed]
- Syn, N.L.; Wang, L.; Chow, E.K.; Lim, C.T.; Goh, B.C. Exosomes in Cancer Nanomedicine and Immunotherapy: Prospects and Challenges. Trends Biotechnol. 2017, 35, 665–676. [Google Scholar] [CrossRef]
| sRNAs | Origination | Interaction | Target Genes | Reference |
|---|---|---|---|---|
| siR1310 | Arabidopsis | Arabidopsis-Phytophthora | fungal virulence gene | [55] |
| miR166 | Gossypium | Gossypium-Verticillium | fungal virulence gene | [65] |
| miR159 | Gossypium | Gossypium-Verticillium | fungal virulence gene | [65] |
| TAS1c-siR483 | Arabidopsis | Arabidopsis-Botrytis | fungal virulence gene | [66] |
| TAS2-siR453 | Arabidopsis | Arabidopsis-Botrytis | fungal virulence gene | [66] |
| IGN-siR1 | Arabidopsis | Arabidopsis-Botrytis | Unknown | [66] |
| BcsiR3.1 | Botrytis | Arabidopsis-Botrytis | host PRXIIF gene | [24] |
| BcsiR3.2 | Botrytis | Arabidopsis-Botrytis | host MPK1/MPK2 genes | [24] |
| BcsiR5 | Botrytis | Arabidopsis-Botrytis | host WAK gene | [24] |
| BcsiR37 | Botrytis | Arabidopsis-Botrytis | Eight host genes | [68] |
| Pst-milR1 | Puccinia | Triticum-Puccinia | PR2 gene | [69] |
| miR12495 | Cuscuta | Arabidopsis-Cuscuta | host BIK1 gene | [67] |
| miR12497a | Cuscuta | Arabidopsis-Cuscuta | host TIR1/AFB2/AFB3 genes | [67] |
| miR12463b | Cuscuta | Arabidopsis-Cuscuta | host BIK1 gene | [67] |
| miR12480 | Cuscuta | Arabidopsis-Cuscuta | host SEOR1 gene | [67] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, C.; Liu, T.; Chang, Y.-N.; Duan, C.-G. Small RNA Functions as a Trafficking Effector in Plant Immunity. Int. J. Mol. Sci. 2019, 20, 2816. https://doi.org/10.3390/ijms20112816
Zhu C, Liu T, Chang Y-N, Duan C-G. Small RNA Functions as a Trafficking Effector in Plant Immunity. International Journal of Molecular Sciences. 2019; 20(11):2816. https://doi.org/10.3390/ijms20112816
Chicago/Turabian StyleZhu, Chen, Ting Liu, Ya-Nan Chang, and Cheng-Guo Duan. 2019. "Small RNA Functions as a Trafficking Effector in Plant Immunity" International Journal of Molecular Sciences 20, no. 11: 2816. https://doi.org/10.3390/ijms20112816
APA StyleZhu, C., Liu, T., Chang, Y.-N., & Duan, C.-G. (2019). Small RNA Functions as a Trafficking Effector in Plant Immunity. International Journal of Molecular Sciences, 20(11), 2816. https://doi.org/10.3390/ijms20112816





