Proteolytic Regulation of Parathyroid Hormone-Related Protein: Functional Implications for Skeletal Malignancy
Abstract
1. Introduction
1.1. PTHrP in Development and Skeletal Biology
1.2. PTHrP in Cancer
1.3. PTHrP Protein Structure and Susceptibility to Cleavage
2. Proteolytic PTHrP Products
2.1. N-terminal Derived Peptides
2.2. Mid-region and C-terminal Derived Peptides
2.3. PTHrP Fragments as Biomarkers of Disease
2.4. How Do Novel PTHrP Fragments Mediate Their Effects?
3. Proteolytic Control of PTHrP Function
3.1. Proprotein Convertases
3.2. Prostate Specific Antigen
3.3. Metalloproteases
3.4. Additional Proteases Capable of Cleaving PTHrP
3.5. Biological Considerations for Studying PTHrP Post-Translation Modification
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Suva, L.J.; Winslow, G.A.; Wettenhall, R.E.; Hammonds, R.G.; Moseley, J.M.; Diefenbach-Jagger, H.; Rodda, C.P.; Kemp, B.E.; Rodriguez, H.; Chen, E.Y.; et al. A parathyroid hormone-related protein implicated in malignant hypercalcemia: Cloning and expression. Science 1987, 237, 893–896. [Google Scholar] [CrossRef] [PubMed]
- Burtis, W.J. Parathyroid hormone-related protein: Structure, function, and measurement. Clin. Chem. 1992, 38, 2171–2183. [Google Scholar] [PubMed]
- McCauley, L.K.; Martin, T.J. Twenty-five years of PTHrP progress: From cancer hormone to multifunctional cytokine. J. Bone Miner. Res. 2012, 27, 1231–1239. [Google Scholar] [CrossRef] [PubMed]
- Martin, T.J. Parathyroid hormone-related protein, its regulation of cartilage and bone development, and role in treating bone diseases. Physiol. Rev. 2016, 96, 831–871. [Google Scholar] [CrossRef] [PubMed]
- Philbrick, W.M.; Wysolmerski, J.J.; Galbraith, S.; Holt, E.; Orloff, J.J.; Yang, K.H.; Vasavada, R.C.; Weir, E.C.; Broadus, A.E.; Stewart, A.F. Defining the roles of parathyroid hormone-related protein in normal physiology. Physiol. Rev. 1996, 76, 127–173. [Google Scholar] [CrossRef] [PubMed]
- Behar, V.; Pines, M.; Nakamoto, C.; Greenberg, Z.; Bisello, A.; Stueckle, S.M.; Bessalle, R.; Usdin, T.B.; Chorev, M.; Rosenblatt, M.; et al. The human PTH2 receptor: Binding and signal transduction properties of the stably expressed recombinant receptor. Endocrinology 1996, 137, 2748–2757. [Google Scholar] [CrossRef] [PubMed]
- Sheikh, S.P.; Vilardarga, J.P.; Baranski, T.J.; Lichtarge, O.; Iiri, T.; Meng, E.C.; Nissenson, R.A.; Bourne, H.R. Similar structures and shared switch mechanisms of the beta2-adrenoceptor and the parathyroid hormone receptor. Zn(ii) bridges between helices iii and vi block activation. J. Biol. Chem. 1999, 274, 17033–17041. [Google Scholar] [CrossRef]
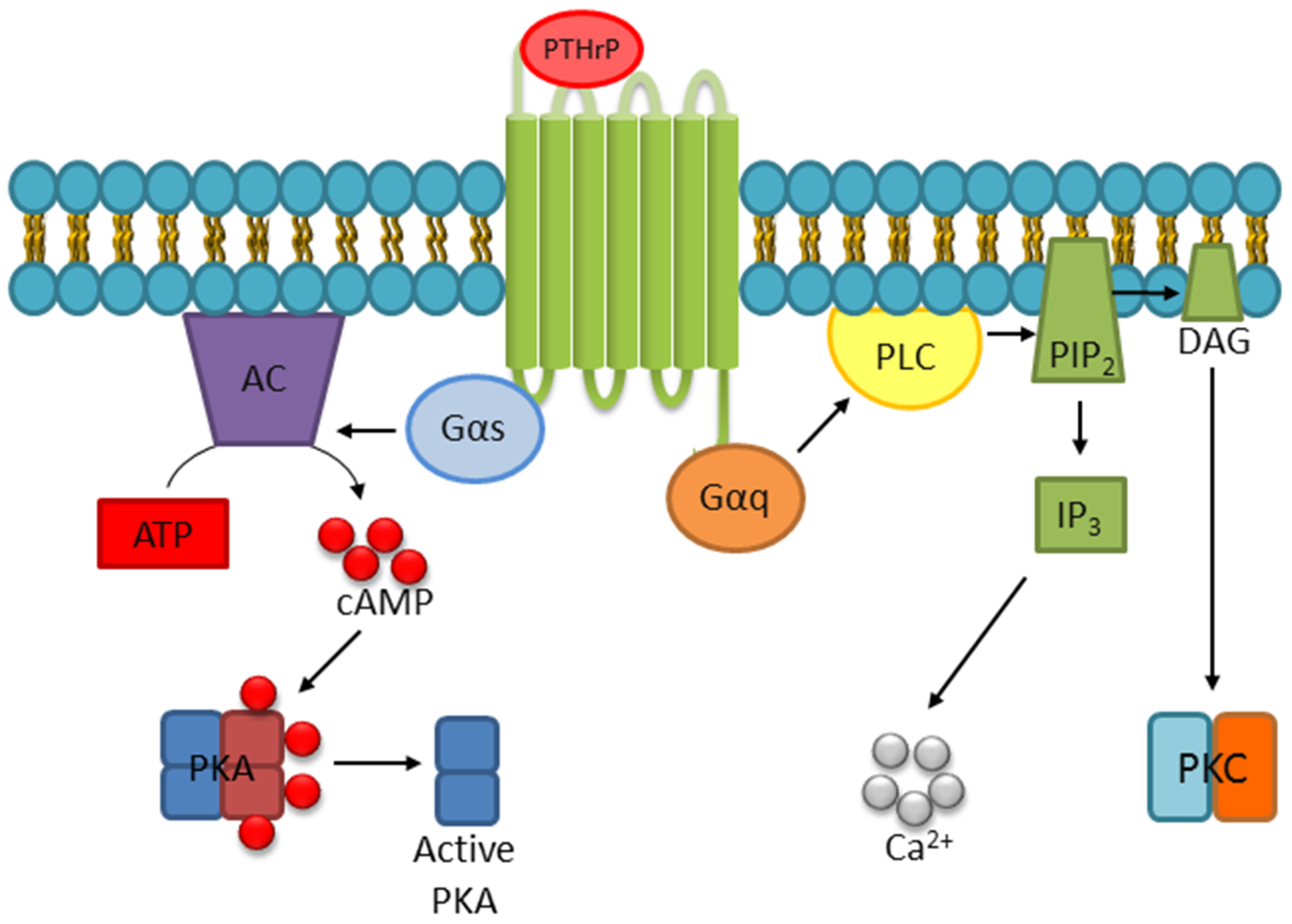
- Abou-Samra, A.B.; Juppner, H.; Force, T.; Freeman, M.W.; Kong, X.F.; Schipani, E.; Urena, P.; Richards, J.; Bonventre, J.V.; Potts, J.T., Jr.; et al. Expression cloning of a common receptor for parathyroid hormone and parathyroid hormone-related peptide from rat osteoblast-like cells: A single receptor stimulates intracellular accumulation of both camp and inositol trisphosphates and increases intracellular free calcium. Proc. Natl. Acad. Sci. USA 1992, 89, 2732–2736. [Google Scholar]
- Cupp, M.E.; Nayak, S.K.; Adem, A.S.; Thomsen, W.J. Parathyroid hormone (PTH) and PTH-related peptide domains contributing to activation of different PTH receptor-mediated signaling pathways. J. Pharmacol. Exp. Ther. 2013, 345, 404–418. [Google Scholar] [CrossRef]
- Datta, N.S.; Samra, T.A.; Mahalingam, C.D.; Datta, T.; Abou-Samra, A.B. Role of PTH1r internalization in osteoblasts and bone mass using a phosphorylation-deficient knock-in mouse model. J. Endocrinol. 2010, 207, 355–365. [Google Scholar] [CrossRef]
- Singh, A.T.; Bhattacharyya, R.S.; Radeff, J.M.; Stern, P.H. Regulation of parathyroid hormone-stimulated phospholipase d in umr-106 cells by calcium, map kinase, and small g proteins. J. Bone Miner. Res. 2003, 18, 1453–1460. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.T.; Gilchrist, A.; Voyno-Yasenetskaya, T.; Radeff-Huang, J.M.; Stern, P.H. G alpha12/g alpha13 subunits of heterotrimeric g proteins mediate parathyroid hormone activation of phospholipase d in umr-106 osteoblastic cells. Endocrinology 2005, 146, 2171–2175. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.T.; Kunnel, J.G.; Strieleman, P.J.; Stern, P.H. Parathyroid hormone (PTH)-(1-34), (Nle(8,18),tyr34) PTH-(3-34) amide, PTH-(1-31) amide, and PTH-related peptide-(1-34) stimulate phosphatidylcholine hydrolysis in umr-106 osteoblastic cells: Comparison with effects of phorbol 12,13-dibutyrate. Endocrinology 1999, 140, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Sneddon, W.B.; Friedman, P.A. Beta-arrestin-dependent parathyroid hormone-stimulated extracellular signal-regulated kinase activation and parathyroid hormone type 1 receptor internalization. Endocrinology 2007, 148, 4073–4079. [Google Scholar] [CrossRef] [PubMed]
- Gesty-Palmer, D.; Chen, M.; Reiter, E.; Ahn, S.; Nelson, C.D.; Wang, S.; Eckhardt, A.E.; Cowan, C.L.; Spurney, R.F.; Luttrell, L.M.; et al. Distinct beta-arrestin- and g protein-dependent pathways for parathyroid hormone receptor-stimulated erk1/2 activation. J. Biol. Chem. 2006, 281, 10856–10864. [Google Scholar] [CrossRef] [PubMed]
- Syme, C.A.; Friedman, P.A.; Bisello, A. Parathyroid hormone receptor trafficking contributes to the activation of extracellular signal-regulated kinases but is not required for regulation of camp signaling. J. Biol. Chem. 2005, 280, 11281–11288. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, G.; Bednenko, J.; Gillespie, M.T.; Gerace, L. Molecular basis for the recognition of a nonclassical nuclear localization signal by importin beta. Mol. Cell 2002, 10, 1345–1353. [Google Scholar] [CrossRef]
- Liu, B.; Goltzman, D.; Rabbani, S.A. Processing of pro-PTHrP by the prohormone convertase, furin: Effect on biological activity. Am. J. Physiol. 1995, 268, E832–E838. [Google Scholar] [CrossRef] [PubMed]
- Francini, G.; Maioli, E.; Petrioli, R.; Paffetti, P.; Gonnelli, S.; Aquino, A. Production of parathyroid hormone and parathyroid-hormone-related protein by breast cancer cells in culture. J. Cancer Res. Clin. Oncol. 1993, 119, 421–425. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, K.; Weir, E.C.; Mangin, M.; Dannies, P.S.; Kinder, B.; Deftos, L.J.; Brown, E.M.; Broadus, A.E. Expression of messenger ribonucleic acids encoding a parathyroid hormone-like peptide in normal human and animal tissues with abnormal expression in human parathyroid adenomas. Mol. Endocrinol. 1988, 2, 1230–1236. [Google Scholar] [CrossRef] [PubMed]
- Dittmer, J.; Gitlin, S.D.; Reid, R.L.; Brady, J.N. Transactivation of the p2 promoter of parathyroid hormone-related protein by human t-cell lymphotropic virus type i tax1: Evidence for the involvement of transcription factor ets1. J. Virol. 1993, 67, 6087–6095. [Google Scholar] [PubMed]
- Dittmer, J.; Pise-Masison, C.A.; Clemens, K.E.; Choi, K.S.; Brady, J.N. Interaction of human t-cell lymphotropic virus type i tax, ets1, and sp1 in transactivation of the PTHrP p2 promoter. J. Biol. Chem. 1997, 272, 4953–4958. [Google Scholar] [CrossRef] [PubMed]
- Foley, J.; Wysolmerski, J.J.; Missero, C.; King, C.S.; Philbrick, W.M. Regulation of parathyroid hormone-related protein gene expression in murine keratinocytes by e1a isoforms: A role for basal promoter and ets-1 site. Mol. Cell Endocrinol. 1999, 156, 13–23. [Google Scholar] [CrossRef]
- Nishishita, T.; Okazaki, T.; Ishikawa, T.; Igarashi, T.; Hata, K.; Ogata, E.; Fujita, T. A negative vitamin d response DNA element in the human parathyroid hormone-related peptide gene binds to vitamin d receptor along with ku antigen to mediate negative gene regulation by vitamin d. J. Biol. Chem. 1998, 273, 10901–10907. [Google Scholar] [CrossRef] [PubMed]
- Chilco, P.J.; Leopold, V.; Zajac, J.D. Differential regulation of the parathyroid hormone-related protein gene p1 and p3 promoters by camp. Mol. Cell Endocrinol 1998, 138, 173–184. [Google Scholar] [CrossRef]
- Heath, J.K.; Southby, J.; Fukumoto, S.; O’Keeffe, L.M.; Martin, T.J.; Gillespie, M.T. Epidermal growth factor-stimulated parathyroid hormone-related protein expression involves increased gene transcription and mrna stability. Biochem. J. 1995, 307, 159–167. [Google Scholar] [CrossRef]
- Cho, Y.M.; Lewis, D.A.; Koltz, P.F.; Richard, V.; Gocken, T.A.; Rosol, T.J.; Konger, R.L.; Spandau, D.F.; Foley, J. Regulation of parathyroid hormone-related protein gene expression by epidermal growth factor-family ligands in primary human keratinocytes. J. Endocrinol. 2004, 181, 179–190. [Google Scholar] [CrossRef]
- Johnson, R.W.; Nguyen, M.P.; Padalecki, S.S.; Grubbs, B.G.; Merkel, A.R.; Oyajobi, B.O.; Matrisian, L.M.; Mundy, G.R.; Sterling, J.A. Tgf-beta promotion of gli2-induced expression of parathyroid hormone-related protein, an important osteolytic factor in bone metastasis, is independent of canonical hedgehog signaling. Cancer Res. 2011, 71, 822–831. [Google Scholar] [CrossRef]
- Gilmore, J.L.; Scott, J.A.; Bouizar, Z.; Robling, A.; Pitfield, S.E.; Riese, D.J., 2nd; Foley, J. Amphiregulin-egfr signaling regulates PTHrP gene expression in breast cancer cells. Breast Cancer Res. Treat. 2008, 110, 493–505. [Google Scholar] [CrossRef]
- McCauley, L.K.; Beecher, C.A.; Melton, M.E.; Werkmeister, J.R.; Juppner, H.; Abou-Samra, A.B.; Segre, G.V.; Rosol, T.J. Transforming growth factor-beta1 regulates steady-state PTH/PTHrP receptor mrna levels and PTHrP binding in ros 17/2.8 osteosarcoma cells. Mol. Cell Endocrinol. 1994, 101, 331–336. [Google Scholar] [CrossRef]
- Merryman, J.I.; DeWille, J.W.; Werkmeister, J.R.; Capen, C.C.; Rosol, T.J. Effects of transforming growth factor-beta on parathyroid hormone-related protein production and ribonucleic acid expression by a squamous carcinoma cell line in vitro. Endocrinology 1994, 134, 2424–2430. [Google Scholar] [CrossRef]
- Wang, N.; Li, P.; Liu, W.; Wang, N.; Lu, Z.; Feng, J.; Zeng, X.; Yang, J.; Wang, Y.; Zhao, W. Mir-141-3p suppresses proliferation and promotes apoptosis by targeting gli2 in osteosarcoma cells. Oncol Rep. 2018, 39, 747–754. [Google Scholar] [CrossRef]
- Karaplis, A.C.; Luz, A.; Glowacki, J.; Bronson, R.T.; Tybulewicz, V.L.; Kronenberg, H.M.; Mulligan, R.C. Lethal skeletal dysplasia from targeted disruption of the parathyroid hormone-related peptide gene. Genes Dev. 1994, 8, 277–289. [Google Scholar] [CrossRef]
- Wysolmerski, J.J.; Philbrick, W.M.; Dunbar, M.E.; Lanske, B.; Kronenberg, H.; Broadus, A.E. Rescue of the parathyroid hormone-related protein knockout mouse demonstrates that parathyroid hormone-related protein is essential for mammary gland development. Development 1998, 125, 1285–1294. [Google Scholar]
- Philbrick, W.M.; Dreyer, B.E.; Nakchbandi, I.A.; Karaplis, A.C. Parathyroid hormone-related protein is required for tooth eruption. Proc. Natl. Acad. Sci. USA 1998, 95, 11846–11851. [Google Scholar] [CrossRef]
- Foley, J.; Longely, B.J.; Wysolmerski, J.J.; Dreyer, B.E.; Broadus, A.E.; Philbrick, W.M. PTHrP regulates epidermal differentiation in adult mice. J. Invest. Dermatol 1998, 111, 1122–1128. [Google Scholar] [CrossRef]
- Amizuka, N.; Karaplis, A.C.; Henderson, J.E.; Warshawsky, H.; Lipman, M.L.; Matsuki, Y.; Ejiri, S.; Tanaka, M.; Izumi, N.; Ozawa, H.; et al. Haploinsufficiency of parathyroid hormone-related peptide (PTHrP) results in abnormal postnatal bone development. Dev. Biol. 1996, 175, 166–176. [Google Scholar] [CrossRef]
- Lanske, B.; Amling, M.; Neff, L.; Guiducci, J.; Baron, R.; Kronenberg, H.M. Ablation of the PTHrP gene or the PTH/PTHrP receptor gene leads to distinct abnormalities in bone development. J. Clin. Invest. 1999, 104, 399–407. [Google Scholar] [CrossRef]
- Vortkamp, A.; Lee, K.; Lanske, B.; Segre, G.V.; Kronenberg, H.M.; Tabin, C.J. Regulation of rate of cartilage differentiation by indian hedgehog and PTH-related protein. Science 1996, 273, 613–622. [Google Scholar] [CrossRef]
- Martin, T.J. Osteoblast-derived PTHrP is a physiological regulator of bone formation. J. Clin. Invest. 2005, 115, 2322–2324. [Google Scholar] [CrossRef]
- Horwitz, M.J.; Tedesco, M.B.; Garcia-Ocana, A.; Sereika, S.M.; Prebehala, L.; Bisello, A.; Hollis, B.W.; Gundberg, C.M.; Stewart, A.F. Parathyroid hormone-related protein for the treatment of postmenopausal osteoporosis: Defining the maximal tolerable dose. J. Clin. Endocrinol. Metab. 2010, 95, 1279–1287. [Google Scholar] [CrossRef]
- Horwitz, M.J.; Tedesco, M.B.; Gundberg, C.; Garcia-Ocana, A.; Stewart, A.F. Short-term, high-dose parathyroid hormone-related protein as a skeletal anabolic agent for the treatment of postmenopausal osteoporosis. J. Clin. Endocrinol. Metab. 2003, 88, 569–575. [Google Scholar] [CrossRef]
- Stewart, A.F.; Cain, R.L.; Burr, D.B.; Jacob, D.; Turner, C.H.; Hock, J.M. Six-month daily administration of parathyroid hormone and parathyroid hormone-related protein peptides to adult ovariectomized rats markedly enhances bone mass and biomechanical properties: A comparison of human parathyroid hormone 1-34, parathyroid hormone-related protein 1-36, and sdz-parathyroid hormone 893. J. Bone Miner. Res. 2000, 15, 1517–1525. [Google Scholar]
- Miller, P.D.; Hattersley, G.; Riis, B.J.; Williams, G.C.; Lau, E.; Russo, L.A.; Alexandersen, P.; Zerbini, C.A.; Hu, M.Y.; Harris, A.G.; et al. Effect of abaloparatide vs placebo on new vertebral fractures in postmenopausal women with osteoporosis: A randomized clinical trial. JAMA 2016, 316, 722–733. [Google Scholar] [CrossRef]
- Leder, B.Z.; O’Dea, L.S.; Zanchetta, J.R.; Kumar, P.; Banks, K.; McKay, K.; Lyttle, C.R.; Hattersley, G. Effects of abaloparatide, a human parathyroid hormone-related peptide analog, on bone mineral density in postmenopausal women with osteoporosis. J. Clin. Endocrinol. Metab. 2015, 100, 697–706. [Google Scholar] [CrossRef]
- Chew, C.K.; Clarke, B.L. Abaloparatide: Recombinant human PTHrP (1-34) anabolic therapy for osteoporosis. Maturitas 2017, 97, 53–60. [Google Scholar] [CrossRef]
- Reginster, J.Y.; Hattersley, G.; Williams, G.C.; Hu, M.Y.; Fitzpatrick, L.A.; Lewiecki, E.M. Abaloparatide is an effective treatment option for postmenopausal osteoporosis: Review of the number needed to treat compared with teriparatide. Calcif. Tissue Int. 2018, 103, 540–545. [Google Scholar] [CrossRef]
- Southby, J.; Kissin, M.W.; Danks, J.A.; Hayman, J.A.; Moseley, J.M.; Henderson, M.A.; Bennett, R.C.; Martin, T.J. Immunohistochemical localization of parathyroid hormone-related protein in human breast cancer. Cancer Res. 1990, 50, 7710–7716. [Google Scholar]
- Powell, G.J.; Southby, J.; Danks, J.A.; Stillwell, R.G.; Hayman, J.A.; Henderson, M.A.; Bennett, R.C.; Martin, T.J. Localization of parathyroid hormone-related protein in breast cancer metastases: Increased incidence in bone compared with other sites. Cancer Res. 1991, 51, 3059–3061. [Google Scholar]
- Yoneda, T.; Sasaki, A.; Mundy, G.R. Osteolytic bone metastasis in breast cancer. Breast Cancer Research and Treatment 1994, 32, 73–84. [Google Scholar] [CrossRef]
- Ash, P.; Loutit, J.F.; Townsend, K.M. Osteoclasts derived from haematopoietic stem cells. Nature 1980, 283, 669–670. [Google Scholar] [CrossRef]
- Guise, T.A.; Yin, J.J.; Taylor, S.D.; Kumagai, Y.; Dallas, M.; Boyce, B.F.; Yoneda, T.; Mundy, G.R. Evidence for a causal role of parathyroid hormone-related protein in the pathogenesis of human breast cancer-mediated osteolysis. J. Clin. Invest. 1996, 98, 1544–1549. [Google Scholar] [CrossRef]
- Kremer, R.; Goltzman, D.; Amizuka, N.; Webber, M.M.; Rhim, J.S. Ras activation of human prostate epithelial cells induces overexpression of parathyroid hormone-related peptide. Clin. Cancer Res. 1997, 3, 855–859. [Google Scholar]
- Liao, J.; Li, X.; Koh, A.J.; Berry, J.E.; Thudi, N.; Rosol, T.J.; Pienta, K.J.; McCauley, L.K. Tumor expressed PTHrP facilitates prostate cancer-induced osteoblastic lesions. Int. J. Cancer 2008, 123, 2267–2278. [Google Scholar] [CrossRef]
- Orloff, J.J.; Reddy, D.; de Papp, A.E.; Yang, K.H.; Soifer, N.E.; Stewart, A.F. Parathyroid hormone-related protein as a prohormone: Posttranslational processing and receptor interactions. Endocr Rev. 1994, 15, 40–60. [Google Scholar]
- Soifer, N.E.; Dee, K.E.; Insogna, K.L.; Burtis, W.J.; Matovcik, L.M.; Wu, T.L.; Milstone, L.M.; Broadus, A.E.; Philbrick, W.M.; Stewart, A.F. Parathyroid hormone-related protein. Evidence for secretion of a novel mid-region fragment by three different cell types. J. Biol. Chem. 1992, 267, 18236–18243. [Google Scholar]
- Hook, V.Y.; Burton, D.; Yasothornsrikul, S.; Hastings, R.H.; Deftos, L.J. Proteolysis of proPTHrP(1-141) by "prohormone thiol protease" at multibasic residues generates PTHrP-related peptides: Implications for PTHrP peptide production in lung cancer cells. Biochem. Biophys Res. Commun. 2001, 285, 932–938. [Google Scholar] [CrossRef]
- Miao, D.; He, B.; Jiang, Y.; Kobayashi, T.; Soroceanu, M.A.; Zhao, J.; Su, H.; Tong, X.; Amizuka, N.; Gupta, A.; et al. Osteoblast-derived PTHrP is a potent endogenous bone anabolic agent that modifies the therapeutic efficacy of administered PTH 1-34. J. Clin. Invest. 2005, 115, 2402–2411. [Google Scholar] [CrossRef]
- Chen, H.L.; Demiralp, B.; Schneider, A.; Koh, A.J.; Silve, C.; Wang, C.Y.; McCauley, L.K. Parathyroid hormone and parathyroid hormone-related protein exert both pro- and anti-apoptotic effects in mesenchymal cells. J. Biol. Chem. 2002, 277, 19374–19381. [Google Scholar] [CrossRef]
- Fiaschi-Taesch, N.M.; Stewart, A.F. Minireview: Parathyroid hormone-related protein as an intracrine factor--trafficking mechanisms and functional consequences. Endocrinology 2003, 144, 407–411. [Google Scholar] [CrossRef]
- Schluter, K.D.; Katzer, C.; Piper, H.M. A n-terminal PTHrP peptide fragment void of a PTH/PTHrP-receptor binding domain activates cardiac et(a) receptors. Br. J. Pharmacol. 2001, 132, 427–432. [Google Scholar] [CrossRef]
- Frieling, J.S.; Shay, G.; Izumi, V.; Aherne, S.T.; Saul, R.G.; Budzevich, M.; Koomen, J.; Lynch, C.C. Matrix metalloproteinase processing of PTHrP yields a selective regulator of osteogenesis, PTHrP1-17. Oncogene 2017. [Google Scholar] [CrossRef]
- Cramer, S.D.; Chen, Z.; Peehl, D.M. Prostate specific antigen cleaves parathyroid hormone-related protein in the PTH-like domain: Inactivation of PTHrP-stimulated camp accumulation in mouse osteoblasts. J. Urol 1996, 156, 526–531. [Google Scholar] [CrossRef]
- Ruchon, A.F.; Marcinkiewicz, M.; Ellefsen, K.; Basak, A.; Aubin, J.; Crine, P.; Boileau, G. Cellular localization of neprilysin in mouse bone tissue and putative role in hydrolysis of osteogenic peptides. J. Bone Miner. Res. 2000, 15, 1266–1274. [Google Scholar] [CrossRef]
- Chirgwin, J.M.; Mohammad, K.S.; Guise, T.A. Tumor-bone cellular interactions in skeletal metastases. J. Musculoskelet Neuronal. Interact. 2004, 4, 308–318. [Google Scholar]
- Weir, E.C.; Terwilliger, G.; Sartori, L.; Insogna, K.L. Synthetic parathyroid hormone-like protein (1-74) is anabolic for bone in vivo. Calcif. Tissue Int. 1992, 51, 30–34. [Google Scholar] [CrossRef]
- Burtis, W.J.; Dann, P.; Gaich, G.A.; Soifer, N.E. A high abundance midregion species of parathyroid hormone-related protein: Immunological and chromatographic characterization in plasma. J. Clin. Endocrinol. Metab. 1994, 78, 317–322. [Google Scholar]
- Abbas, S.K.; Pickard, D.W.; Rodda, C.P.; Heath, J.A.; Hammonds, R.G.; Wood, W.I.; Caple, I.W.; Martin, T.J.; Care, A.D. Stimulation of ovine placental calcium transport by purified natural and recombinant parathyroid hormone-related protein (PTHrP) preparations. Q J. Exp. Physiol. 1989, 74, 549–552. [Google Scholar] [CrossRef]
- Edwards, R.C.; Ratcliffe, W.A.; Walls, J.; Morrison, J.M.; Ratcliffe, J.G.; Holder, R.; Bundred, N.J. Parathyroid hormone-related protein (PTHrP) in breast cancer and benign breast tissue. Eur. J. Cancer 1995, 31A, 334–339. [Google Scholar] [CrossRef]
- Bowden, S.J.; Emly, J.F.; Hughes, S.V.; Powell, G.; Ahmed, A.; Whittle, M.J.; Ratcliffe, J.G.; Ratcliffe, W.A. Parathyroid hormone-related protein in human term placenta and membranes. J. Endocrinol. 1994, 142, 217–224. [Google Scholar] [CrossRef]
- Suehiro, M.; Murakami, M.; Fukuchi, M. Circulating forms of immunoreactive parathyroid hormone-related protein for identifying patients with humoral hypercalcemia of malignancy: A comparative study with C-terminal(109-141)- and N-terminal(1-86)-region-specific PTHrP radioassay. Ann. Nucl. Med. 1994, 8, 231–237. [Google Scholar] [CrossRef]
- Zhang, K.; Zhang, F.J.; Zhao, W.J.; Xing, G.S.; Bai, X.; Wang, Y. Effects of parathyroid hormone-related protein on osteogenic and adipogenic differentiation of human mesenchymal stem cells. Eur. Rev. Med. Pharmacol. Sci. 2014, 18, 1610–1617. [Google Scholar]
- Ratcliffe, W.A.; Bowden, S.J.; Dunne, F.P.; Hughes, S.; Emly, J.F.; Baker, J.T.; Pye, J.K.; Williams, C.P. Expression and processing of parathyroid hormone-related protein in a pancreatic endocrine cell tumour associated with hypercalcaemia. Clin. Endocrinol. 1994, 40, 679–686. [Google Scholar] [CrossRef]
- Washam, C.L.; Byrum, S.D.; Leitzel, K.; Ali, S.M.; Tackett, A.J.; Gaddy, D.; Sundermann, S.E.; Lipton, A.; Suva, L.J. Identification of PTHrP(12-48) as a plasma biomarker associated with breast cancer bone metastasis. Cancer Epidemiol. Biomarkers Prev. 2013, 22, 972–983. [Google Scholar] [CrossRef]
- Kamalakar, A.; Washam, C.L.; Akel, N.S.; Allen, B.J.; Williams, D.K.; Swain, F.L.; Leitzel, K.; Lipton, A.; Gaddy, D.; Suva, L.J. PTHrP(12-48) modulates the bone marrow microenvironment and suppresses human osteoclast differentiation and lifespan. J. Bone Miner. Res. 2017, 32, 1421–1431. [Google Scholar] [CrossRef]
- Care, A.D.; Abbas, S.K.; Pickard, D.W.; Barri, M.; Drinkhill, M.; Findlay, J.B.; White, I.R.; Caple, I.W. Stimulation of ovine placental transport of calcium and magnesium by mid-molecule fragments of human parathyroid hormone-related protein. Exp. Physiol. 1990, 75, 605–608. [Google Scholar] [CrossRef]
- Hastings, R.H.; Asirvatham, A.; Quintana, R.; Sandoval, R.; Dutta, R.; Burton, D.W.; Deftos, L.J. Parathyroid hormone-related protein-(38-64) regulates lung cell proliferation after silica injury. Am. J. Physiol. Lung Cell Mol. Physiol. 2002, 283, L12–L21. [Google Scholar] [CrossRef]
- Luparello, C.; Romanotto, R.; Tipa, A.; Sirchia, R.; Olmo, N.; Lopez de Silanes, I.; Turnay, J.; Lizarbe, M.A.; Stewart, A.F. Midregion parathyroid hormone-related protein inhibits growth and invasion in vitro and tumorigenesis in vivo of human breast cancer cells. J. Bone Miner. Res. 2001, 16, 2173–2181. [Google Scholar] [CrossRef]
- Luparello, C.; Sirchia, R.; Lo Sasso, B. Midregion PTHrP regulates rip1 and caspase expression in mda-mb231 breast cancer cells. Breast Cancer Res. Treat. 2008, 111, 461–474. [Google Scholar] [CrossRef]
- Wu, T.L.; Vasavada, R.C.; Yang, K.; Massfelder, T.; Ganz, M.; Abbas, S.K.; Care, A.D.; Stewart, A.F. Structural and physiologic characterization of the mid-region secretory species of parathyroid hormone-related protein. J. Biol. Chem. 1996, 271, 24371–24381. [Google Scholar] [CrossRef]
- Strid, H.; Care, A.; Jansson, T.; Powell, T. Parathyroid hormone-related peptide (38-94) amide stimulates atp-dependent calcium transport in the basal plasma membrane of the human syncytiotrophoblast. J. Endocrinol. 2002, 175, 517–524. [Google Scholar] [CrossRef][Green Version]
- Philbrick, W. Parathyroid hormone-related protein: Gene structure, biosynthesis, metabolism, and regulation. In The Parathyroids; Academic Press: Cambridge, MA, USA, 2001; pp. 31–51. ISBN 978-0-12-098651-4. [Google Scholar]
- Luparello, C.; Sirchia, R.; Pupello, D. PTHrP [67-86] regulates the expression of stress proteins in breast cancer cells inducing modifications in urokinase-plasminogen activator and mmp-1 expression 5. J. Cell Sci. 2003, 116, 2421–2430. [Google Scholar] [CrossRef]
- Kovacs, C.S.; Lanske, B.; Hunzelman, J.L.; Guo, J.; Karaplis, A.C.; Kronenberg, H.M. Parathyroid hormone-related peptide (PTHrP) regulates fetal-placental calcium transport through a receptor distinct from the PTH/PTHrP receptor. Proc. Natl. Acad. Sci. USA 1996, 93, 15233–15238. [Google Scholar] [CrossRef]
- Hastings, R.H.; Quintana, R.A.; Sandoval, R.; Duey, D.; Rascon, Y.; Burton, D.W.; Deftos, L.J. Proapoptotic effects of parathyroid hormone-related protein in type ii pneumocytes. Am. J. Respir. Cell Mol. Biol. 2003, 29, 733–742. [Google Scholar] [CrossRef]
- Rihani-Basharat, S.; Lewinson, D. PTHrP(107-111) inhibits in vivo resorption that was stimulated by PTHrP(1-34) when applied intermittently to neonatal mice. Calcif. Tissue Int. 1997, 61, 426–428. [Google Scholar] [CrossRef]
- Kaji, H.; Sugimoto, T.; Kanatani, M.; Fukase, M.; Chihara, K. Carboxyl-terminal peptides from parathyroid hormone-related protein stimulate osteoclast-like cell formation. Endocrinology 1995, 136, 842–848. [Google Scholar] [CrossRef]
- Fenton, A.J.; Kemp, B.E.; Hammonds, R.G., Jr.; Mitchelhill, K.; Moseley, J.M.; Martin, T.J.; Nicholson, G.C. A potent inhibitor of osteoclastic bone resorption within a highly conserved pentapeptide region of parathyroid hormone-related protein; PTHrP[107-111]. Endocrinology 1991, 129, 3424–3426. [Google Scholar] [CrossRef]
- Conlan, L.A.; Martin, T.J.; Gillespie, M.T. The cooh-terminus of parathyroid hormone-related protein (PTHrP) interacts with beta-arrestin 1b. FEBS Lett 2002, 527, 71–75. [Google Scholar] [CrossRef]
- Burtis, W.J.; Brady, T.G.; Orloff, J.J.; Ersbak, J.B.; Warrell, R.P., Jr.; Olson, B.R.; Wu, T.L.; Mitnick, M.E.; Broadus, A.E.; Stewart, A.F. Immunochemical characterization of circulating parathyroid hormone-related protein in patients with humoral hypercalcemia of cancer. N. Engl. J. Med. 1990, 322, 1106–1112. [Google Scholar] [CrossRef]
- Plawner, L.L.; Philbrick, W.M.; Burtis, W.J.; Broadus, A.E.; Stewart, A.F. Cell type-specific secretion of parathyroid hormone-related protein via the regulated versus the constitutive secretory pathway. J. Biol. Chem. 1995, 270, 14078–14084. [Google Scholar] [CrossRef]
- Nguyen, M.; He, B.; Karaplis, A. Nuclear forms of parathyroid hormone-related peptide are translated from non-aug start sites downstream from the initiator methionine. Endocrinology 2001, 142, 694–703. [Google Scholar] [CrossRef]
- Lam, M.H.; Briggs, L.J.; Hu, W.; Martin, T.J.; Gillespie, M.T.; Jans, D.A. Importin beta recognizes parathyroid hormone-related protein with high affinity and mediates its nuclear import in the absence of importin alpha. J. Biol. Chem. 1999, 274, 7391–7398. [Google Scholar] [CrossRef]
- Truant, R.; Cullen, B.R. The arginine-rich domains present in human immunodeficiency virus type 1 tat and rev function as direct importin beta-dependent nuclear localization signals. Mol. Cell Biol. 1999, 19, 1210–1217. [Google Scholar] [CrossRef]
- Gu, Z.; Liu, Y.; Zhang, Y.; Jin, S.; Chen, Q.; Goltzman, D.; Karaplis, A.; Miao, D. Absence of PTHrP nuclear localization and carboxyl terminus sequences leads to abnormal brain development and function. PLoS One 2012, 7, e41542. [Google Scholar] [CrossRef]
- Fenton, A.J.; Martin, T.J.; Nicholson, G.C. Long-term culture of disaggregated rat osteoclasts: Inhibition of bone resorption and reduction of osteoclast-like cell number by calcitonin and PTHrP[107-139]. J. Cell Physiol. 1993, 155, 1–7. [Google Scholar] [CrossRef]
- Cornish, J.; Callon, K.E.; Nicholson, G.C.; Reid, I.R. Parathyroid hormone-related protein-(107-139) inhibits bone resorption in vivo. Endocrinology 1997, 138, 1299–1304. [Google Scholar] [CrossRef]
- de Castro, L.F.; Lozano, D.; Portal-Nunez, S.; Maycas, M.; De la Fuente, M.; Caeiro, J.R.; Esbrit, P. Comparison of the skeletal effects induced by daily administration of PTHrP (1-36) and PTHrP (107-139) to ovariectomized mice. J. Cell Physiol. 2012, 227, 1752–1760. [Google Scholar] [CrossRef]
- Donovan, P.J.; Achong, N.; Griffin, K.; Galligan, J.; Pretorius, C.J.; McLeod, D.S. PTHrP-mediated hypercalcemia: Causes and survival in 138 patients. J. Clin. Endocrinol. Metab. 2015, 100, 2024–2029. [Google Scholar] [CrossRef]
- Pioszak, A.A.; Parker, N.R.; Gardella, T.J.; Xu, H.E. Structural basis for parathyroid hormone-related protein binding to the parathyroid hormone receptor and design of conformation-selective peptides. J. Biol. Chem. 2009, 284, 28382–28391. [Google Scholar] [CrossRef]
- Gardella, T.J.; Juppner, H. Molecular properties of the PTH/PTHrP receptor. Trends Endocrinol Metab 2001, 12, 210–217. [Google Scholar] [CrossRef]
- Errecart, M.T.; Ross, J.G.; Robb, W.; Warren, C.W.; Kann, L.; Collins, J.L.; Pateman, B.C.; Small, M.L.; Sundberg, E.C. Methodology. J. Sch. Health 1995, 65, 295–301. [Google Scholar] [CrossRef]
- Valin, A.; Garcia-Ocana, A.; De Miguel, F.; Sarasa, J.L.; Esbrit, P. Antiproliferative effect of the c-terminal fragments of parathyroid hormone-related protein, PTHrP-(107-111) and (107-139), on osteoblastic osteosarcoma cells. J. Cell Physiol. 1997, 170, 209–215. [Google Scholar] [CrossRef]
- Whitfield, J.F.; Isaacs, R.J.; Chakravarthy, B.R.; Durkin, J.P.; Morley, P.; Neugebauer, W.; Williams, R.E.; Willick, G.; Rixon, R.H. C-terminal fragments of parathyroid hormone-related protein, PTHrP-(107-111) and (107-139), and the n-terminal PTHrP-(1-40) fragment stimulate membrane-associated protein kinase c activity in rat spleen lymphocytes. J. Cell Physiol. 1994, 158, 518–522. [Google Scholar] [CrossRef]
- Esbrit, P.; Alvarez-Arroyo, M.V.; De Miguel, F.; Martin, O.; Martinez, M.E.; Caramelo, C. C-terminal parathyroid hormone-related protein increases vascular endothelial growth factor in human osteoblastic cells. J. Am. Soc. Nephrol 2000, 11, 1085–1092. [Google Scholar]
- Boileau, G.; Tenenhouse, H.S.; Desgroseillers, L.; Crine, P. Characterization of phex endopeptidase catalytic activity: Identification of parathyroid-hormone-related peptide107-139 as a substrate and osteocalcin, ppi and phosphate as inhibitors. Biochem. J. 2001, 355, 707–713. [Google Scholar] [CrossRef]
- Mohammad, K.S.; Guise, T.A. Mechanisms of osteoblastic metastases: Role of endothelin-1. Clin. Orthop. Relat. Res. 2003, S67–S74. [Google Scholar] [CrossRef]
- Langlois, C.; Letourneau, M.; Turcotte, K.; Detheux, M.; Fournier, A. PTHrP fragments 1-16 and 1-23 do not bind to either the eta or the etb endothelin receptors. Peptides 2005, 26, 1436–1440. [Google Scholar] [CrossRef]
- Lam, M.H.; Thomas, R.J.; Loveland, K.L.; Schilders, S.; Gu, M.; Martin, T.J.; Gillespie, M.T.; Jans, D.A. Nuclear transport of parathyroid hormone (PTH)-related protein is dependent on microtubules. Mol. Endocrinol. 2002, 16, 390–401. [Google Scholar] [CrossRef]
- Lopez-Otin, C.; Bond, J.S. Proteases: Multifunctional enzymes in life and disease. J. Biol. Chem. 2008, 283, 30433–30437. [Google Scholar] [CrossRef]
- Fingleton, B.; Lynch, C.C. A new dress code for mmps: Cleavage optional. Dev. Cell 2010, 18, 3–4. [Google Scholar] [CrossRef]
- Lopez-Otin, C.; Overall, C.M. Protease degradomics: A new challenge for proteomics. Nat. Rev. Mol. Cell Biol. 2002, 3, 509–519. [Google Scholar] [CrossRef]
- Turk, B. Targeting proteases: Successes, failures and future prospects. Nat. Rev. Drug Discov. 2006, 5, 785–799. [Google Scholar] [CrossRef]
- Fortelny, N.; Cox, J.H.; Kappelhoff, R.; Starr, A.E.; Lange, P.F.; Pavlidis, P.; Overall, C.M. Network analyses reveal pervasive functional regulation between proteases in the human protease web. PLoS Biol. 2014, 12, e1001869. [Google Scholar] [CrossRef]
- Kappelhoff, R.; Puente, X.S.; Wilson, C.H.; Seth, A.; Lopez-Otin, C.; Overall, C.M. Overview of transcriptomic analysis of all human proteases, non-proteolytic homologs and inhibitors: Organ, tissue and ovarian cancer cell line expression profiling of the human protease degradome by the clip-chip DNA microarray. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 2210–2219. [Google Scholar] [CrossRef]
- Hendy, G.N.; Bennett, H.P.; Gibbs, B.F.; Lazure, C.; Day, R.; Seidah, N.G. Proparathyroid hormone is preferentially cleaved to parathyroid hormone by the prohormone convertase furin. A mass spectrometric study. J. Biol. Chem. 1995, 270, 9517–9525. [Google Scholar] [CrossRef]
- Seidah, N.G.; Prat, A. The biology and therapeutic targeting of the proprotein convertases. Nat. Rev. Drug Discov. 2012, 11, 367–383. [Google Scholar] [CrossRef]
- Diefenbach-Jagger, H.; Brenner, C.; Kemp, B.E.; Baron, W.; McLean, J.; Martin, T.J.; Moseley, J.M. Arg21 is the preferred kexin cleavage site in parathyroid-hormone-related protein. Eur J. Biochem. 1995, 229, 91–98. [Google Scholar] [CrossRef]
- Hara, M.; Koyanagi, Y.; Inoue, T.; Fukuyama, T. some physico-chemical characteristics of " -seminoprotein", an antigenic component specific for human seminal plasma. Forensic immunological study of body fluids and secretion. Vii. Nihon Hoigaku Zasshi 1971, 25, 322–324. [Google Scholar]
- Balk, S.P.; Ko, Y.J.; Bubley, G.J. Biology of prostate-specific antigen. J. Clin. Oncol. 2003, 21, 383–391. [Google Scholar] [CrossRef]
- Mattsson, J.M.; Ravela, S.; Hekim, C.; Jonsson, M.; Malm, J.; Narvanen, A.; Stenman, U.H.; Koistinen, H. Proteolytic activity of prostate-specific antigen (psa) towards protein substrates and effect of peptides stimulating psa activity. PLoS One 2014, 9, e107819. [Google Scholar] [CrossRef]
- Hong, S.K. Kallikreins as biomarkers for prostate cancer. Biomed. Res. Int. 2014, 2014, 526341. [Google Scholar] [CrossRef]
- Webber, M.M.; Waghray, A.; Bello, D. Prostate-specific antigen, a serine protease, facilitates human prostate cancer cell invasion. Clin. Cancer Res. 1995, 1, 1089–1094. [Google Scholar]
- Cohen, P.; Graves, H.C.; Peehl, D.M.; Kamarei, M.; Giudice, L.C.; Rosenfeld, R.G. Prostate-specific antigen (psa) is an insulin-like growth factor binding protein-3 protease found in seminal plasma. J. Clin. Endocrinol. Metab. 1992, 75, 1046–1053. [Google Scholar]
- Kanety, H.; Madjar, Y.; Dagan, Y.; Levi, J.; Papa, M.Z.; Pariente, C.; Goldwasser, B.; Karasik, A. Serum insulin-like growth factor-binding protein-2 (igfbp-2) is increased and igfbp-3 is decreased in patients with prostate cancer: Correlation with serum prostate-specific antigen. J. Clin. Endocrinol. Metab. 1993, 77, 229–233. [Google Scholar]
- Williams, S.A.; Singh, P.; Isaacs, J.T.; Denmeade, S.R. Does psa play a role as a promoting agent during the initiation and/or progression of prostate cancer? Prostate 2007, 67, 312–329. [Google Scholar] [CrossRef]
- Iwamura, M.; Hellman, J.; Cockett, A.T.; Lilja, H.; Gershagen, S. Alteration of the hormonal bioactivity of parathyroid hormone-related protein (PTHrP) as a result of limited proteolysis by prostate-specific antigen. Urology 1996, 48, 317–325. [Google Scholar] [CrossRef]
- Yu, H.; Diamandis, E.P.; Sutherland, D.J. Immunoreactive prostate-specific antigen levels in female and male breast tumors and its association with steroid hormone receptors and patient age. Clin. Biochem. 1994, 27, 75–79. [Google Scholar] [CrossRef]
- Papotti, M.; Paties, C.; Peveri, V.; Moscuzza, L.; Bussolati, G. Immunocytochemical detection of prostate-specific antigen (psa) in skin adnexal and breast tissues and tumors. Basic Appl Histochem 1989, 33, 25–29. [Google Scholar]
- Chang, C.; Werb, Z. The many faces of metalloproteases: Cell growth, invasion, angiogenesis and metastasis. Trends Cell Biol. 2001, 11, S37–S43. [Google Scholar] [CrossRef]
- Roques, B.P.; Noble, F.; Dauge, V.; Fournie-Zaluski, M.C.; Beaumont, A. Neutral endopeptidase 24.11: Structure, inhibition, and experimental and clinical pharmacology. Pharmacol Rev. 1993, 45, 87–146. [Google Scholar]
- Turner, A.J.; Tanzawa, K. Mammalian membrane metallopeptidases: Nep, ece, kell, and pex. FASEB J. 1997, 11, 355–364. [Google Scholar] [CrossRef]
- Maguer-Satta, V.; Besancon, R.; Bachelard-Cascales, E. Concise review: Neutral endopeptidase (cd10): A multifaceted environment actor in stem cells, physiological mechanisms, and cancer. Stem Cells 2011, 29, 389–396. [Google Scholar] [CrossRef]
- Nagase, H.; Visse, R.; Murphy, G. Structure and function of matrix metalloproteinases and timps. Cardiovasc Res. 2006, 69, 562–573. [Google Scholar] [CrossRef]
- Thiolloy, S.; Edwards, J.R.; Fingleton, B.; Rifkin, D.B.; Matrisian, L.M.; Lynch, C.C. An osteoblast-derived proteinase controls tumor cell survival via tgf-beta activation in the bone microenvironment. PLoS One 2012, 7, e29862. [Google Scholar] [CrossRef]
- Bruni-Cardoso, A.; Johnson, L.C.; Vessella, R.L.; Peterson, T.E.; Lynch, C.C. Osteoclast-derived matrix metalloproteinase-9 directly affects angiogenesis in the prostate tumor-bone microenvironment. Mol. Cancer Res. 2010, 8, 459–470. [Google Scholar] [CrossRef]
- Lynch, C.C.; Hikosaka, A.; Acuff, H.B.; Martin, M.D.; Kawai, N.; Singh, R.K.; Vargo-Gogola, T.C.; Begtrup, J.L.; Peterson, T.E.; Fingleton, B.; et al. Mmp-7 promotes prostate cancer-induced osteolysis via the solubilization of rankl. Cancer Cell 2005, 7, 485–496. [Google Scholar] [CrossRef]
- Toribio, R.E.; Kohn, C.W.; Capen, C.C.; Rosol, T.J. Parathyroid hormone (PTH) secretion, PTH mrna and calcium-sensing receptor mrna expression in equine parathyroid cells, and effects of interleukin (il)-1, il-6, and tumor necrosis factor-alpha on equine parathyroid cell function. J. Mol. Endocrinol 2003, 31, 609–620. [Google Scholar] [CrossRef]
- The human protein atlas. Available online: https://www.proteinatlas.org/ENSG00000140564-FURIN/tissue/parathyroid+gland (accessed on 5 June 2019).
- Khan, A.R.; James, M.N. Molecular mechanisms for the conversion of zymogens to active proteolytic enzymes. Protein Sci. 1998, 7, 815–836. [Google Scholar] [CrossRef]
- Kawashima-Ohya, Y.; Satakeda, H.; Kuruta, Y.; Kawamoto, T.; Yan, W.; Akagawa, Y.; Hayakawa, T.; Noshiro, M.; Okada, Y.; Nakamura, S.; et al. Effects of parathyroid hormone (PTH) and PTH-related peptide on expressions of matrix metalloproteinase-2, -3, and -9 in growth plate chondrocyte cultures. Endocrinology 1998, 139, 2120–2127. [Google Scholar] [CrossRef]
- Yoon, H.; Blaber, S.I.; Li, W.; Scarisbrick, I.A.; Blaber, M. Activation profiles of human kallikrein-related peptidases by matrix metalloproteinases. Biol. Chem. 2013, 394, 137–147. [Google Scholar] [CrossRef]
- Gonnelli, S.; Caffarelli, C. Abaloparatide. Clin. Cases Miner. Bone Metab. 2016, 13, 106–109. [Google Scholar] [CrossRef]
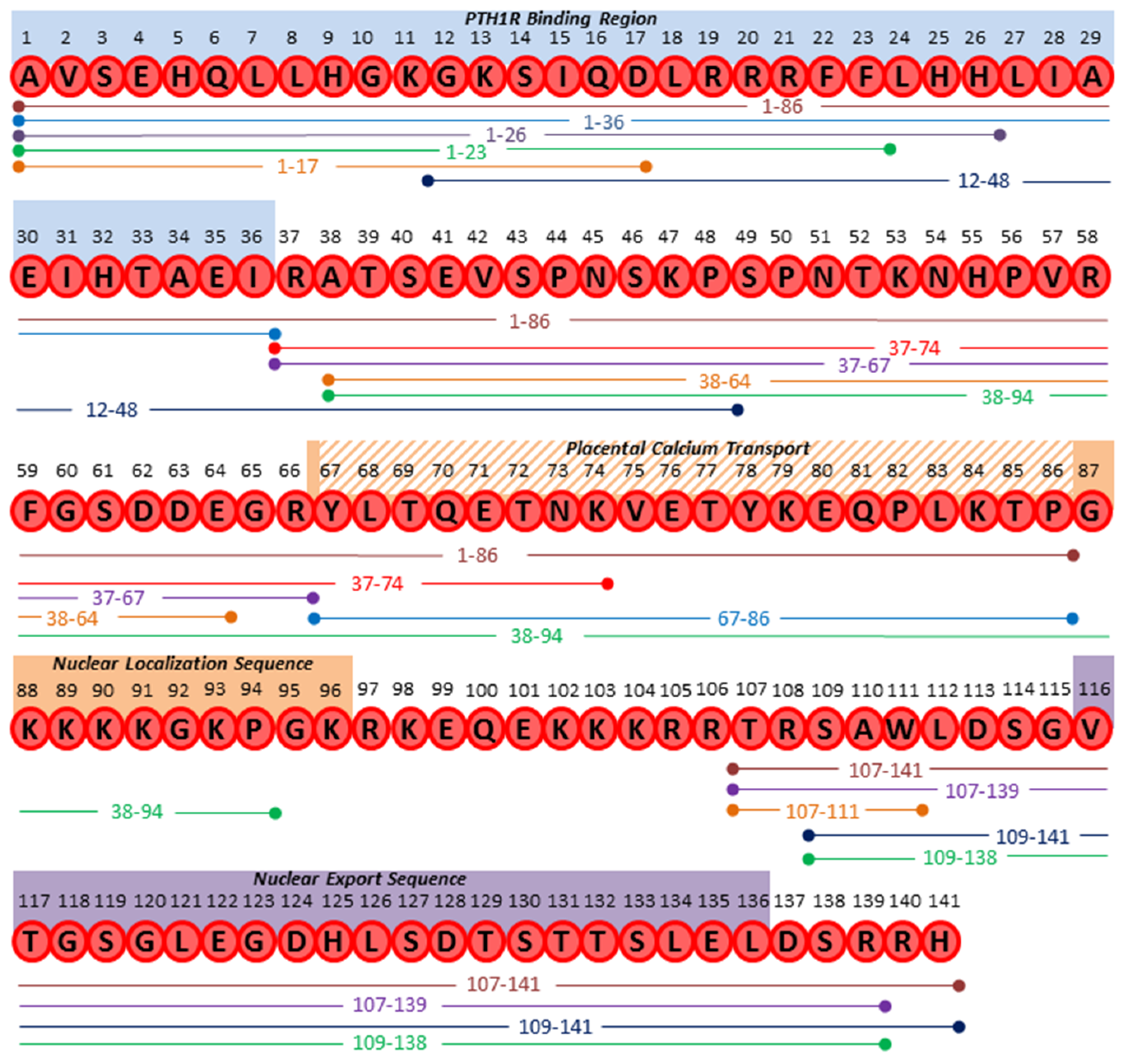
| Product | Putative Proteases | Proposed Proteases | Function | Reference |
|---|---|---|---|---|
| 1–139/141/173 | n/a | Full Length Protein | ||
| 1–36 | Prohormone Thiol Protease (PTP), Furin | PC1, 2, 4, 5, PACE4 | “Mature PTHrP;” bone homeostasis and developmental roles; ubiquitous tissue expression | [4,18,60] |
| 1–16 | Unknown | n/a | Cardiomyocyte contractility; activation of cardiac endothelin-A receptor | [61] |
| 1–17 | MMP-2, 3, 7, 9 | n/a | Stimulates osteogenesis in vitro and in vivo but does not induce osteolysis; secreted by prostate cancer and osteosarcoma cell lines | [62] |
| 1–23 | Neprilysin; Prostate Specific Antigen (PSA) | n/a | Abolished cAMP induction in vitro in mouse osteoblasts; new bone formation in calvaria | [63,64,65] |
| 1–26 | Neprilysin | n/a | Unknown | [64] |
| 1–74 | Unknown | n/a | Anabolic effects in bone at low dosage in vivo (synthetic peptide) | [66,67] |
| 1–84 | Unknown | Trypsin-like or carboxypeptidase B-like enzymes? | Stimulates calcium transport across placenta (in sheep) | [68] |
| 1–86 | Unknown | Trypsin-like or carboxypeptidase B-like enzymes? | Promotes osteogenesis and inhibits adipogenesis of human MSCs; positive correlation with humoral hypercalcemia of malignancy; detected in circulation from patients and in breast cancer tissue extracts | [69,70,71,72,73] |
| 1–108 | Unknown | n/a | Stimulates calcium transport across placenta (in sheep) | [68] |
| 12–48 | Prolyl oligopeptidase | PSA; Dipeptidyl peptidase | Suppression of osteoclast differentiation and survival in vitro; prognostic for bone metastases in breast cancer patient plasma | [4,74,75] |
| 37–67 | Unknown | Furin; Prohormone thiol protease (PTP) | Functions unclear; immunoreactivity within amnion covering placenta; Present in pancreatic and breast cancer tissue extracts | [67,69,70,73] |
| 37–74 | Unknown | Furin; Prohormone thiol protease (PTP) | Functions unclear; present in plasma from patients with humoral hypercalcemia of malignancy | [67,73] |
| 37–94 | Unknown | Furin; Prohormone thiol protease (PTP) | Stimulates calcium transport across placenta (in sheep) | [76] |
| 38–64 | Unknown | n/a | Promotes type II lung cell growth and repair | [77] |
| 38–94 | Unknown | Peptidyl α-amidating mono-oxygenase | Inhibition of growth, invasion, and viability in breast cancer cell lines in vitro; attenuated breast cancer tumorigenesis in vivo; translocation to nucleoplasm and binding of chromatin; potent activator of intracellular calcium signaling pathway; placental calcium transport; secreted in vitro by RIN cells transfected to express PTHrP | [78,79,80,81] |
| 38–95 | Unknown | n/a | Functions unclear; secreted in vitro by RIN cells transfected to express PTHrP | [80] |
| 38–101 | Unknown | n/a | Functions unclear; secreted in vitro by RIN cells transfected to express PTHrP | [80] |
| 38–111 | Unknown | n/a | Functions unclear; detected in circulation | [82] |
| 67–86 | Unknown | Trypsin-like or carboxypeptidase B-like enzymes | Stimulates calcium transport across placenta from maternal to fetal circulation (in sheep); stimulates intracellular calcium and inositol phosphates in human squamous carcinoma cells; inhibit mitogenesis but stimulate metastatic potential of human breast cancer cell line 8701-BC; proapoptotic in type II lung cells | [55,76,83,84,85] |
| 75–84 | Unknown | Trypsin-like or carboxypeptidase B-like enzymes | Stimulates calcium transport across placenta from maternal to fetal circulation (in sheep) | [76] |
| 75–86 | Unknown | Trypsin-like or carboxypeptidase B-like enzymes | Stimulates calcium transport across placenta from maternal to fetal circulation (in sheep) | [68] |
| 107–111 | Unknown | Furin; PHEX | Inhibition of osteoclast resorption in vivo; bone anabolic effects in vivo | [86,87] |
| 107–139 | Unknown | Furin | Inhibition of osteoclast resorption in vivo; bone anabolic effects in vivo; osteogenic differentiation in vitro | [88] |
| 107–141 | Unknown | Furin | Interacts with cytoplasmic β-arrestin in yeast two-hybrid system (function unclear) | [89] |
| 109–138 | Unknown | Furin | Secreted by RIN cells transfected to express PTHrP; elevated levels in detected in plasma from patients with humoral hypercalcemia of malignancy | [90,91] |
| 109–141 | Unknown | Furin | Detected in patients with humoral hypercalcemia of malignancy | [71] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Frieling, J.S.; Lynch, C.C. Proteolytic Regulation of Parathyroid Hormone-Related Protein: Functional Implications for Skeletal Malignancy. Int. J. Mol. Sci. 2019, 20, 2814. https://doi.org/10.3390/ijms20112814
Frieling JS, Lynch CC. Proteolytic Regulation of Parathyroid Hormone-Related Protein: Functional Implications for Skeletal Malignancy. International Journal of Molecular Sciences. 2019; 20(11):2814. https://doi.org/10.3390/ijms20112814
Chicago/Turabian StyleFrieling, Jeremy S., and Conor C. Lynch. 2019. "Proteolytic Regulation of Parathyroid Hormone-Related Protein: Functional Implications for Skeletal Malignancy" International Journal of Molecular Sciences 20, no. 11: 2814. https://doi.org/10.3390/ijms20112814
APA StyleFrieling, J. S., & Lynch, C. C. (2019). Proteolytic Regulation of Parathyroid Hormone-Related Protein: Functional Implications for Skeletal Malignancy. International Journal of Molecular Sciences, 20(11), 2814. https://doi.org/10.3390/ijms20112814




