Abstract
The house swine (Sus scrofa domestica Linnaeus 1758) is an important model organism regarding the study of neurodegenerative diseases, especially ocular neuropathies such as glaucoma. This is due to the high comparability of the porcine and human eye regarding anatomy and molecular features. In the pathogenesis of glaucoma, the trabecular meshwork (TM) forms a key ocular component in terms of intraocular pressure (IOP) elevation. Thereby, functional TM abnormalities are correlated with distinct proteomic alterations. However, a detailed analysis of the TM proteome has not been realized so far. Since the porcine eye has high potential as a model system to study ocular diseases such as glaucoma, the present study focuses on the in-depth analysis of the porcine TM proteome. By use of a bottom-up (BU) mass spectrometric (MS) platform utilizing electrospray ionization liquid chromatography tandem MS (LC-ESI-MS/MS) considering database-dependent and peptide de novo sequencing, more than 3000 TM proteins were documented with high confidence (FDR < 1%). A distinct number of proteins with neuronal association were revealed. To the best to our knowledge, many of these protein species have not been reported for TM tissue before such as reelin, centlein and high abundant neuroblast differentiation-associated protein AHNAK (AHNAK). Thereby, AHNAK might play a superordinate role in the TM regarding proposed tissue involvement in barrier function. Also, a high number of secretory proteins could be identified. The generated TM proteomic landscape underlines a multifunctional character of the TM beyond representing a simple drainage system. Finally, the protein catalogue of the porcine TM provides an in-depth view of the TM molecular landscape and will serve as an important reference map in terms of glaucoma research utilizing porcine animal models, porcine TM tissues and/or cultured TM cells.
1. Introduction
The trabecular meshwork (TM) is a highly specialized eye tissue responsible for the regulation of the anterior eye chamber aqueous humor (AH) outflow and the control of the intraocular pressure (IOP) [1,2]. Since an elevated IOP represents the predominant risk factor for primary open angle glaucoma (POAG) in humans [3,4], TM tissue and disease-associated structural, molecular and functional alterations are important structures in glaucoma research. Pescosolido et al. (2012) [5], for instance, postulated that particular glycosaminoglycans undergo specific pathogenic structural changes in the TM tissue of glaucoma patients and may responsible for IOP elevation. However, distinctive proteomic alterations of the TM tissue were correlated with glaucomatous conditions as recently reviewed in detail by our group [6]. The contractile TM, which is located at the corneoscleral transition zone, consists of different cellular components encompassing endothelial, stem and smooth muscle cells as well as the extracellular matrix (ECM). All components are organized in a functional system comprising the Schlemm‘s canal [2,7,8,9], which is represented by a multiple vessel system in most non-primate mammals including the domestic pig [10]. Considering the anatomical ocular architecture and neuronal comparability of pigs and humans, the house swine (Sus scrofa domestica Linnaeus 1758) represents an attractive model organism, especially for the study of ocular neuropathies including glaucoma [11,12,13,14]. Thereby, an important advantage is the unproblematic availability of porcine ocular tissue material for the establishment of an ocular organ culture [15]. With regard to the limited TM tissue amount which can be obtained from a single eye ball, the easy accessibility of porcine material is highly advantageous to source appropriate protein amount for in-depth discovery and other proteomic profiling studies. Regarding TM morphology, in comparison with other non-primate species, the domestic pig TM shows the highest similarity to the human TM [10,16]. Since TM structural and functional changes are associated with aberrant proteomic alterations [6] only a few works have characterized the complex TM proteome with high sensitivity, and a detailed protein catalogue of the porcine TM is still missing. Accordingly, the aim of the present work was to analyze the TM proteome of an important study animal by use of an established “bottom up” high performance liquid chromatography tandem mass spectrometry (BULCMS) workflow and to provide a detailed protein map, which can serve as a reference in terms of ocular proteomics in future. Sensitive TM proteomic analysis should importantly contribute to the molecular understanding of this highly specialized tissue. Finally, in correspondence with research on anatomical features of the porcine eye [14], the present proteomic work should assist with the establishment of the house swine as a reliable model system organism for ocular diseases with a special focus on glaucoma on the molecular level.
2. Results
Excised trabecular meshwork (TM) tissues showed a high degree of purity. No connective tissue contaminants could be indicated regarding the microscopic inspections (Figure 1A–D). TM extracts displayed distinctive 1D SDS PAGE protein patterns appropriate for bottom-up liquid chromatography-mass spectrometric (BULCMS) analysis (Figure 1E). BULCMS analysis resulted in the identification of more than 3000 proteins considering a high confident protein identification (FDR < 1%) (see Table S1). The highest number of identified proteins (79%) could be achieved by use of a combinatory database-related/de novo peptide sequencing strategy. Regarding the identified TM proteins a distinct degree of congruency could be achieved using both identification procedures (Figure 2). Approximately 93% of all identified proteins could be annotated to cellular components referring to GO analysis. The majority of proteins represented intracellular species, whereby only nearly 6% of annotated TM proteins could be exclusively associated with the extracellular milieu (Figure 3A). Thereby, besides the identification of collagens (I, III, IV), tenascins and laminins, further important extracellular matrix (ECM) proteins comprising lumican, nidigen-1 and 2, sushi nidogen and EGF-like domain-containing protein 1, γ, podocan, fibulin-5, retinoic acid receptor responder protein 2, calreticulin, EGF-containing fibulin-like ECM protein 1, prolyl 3-hydroxylase 1, biglycan, prolargin, matrix metalloprotease 9 and thrombospondins could be catalogued confidently with respect to the current literature concerning the composition of TM ECM subproteomes [17,18,19]. Also, contractile TM elements such as tropomyosin α and β, myosin 9, 10 and 11 as well as cytoskeletal proteins vimentin, actin and tubulin forms could be reported with high identification scores. In terms of the subcellular distribution, most annotated TM proteins were found to be associated with the nucleus (Figure 3C), e.g., histones, nucleolin, serine/arginine-rich splicing factor 3 and heterogeneous nuclear ribonucleoproteins A2/B1. Moreover, the detection of characteristic marker proteins expressed by TM cells including aquaporin-1, α-crystallin B chain, matrix Gla protein and smooth muscle actin [20] support the cellular representation within the whole TM tissue samples. Interestingly, retina-specific copper amine oxidase and further retinal proteins including rhodopsin, interphotoreceptor matrix proteoglycan 1, retinal dehydrogenase 1 and 2, retinaldehyde-binding protein 1 and 2, retinal guanylyl cyclase 2 and retinal-specific ATP-binding cassette transporter could be identified in the TM tissue samples. Moreover, two myelin proteins could be identified with scores indicating distinct abundance in the TM tissue: myelin protein P0 (Proteome Discoverer score = 1700; PEAKS − 10logP = 189) and myelin basic protein (Proteome Discoverer score = 113; PEAKS − 10logP = 67). Approximately 4% of all annotated proteins were found to be associated with the myelin sheet. Also, nearly 14% of the annotated TM proteins were found to be neuronal related (Figure 3D) with respect to the GO database categories “cellular component” and ”molecular function”, e.g., reelin, centlein, neuron navigator 3, neuroblastoma amplified sequence, neurobeaching-like proteins (1, 2), neurochondrin, neurofascin, neuron navigator 1 and neurabin-2 (see Table S2). Numerous neural marker proteins show moderate to high scores in correspondence with both protein identification software programs, e.g., neurofilament light polypeptide (ProtDisc = 1343, PEAKS − 10lgP = 100), glial fibrillary acidic protein (GFAP) (ProtDisc = 862, PEAKS − 10lgP = 154) or neural cell adhesion molecule 1 (ProtDisc = 150, PEAKS − 10lgP = 87), indicating high abundance of nerve-associated proteins in the TM tissues. Among the most abundant proteins with neuronal relevance, neuroblast differentiation-associated protein AHNAK (ProtDisc = 2537; PEAKS − 10lgP = 233) (Figure 4) could be recovered from TM tissues. Also, a high number of typical marker proteins associated with neuronal stress such as huntingtin and chaperones encircling heat shock proteins and crystallins could be detected. Furthermore, 46% of all TM proteins could be GO annotated regarding their molecular function. Also, approximately 11% of these annotated TM proteins were found to be involved in receptor binding including dopamine, type 2A serotonin, acetylcholine, GABA and glutamate receptor binding. Thereby, metabotropic glutamate receptors 1, 3 and 5 could be recovered from TM tissues. Ephrin type-B receptor 2, which is involved in axon guidance and translocator protein, are two exemplary important neuronal proteins recovered from TM tissues referred to in GO molecular function analysis. Furthermore, 13% of all annotated proteins are involved in cytoskeletal protein binding, especially actin binding. A comparison to the porcine retinal proteome [21] showed that approximately 47% of the focused porcine retina proteome was recovered in the TM tissue samples also comprising neuronal proteins. Approximately 66% of the TM proteins were exclusively documented for the TM proteome and have not been reported for the porcine retina proteome, which on the other hand underlines the ocular exclusivity of the TM proteome.
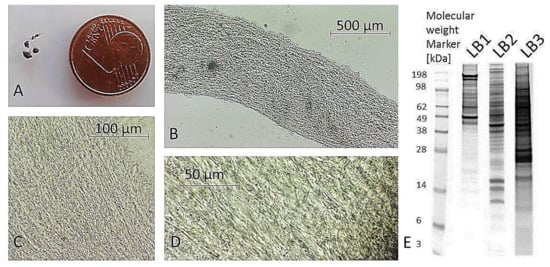
Figure 1.
Processed porcine trabecular meshwork (TM). (A) TM sample acquisition is difficult due to small size of processed TM tissue as indicated by the coin comparison. (B) Isolated TM displays no connective tissue remains indicating the purity of TM tissue sample (5× magnification). TM sample shows a typical mesh-like structure ((C) 10× magnification, (D) 40× magnification), (E) TM corresponding exemplary extracts fractionated by 1D-SDS PAGE using three different lysis protocols (LB1 = 2 M thiourea, 7 M urea, 30 mM tris-HCl, 4% CHAPS, 2% ASB-14 (8 µg protein); LB2 = 50 mM tris-HCl, 8 mM Na2HPO4, 137 mM NaCl, 3 mM KCl, 0,5% SDS, 0,5% triton X-100, 0,5% DDM (8 µg protein); LB3 = 0.5% DDM (50 µg)); all different extracts were used for combinatory BULCMS TM proteome analysis.
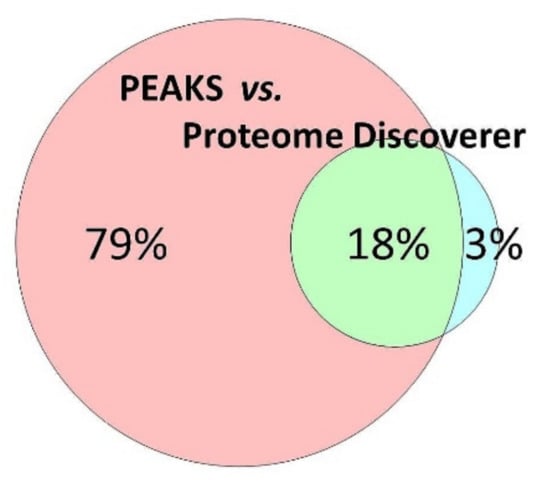
Figure 2.
The majority of the porcine trabecular meshwork (TM) could be identified by use of the PEAKS combinatory de novo peptide sequencing/database related protein identification strategy. In summary, more 3747 proteins could be documented for the porcine TM.
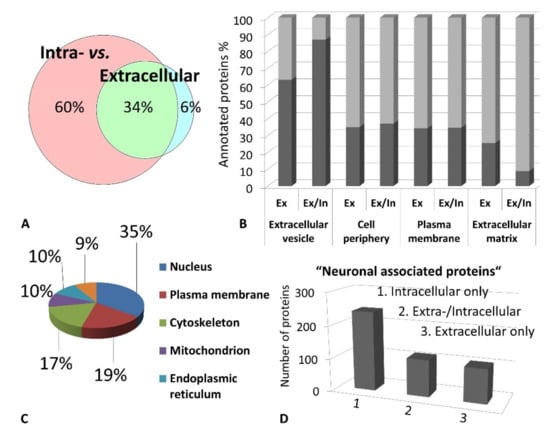
Figure 3.
(A). The majority of identified proteins correspond to the intracellular fraction of the trabecular meshwork (TM) proteome inferred from gene ontology (GO) annotation analysis. (B) Exclusively extracellular proteins (Ex) as well as proteins with localization in the extra-and intracellular milieu (Ex/In) correspond predominantly to extracellular vesicles indicating secretory processes. (C) Most intracellular TM proteins correspond to the nucleus followed by the cellular plasma membrane. (D) The majority of “neuronal associated proteins” are intracellular protein species.
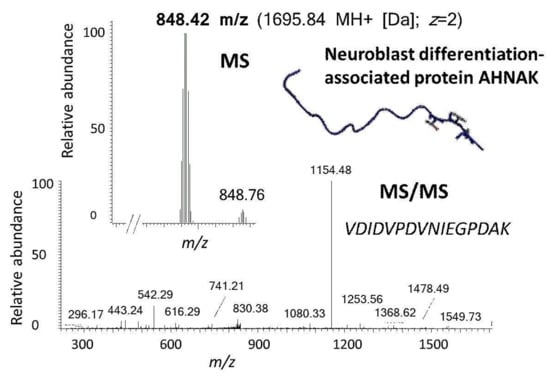
Figure 4.
Exemplary identification of a prominent barrier function-related protein of neuronal relevance in the porcine trabecular meshwork (TM) by use of the established BULCMS workflow. A unique reporter peptide peak 848.42 m/z (MS) was fragmented by collision-induced decay (CID) (MS/MS) leading to the stringent identification of neuroblast differentiation-associated protein AHNAK (FDR < 1%) and was supported by both, database-related and de novo peptide sequencing analysis. (AHNAK image from the RCSB PDB (rcsb.org) of PDB ID 4FTG [22]).
3. Discussion
The present work reports the largest trabecular meshwork (TM) protein catalogue to date, comprising more than 3000 highly confident TM-associated proteins. Moreover, extracellular as well as intracellular TM protein species could be recovered providing an in-depth view into the TM extracellular matrix (ECM) and TM cellular proteome of the house swine. Compared to former proteomic TM cell-related studies, a high congruency could be reported regarding studies cataloguing 235 [23], 718 [24], 853 [25] and 1644 proteins [26]. For the whole TM tissue specimen, 368 proteins have been recovered so far [27]. To our best knowledge, no study has been done on porcine TM tissues yet and therefore the protein map generated in the present work can serve as an important reference list in terms of ocular proteomics, especially suitable for glaucoma research using the domestic pig as a model organism. Based on similarities in architecture and molecular features the porcine eye has a high potential for the use as a model system in glaucoma [11,16] and recently our group provided a porcine retina organ culture for the study of glaucoma [15]. Regarding a comparison between the porcine retina and TM, numerous neuronal proteins are shared in both ocular sites; however, a high portion of TM proteins could be exclusively documented for the TM and has not been detected in the retina referring to a recent study [21], which might reflect the functional specialization but considering the neural origin. An important reason for the observed tissue specificity of the TM proteome most likely can be found in the TM ECM. Interestingly, TM ECM is highly comparative to the optic nerve head (ONH) lamina cribrosa ECM based on the proteomic level [28]. ECM and also its crosstalk with cell cytoskeleton play important roles in terms of the structural dynamic and the function of the TM [2]. A distinct recovery of TM ECM-specific proteins including tenascin (TNC), smooth muscle actin, fibronectin, integrins, collagen IV, laminin [1,29,30,31,32,33] support the proper illustration of the TM ECM proteome in the present work. Despite the fact that numerous ECM proteins are characteristic for TM ECM and neuronal ECM comprising laminins, collagens (I, IV), fibronectin and tenascin C [34], neurocan core protein, a neuronal specific ECM chrondoitin sulfate proteoglycan [35] was identified in the porcine TM in the present study. Also, the identification of many further neuronal-related proteins, such as highly abundant neuroblast differentiation-associated protein AHNAK in the TM is in confidence with the neural crest and the cranial paraxial mesodermal origin of the TM [36,37,38]. AHNAK is highly expressed in brain endothelial cells conciliating blood-brain barrier properties [39]. TM tissue-related barrier functions are important regarding the aqueous outflow channel isolation in the TM cribriform region with endothelial participation [8,40]. In light of this, it is highly thinkable, that referring to AHNAK TM tissue prominence, the protein plays a crucial role in maintaining this barrier function and therefore should be considered in future glaucoma studies with a special focus on TM. However, with respect to several neuronal proteins of lower abundance, their origin from TM surrounding retinal tissues might be assumed, and these proteins might represent tissue processing artifacts. Nevertheless, microscopic TM tissue quality inspection and high abundance of recovered key neuronal proteins, e.g., neurofilament polypeptides, indicate their TM-specific tissue origin. Also, traces of neuronal proteins could be sourced by scleral spur nerve endings suggested to function as afferent TM mechanoreceptors [41]. Interestingly, myelinated and unmyelinated nerves within the TM were supported for guinea pigs [42]. Also, innervation of the anterior eye outflow region was demonstrated in the house swine [43] and the presence of neuroregulatory cells was suggested for the primate TM [44]. Interestingly, TM cells display a similar CD44 cytotoxicity [45] such as neuroretinal cells, which supports the idea of a neuronal TM in accordance. This hypothesis is in confidence with the recovery of myelin basic, myelin protein P0 and further myelin associated proteins from the porcine TM tissues in the present study. Also, the detection of neuronal RNA binding proteins comprising RNA-binding protein Musashi homolog 1 and calcitonin [46] support the neuronal character of the TM tissue. Accordingly, TM proteomic data support the hypothesis of the TM-specific neuronal features. Despite the limitation to provide a subcellular proteomic view, but given the high number of recovered neural proteins, the present work contributes to the question of whether neural molecular features of the TM represent a contact point in glaucoma pathology. This thought is accompanied by the finding that comparable molecular events in TM and ONH drive neurodegeneration processes in glaucoma [47,48]. Regarding glaucoma related proteins, many of them could be identified in the porcine TM in this study, encompassing Toll-like receptor 4 [49], huntingtin [50], BMP-2-inducible protein [51], TGF-β-activated kinase 1 and MAP3K7-binding protein 1 [24], ADP/ATP translocase 3, heat shock proteins (e.g., HSP90β, HSP70) [52], crystallins [53,54,55,56] and integrins [57]. Also, retinol-binding protein 3, retinal dehydrogenase 2 and retinoic acid receptor responder protein 2 could be identified, which is interesting since TM expression of glaucoma-related myocilin [58] was demonstrated to be regulated by retinoic acid [59]. The recovery of glaucoma-associated proteins from the porcine TM tissue supports the house swine as an appropriate model system in terms of glaucoma TM proteomics. In conclusion, the present study provides a detailed view into the complex porcine TM proteome as an important reference for ocular studies, especially in terms of glaucoma research. It also represents an important contribution regarding “farm animal proteomics” [60], while strengthening the house swine as an attractive model organism in terms of ocular clinical proteomics. Also, the identification of neuronal-related proteins in the TM provides new input for the debate on TM origin and function.
4. Materials and Methods
4.1. Sample Preparation
Trabecular meshwork (TM) tissues were prepared from freshly enucleated eye bulbs (n = 30) from house swine Sus scrofa domestica individuals (sacrificed at 3–6 month, female: male = 3:2) provided by local slaughterhouses (Landmetzgerei Harth, Stadecken-Elsheim, Germany; Metzgerei Köppel, Mainz, Germany). The application of animal by-products for research purposes was approved by the Kreisverwaltung Mainz-Bingen in Germany (Identification Code: DE 07 315 0006 21, approved on 13 January 2014). Eye bulbs were equatorially opened with a scalpel removing lens, vitreous, iris and ciliary body. TM surrounding the porcine specific serial collector vessel “Schlemm‘s canal” representative [10] was carefully probed and isolated under a SZ 61 binocular microscope (Olympus, Tokyo, Japan), whereby tissue purity was visually inspected considering 5–40× magnification. TM tissue was immediately stored at −80 °C. TM tissues were homogenized by mechanical scratching using a 1.8 mm diamond milling head (LUX-TOOLS, Wermelskirchen, Germany), five freeze/thawing cycles (room temperature vs. −20 °C) followed by sonication. Homogenate aliquots corresponding to majority of eye bulbs (n = 24) were scheduled for one-step detergent protein extraction using separate detergents (detergent: homogenate = 1:1, v/v, (0.5% dodecyl-β-maltoside (DDM; Sigma-Aldrich, Steinheim, Germany), 0.1% sodium dodecyl sulfate (SDS; Sigma-Aldrich, Steinheim, Germany), 0.5% Tween 20 (Carl Roth, Karlsruhe, Germany), Triton-X-100 (Sigma-Aldrich, Steinheim, Germany)) and corresponding detergent mixture. Additionally sequential extraction was used, starting with 0.5% DDM followed by 0.1% SDS and finally 1% trifluoroacetic acid (TFA, Sigma-Aldrich, Steinheim, Germany) /20% acetonitrile (ACN; Applichem, Darmstadt, Germany). Additionally, TM samples from a few eye bulbs (n = 6) were extracted by use of two combinatory lysis buffers (buffer 1: 2M thiourea (Sigma-Aldrich, Steinheim, Germany), 7M urea (Carl Roth, Karlsruhe, Germany), 30mM Tris-HCl (Carl Roth, Karlsruhe, Germany) pH 8.5, 4% CHAPS (Sigma-Aldrich, Steinheim, Germany), 2% ASB14 (Sigma-Aldrich, Steinheim, Germany); buffer 2: 8mM Na2HPO4, 50mM Tris-HCl pH7, 137 mM natrium chloride (NaCl; Merck, Darmstadt, Germany), 3mM potassium chloride (KCl; Carl Roth, Karlsruhe, Germany), 0.5% SDS, 0.5% Triton-X-100, 0.5% DDM). Samples were centrifuged (2000× g, 15 min, 4°C) and extract supernatant aliquots were stored at −80°C for further BULCMS analysis.
4.2. Proteomic Analysis of TM Tissue
Protein concentration analysis of extracts was realized by use of the Pierce BCA protein assay kit (Thermo Fisher Scientific, Rockford, IL, USA) recorded by a Multiscan Ascent plate reader (Thermo Electron Corporation, Thermo Fisher Scientific, Rockford, IL, USA) at 570 nm according to the manufacturer‘s instructions. The TM samples (50 µg/sample) were run on 10-well NuPAGE 12% Bis-Tris minigels (Invitrogen, Carlsbad, CA, USA) for 1 h at 150 V under reduced conditions. Gels were fixed and stained using the Novex Colloidal Blue Staining Kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer‘s instructions followed by scanning on a DCP-9042 CDN bench top scanner (Brother Industries Ltd., Nagoya, Japan) at 1200 dpi. For tryptic in-gel digestion, a modified protocol of Shevchenkov and colleagues [61] was used. Solid phase extraction (SPE)-based peptide purification was carried out using C18 ZIPTIP pipette tips (Millipore, Billerica, MA, USA) according to the supplier’s protocol. For LC-MS/MS analysis, resolubilized (10 µL 0.1% TFA) peptides were fractionated using a Rheos Allegro (Thermo Fisher Scientific, Rockford, IL, USA) capillary HPLC system (BioBasic C18 pre (30 × 0.5 mm) & analytical column (150 × 0.5 mm); flow rate: 6.7 ± 0.03 μL/min) coupled to a LTQ Orbitrap XL system (Thermo Fisher Scientific, Rockford, IL, USA) with an electro spray ionization (ESI) source. For each peptide aliquot, an elution and MS record period of 50 min were set. The gradient program was set as follows: Buffers: A = 98% H2O, 1.94% ACN, 0.06% methanol, 0.05% formic acid; B = 95% ACN, 3% methanol, 2% H2O, 0.05% formic acid; gradient program: 15–20% B (0–2 min), 20–60% B (2–35 min), 60–100% B (35–40 min), 100–0% B (40–45 min), 0% B (45–50 min)]. MS parameters were set as follows: detection range= 300–2000 m/z, LTQ injection time = 50 ms, FT injection time = 500 ms, normalized energy of collision induced decay (CID) = 35, activation time = 30 ms, activation Q = 0.25, m/z isolation width for fragmentation = 2, dynamic exclusion = 90 s, repeat duration = 30 s, resolution = 30,000, centroid detection, fragmentation selection = top 5 monoisotopic m/z signals (z = 1+, 2+, 3+ and 4+, intensity >500). The BULCMS workflow was established in previous studies analyzing neuroretinal cell extracts [62,63] as well as human and porcine retinal tissues [21,52,64]. All resulting LC MS/MS Thermo RAW data were combined for protein identification using database-dependent protein identification in Proteome Discoverer (version 1.1, Thermo Fisher Scientific, Rockford, IL, USA) implementing the MASCOT search engine (version 2.2.07). A second protein identification search was performed using the combinatory database/de novo peptide sequencing software tool PEAKS (Studio version 7.5 trial; Bioinformatics Solutions Inc., Waterloo, ON, Canada). Mass tolerances were adjusted to 20 ppm (precursor peptides) and 0.8 Da (fragments). Carbamidomethylation (C) was set as fixed modification. Trypsin was chosen as enzyme and a maximum of two missed cleavages were allowed per peptide. For stringent protein identification, output was filtered confidently based on the false discovery rate (FDR) < 1%. In addition, for PEAKS output data, the matrix average local confidence peptide score (ALC) threshold was adjusted to >60%, and for proteins the −10logP score was set >20. For database screening, a combinatory strategy was selected considering Sus scrofa and Homo sapiens as taxonomies. A complemented TM protein list was scheduled for gene ontology (GO) analysis by use of Cytoscape (version 2.8.3 with BINGO 2.44 plugin; www.cytoscape.org). Thereby, GO categories “cellular component” and “molecular function” were screened for potential annotations. Moreover, the combined TM protein list was manually screened for functional protein associations. Protein-protein interaction (PPI) analysis was realized by use of STRING version 10 (http://string-db.org) [65]. For Venn chart analysis, the software package Venny (version 2.1, http://bioinfogp.cnb.csic.es/tools/venny/) [66] was used.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/20/10/2526/s1. The supplementary file contains Tables S1 and S2. Identified proteins are listed in Table S1 and list of neuronal-related proteins is provided in Table S2.
Author Contributions
S.F. developed the study design, organized the experimental set-up and wrote the manuscript. V.M.B. and L.B. performed the experiments, assisted with the mass spectrometric measurements and data analysis, proofread the manuscript and contributed important intellectual content. C.S., N.P. (Natarajan Perumal) and V.Z. participated in the data analysis, reviewed the manuscript and contributed important intellectual information. N.P. (Norbert Pfeiffer) critically proofread the manuscript and provided important intellectual input. F.H.G. performed the study coordination as well as the study design and participated in review and approval of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Vranka, J.A.; Kelley, M.J.; Acott, T.S.; Keller, K.E. Extracellular matrix in the trabecular meshwork: Intraocular pressure regulation and dysregulation in glaucoma. Exp. Eye Res. 2015, 133, 112–125. [Google Scholar] [CrossRef] [PubMed]
- Sacca, S.C.; Gandolfi, S.; Bagnis, A.; Manni, G.; Damonte, G.; Traverso, C.E.; Izzotti, A. The outflow pathway: A tissue with morphological and functional unity. J. Cell. Physiol. 2016, 231, 1876–1893. [Google Scholar] [CrossRef] [PubMed]
- Actis, A.G.; Versino, E.; Brogliatti, B.; Rolle, T. The ocular hypertension treatment study. Open Ophthalmol. J. 2016, 10, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Greco, A.; Rizzo, M.I.; De Virgilio, A.; Gallo, A.; Fusconi, M.; de Vincentiis, M. Emerging concepts in glaucoma and review of the literature. Am. J. Med. 2016. [Google Scholar] [CrossRef] [PubMed]
- Pescosolido, N.; Cavallotti, C.; Rusciano, D.; Nebbioso, M. Trabecular meshwork in normal and pathological eyes. Ultrastruct. Pathol. 2012, 36, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Funke, S.; Perumal, N.; Bell, K.; Pfeiffer, N.; Grus, F.H. The potential impact of recent insights into proteomic changes associated with glaucoma. Expert Rev. Proteom. 2017, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Alverado, J.A.; Yeh, R.F.; Franse-Carman, L.; Marcellino, G.; Brownstein, M.J. Interactions between endothelia of the trabecular meshwork and of schlemm‘s canal: A new insight into the regulation of aqueous outflow in the eye. Trans. Am. Ophthalmol. Soc. 2005, 103, 148–163. [Google Scholar]
- Abu-Hassan, D.W.; Acott, T.S.; Kelley, M.J. The trabecular meshwork: A basic review of form and function. J. Ocul. Biol. 2014, 2, 1–22. [Google Scholar]
- Lepple-Wienhues, A.; Stahl, F.; Wiederholt, M. Differential smooth muscle-like contractile properties of trabecular meshwork and ciliary muscle. Exp. Eye Res. 1991, 53, 33–38. [Google Scholar] [CrossRef]
- McMenamin, P.G.; Steptoe, R.J. Normal anatomy of the aqueous humour outflow system in the domestic pig eye. J. Anat. 1991, 178, 65–77. [Google Scholar] [PubMed]
- Ruiz-Ederra, J.; Garcia, M.; Hernandez, M.; Urcola, H.; Hernandez-Barbachano, E.; Araiz, J.; Vecino, E. The pig eye as a model of glaucoma. Exp. Eye Res. 2005, 81, 561–569. [Google Scholar] [CrossRef]
- Ceciliani, F.; Restelli, L.; Lecchi, C. Proteomics in farm animals models of human diseases. Proteom. Clin. Appl. 2014, 8, 677–688. [Google Scholar] [CrossRef]
- Marino, L.; Colvin, C.M. Thinking pigs: A comparative review of cognition, emotion, and personality in sus domesticus. Int. J. Comp. Psychol. 2015, 28, 1–22. [Google Scholar]
- Sanchez, I.; Martin, R.; Ussa, F.; Fernandez-Bueno, I. The parameters of the porcine eye ball. Graefes Arch. Clin. Exp. Ophthalmol. 2011, 249, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Bell, K.; Wilding, C.; Funke, S.; Perumal, N.; Beck, S.; Wolters, D.; Holz-Müller, J.; Pfeiffer, N.; Grus, F.H. Neuroprotective effects of antibodies on retinal ganglion cells in an adolescent retina organ culture. J. Neurochem. 2016, 139, 256–269. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, B.; Birke, M.; Kook, D.; Eichhorn, M.; Lütjen-Drecoll, E. Ultrastructural and biochemical evaluation of the porcine anteror chamber perfusion model. Investig. Ophthalmol. Vis. Sci. 2006, 47, 2011–2020. [Google Scholar] [CrossRef]
- Murphy, C.G.; Yun, A.J.; Newsome, D.A.; Alvarado, J.A. Localiation of extracellular proteins of the human trabecular meshwork by indirect immunofluorescence. Am. J. Ophthalmol. 1987, 104, 33–43. [Google Scholar] [CrossRef]
- Flügel, C.; Tamm, E.; Lütjen-Drecoll, E. Different cell populations in bovine trabecular meshwork: An ultrastructural and immunohistochemical study. Exp. Eye Res. 1991, 52, 681–690. [Google Scholar] [CrossRef]
- Hiscott, P.; Paraoan, L.; Choudhary, A.; Ordonez, J.L.; Al-Khaier, A.; Armstrong, D.J. Thrombospondin 1, thrombospondin 2 and the eye. Prog. Retin. Eye Res. 2006, 25, 1–18. [Google Scholar] [CrossRef]
- Stamer, W.D.; Clark, A.F. The many faces of the trabecular meshwork cell. Exp. Eye Res. 2017, 158, 112–123. [Google Scholar] [CrossRef]
- Funke, S.; Markowitsch, S.; Schmelter, C.; Perumal, N.; Mwiiri, F.K.; Gabel-Scheurich, S.; Pfeiffer, N.; Grus, F.H. In-depth proteomic analysis of the porcine retina by use of a four step differential extraction bottom up lc ms platform. Mol. Neurobiol. 2016, 54, 7262–7275. [Google Scholar] [CrossRef]
- Ozorowski, G.; Milton, S.; Luecke, H. Structure of a C-terminal AHNAK peptide in a 1:2:2 complex with S100A10 and an acetylated N-terminal peptide of annexin A2. Acta Crystallogr. Sect. D 2013, 69, 92–104. [Google Scholar] [CrossRef]
- Yu, M.B.; Sun, J.; Peng, W.; Chen, Z.Y.; Lin, X.C.; Liu, X.Y.; Li, M.T.; Wu, K.L. Protein expression in human trabecular meshwork: Downregulation of rhogdi by dexamethasone in vitro. Mol. Vis. 2010, 16, 213–223. [Google Scholar]
- Bollinger, K.E.; Crabb, J.S.; Yuan, X.; Putliwala, T.; Clark, A.F.; Crabb, J.W. Proteomic similarities in steroid responsiveness in normal and glaucomatous trabecular meshwork cells. Mol. Vis. 2012, 18, 2001–2011. [Google Scholar]
- Bollinger, K.E.; Crabb, J.S.; Yuan, X.; Putliwala, T.; Clark, A.F.; Crabb, J.W. Quantitative proteomics: Tgfbeta(2) signaling in trabecular meshwork cells. Investig. Ophthalmol. Vis. Sci. 2011, 52, 8287–8294. [Google Scholar] [CrossRef]
- Clark, R.; Nosie, A.; Walker, T.; Faralli, J.A.; Filla, M.S.; Barrett-Wilt, G.; Peters, D.M. Comparative genomic and proteomic analysis of cytoskeletal changes in dexamethasone-treated trabecular meshwork cells. Mol. Cell Proteom. 2013, 12, 194–206. [Google Scholar] [CrossRef]
- Bhattacharya, S.K.; Rockwood, E.J.; Smith, S.D.; Bonilha, V.L.; Crabb, J.S.; Kuchtey, R.W.; Robertson, N.G.; Peachey, N.S.; Morton, C.C.; Crabb, J.W. Proteomics reveal cochlin deposits associated with glaucomatous trabecular meshwork. J. Biol. Chem. 2005, 280, 6080–6084. [Google Scholar] [CrossRef]
- Steely, H.T.; English-Wright, S.L.; Clark, A.F. The similarity of protein expression in trabecular meshwork and lamina cribrosa: Implications for glaucoma. Exp. Eye Res. 2000, 70, 17–30. [Google Scholar] [CrossRef]
- Hann, C.R.; Springett, M.J.; Wang, X.; Johnson, D.H. Ultrastructural localization of collagen iv, fibronectin, and laminin in the trabecular meshwork of normal and glaucomatous eyes. Ophthalmic Res. 2001, 33, 314–324. [Google Scholar] [CrossRef]
- Acott, T.S.; Kelley, M.J. Extracellular matrix in the trabecular meshwork. Exp. Eye Res. 2008, 86, 543–561. [Google Scholar] [CrossRef]
- Rao, V.R.; Maddala, R.; Skiba, N.P. Proteomic analysis of trabecular meshwork extracellular matrix. Investig. Ophthalmol. Vis. Sci. 2013, 54, 3533. [Google Scholar]
- Gagen, D.; Faralli, J.A.; Filla, M.S.; Peters, D.M. The role of integrins in the trabecular meshwork. J. Ocul. Pharm. Ther. 2014, 30, 110–120. [Google Scholar] [CrossRef]
- Vranka, J.A.; Bradley, J.M.; Yang, Y.F.; Keller, K.E.; Acott, T.S. Mapping molecular differences and extracellular matrix gene expression in segmental outflow pathways of the human ocular trabecular meshwork. PLoS ONE 2015, 10, e0122483. [Google Scholar] [CrossRef]
- Vecino, E.; Kwok, J.C.F. Chapter 5: The extracellular matrix in the nervous system: The good and the bad aspects. In Composition and Function of the Extracellular Matrix in the Human Body; Travascio, F., Ed.; Intech: London, UK, 2016. [Google Scholar]
- Friedlander, D.R.; Milev, P.; Karthikeyan, M.R.K.; Margolis, R.U.; Grumet, M. The neuronal chondroitin sulfate proteoglycan neurcan binds to the neuronal cell adhesion molecules ng-cam/l1/nile and n-cam, and inhibits neuronal adhesion and neurite outgrowth. J. Cell Biol. 1994, 125, 669–680. [Google Scholar] [CrossRef]
- Tripathi, B.J.; Tripathi, R.C. Neural crest origin of human trabecular meshwork and its implications for the pathogenesis of glaucoma. Am. J. Ophthalmol. 1989, 107, 583–590. [Google Scholar] [CrossRef]
- Gould, D.B.; Smith, R.S.; John, S.W.M. Anterior segment development relevant to glaucoma. Int. J. Dev. Biol. 2004, 48, 1015–1029. [Google Scholar] [CrossRef] [PubMed]
- Kaiser-Kupfer, M.I. Neural crest origin of trabecular meshwork cells and other structures of the anterior chamber. Am. J. Ophthalmol. 1989, 107, 671–672. [Google Scholar] [CrossRef]
- Gentil, B.J.; Benaud, C.; Delphin, C.; Remy, C.; Berezowski, V.; Cecchelli, R.; Feraud, O.; Vittet, D.; Baudier, J. Specific ahnak expression in brain endothelial cells with barrier properties. J. Cell. Physiol. 2005, 203, 362–371. [Google Scholar] [CrossRef]
- Keller, K.E.; Acott, T.S. The juxtacanalicular region of the ocular trabecular meshwork: A tissue with a unique extracellular matrix and specialized function. J. Ocul. Biol. 2013, 1, 3–15. [Google Scholar]
- Tamm, E.R.; Flügel, C.; Stefani, F.H.; Lütjen-Drecoll, E. Nerve endings with structural characteristics of mechanoreceptors in the human scleral spur. Investig. Ophthalmol. Vis. Sci. 1994, 35, 1157–1166. [Google Scholar] [PubMed]
- Sasamoto, M.; Chen, H.B.; Tsukahara, S. Ultrastructural identification of substance p containing nerves in the guinea pig trabecular meshwork. Opthalmic Res. 1998, 30, 127–134. [Google Scholar] [CrossRef]
- May, C.A.; Skorski, L.M.; Lütjen-Drecoll, E. Inervation of the porcine ciliary muscle and outflow region. J. Anat. 2005, 206, 231–236. [Google Scholar] [CrossRef]
- Stone, A.R.; Kuwayama, Y.; Laties, A.M.; Marangos, P.J. Neuron-specific enolase-containing cells in the rhesus monkey trabecular meshwork. Investig. Ophthalmol. Vis. Sci. 1984, 25, 1332–1334. [Google Scholar]
- Choi, J.; Miller, A.M.; Nolan, M.J.; Yue, B.Y.J.T.; Thotz, S.T.; Clark, A.F.; Agarwal, N.; Knepper, P.A. Soluble cd44 is cytotoxic to trabecular meshwork and retinal ganglion cells in vitro. Investig. Ophthalmol. Vis. Sci. 2005, 46, 214–222. [Google Scholar] [CrossRef][Green Version]
- Pascale, A.; Amadio, M.; Quattrone, A. Defining a neuron: Neuronal elav proteins. Cell. Mol. Life Sci. 2008, 65, 128–140. [Google Scholar] [CrossRef]
- Sacca, S.C.; Izzotti, A. Focus on molecular events in the anterior chamber leading to glaucoma. Cell. Mol. Life Sci. 2014, 71, 2197–2218. [Google Scholar] [CrossRef]
- Sacca, S.C.; Pulliero, A.; Izzotti, A. The dysfunction of the trabecular meshwork during glaucoma course. J. Cell. Physiol. 2015, 230, 510–525. [Google Scholar] [CrossRef]
- Luo, C.; Yang, X.; Kain, A.D.; Powell, D.W.; Kuehn, M.H.; Tezel, G. Glaucomatous tissue stress and the regulation of immune response through glial toll-like receptor signaling. Investig. Ophthalmol. Vis. Sci. 2010, 51, 5697–5707. [Google Scholar] [CrossRef]
- Tezel, G.; Thornton, I.L.; Tong, M.G.; Luo, C.; Yang, X.J.; Cai, J.; Powell, D.W.; Soltau, J.B.; Liebmann, J.M.; Ritch, R. Immunoproteomic analysis of potential serum biomarker candidates in human glaucoma. Investig. Ophthalmol. Vis. Sci. 2012, 53, 8222–8231. [Google Scholar] [CrossRef]
- Wordinger, R.J.; Sharma, T.; Clark, A.F. The role of tgf-β2 and bone morphogenetic proteins in the trabecular meshwork and glaucoma. J. Ocul. Pharm. Ther. 2014, 30, 154–162. [Google Scholar] [CrossRef]
- Funke, S.; Perumal, N.; Beck, S.; Gabel-Scheurich, S.; Schmelter, C.; Teister, J.; Gerbig, C.; Gramlich, O.W.; Pfeiffer, N.; Grus, F.H. Glaucoma related proteomic alterations in human retina samples. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Cavusoglu, N.; Thierse, D.; Mohand-Said, S.; Chalmel, F.; Poch, O.; Van-Dorsselaer, A.; Sahel, J.A.; Leveillard, T. Differential proteomic analysis of the mouse retina: The induction of crystallin proteins by retinal degeneration in the rd1 mouse. Mol. Cell Proteom. 2003, 2, 494–505. [Google Scholar] [CrossRef]
- Anders, F.; Teister, J.; Liu, A.H.; Funke, S.; Grus, F.H.; Thanos, S.; von Pein, H.D.; Pfeiffer, N.; Prokosch, V. Intravitreal injection of β-crystallin b2 improves retinal ganglion cell survival in an experimental animal model of glaucoma. PLoS ONE 2017, 12, e0175451. [Google Scholar] [CrossRef]
- Anders, F.; Teister, J.; Funke, S.; Pfeiffer, N.; Grus, F.; Thanos, S.; Prokosch, V. Proteomic profiling reveals crucial retinal protein alterations in the early phase of an experimental glaucoma model. Graefes Arch. Clin. Exp. Ophthalmol. 2017, 255, 1395–1407. [Google Scholar] [CrossRef]
- Joachim, S.C.; Bruns, K.; Lackner, K.J.; Pfeiffer, N.; Grus, F.H. Antibodies to alpha b-crystallin, vimentin, and heat shock protein 70 in aqueous humor of patients with normal tension glaucoma and igg antibody patterns against retinal antigen in aqueous humor. Curr. Eye Res. 2007, 32, 501–509. [Google Scholar] [CrossRef]
- Zhong, Y.S.; Wang, J.; Luo, X.D. Integrins in trabecular meshwork and optic nerve head: Possible association with the pathogenesis of glaucoma. Biomed. Res. Int. 2013. [Google Scholar] [CrossRef]
- Borras, T. The effects of myocilin expression on functionally relevant trabecular meshwork genes: A mini-review. J. Ocul. Pharm. 2014, 30, 202–212. [Google Scholar] [CrossRef]
- Prat, C.; Belville, C.; Comptour, A.; Marceau, G.; Clairefond, G.; Chiambaretta, F.; Sapin, V.; Blanchon, L. Myocilin expression is regulated by retinoic acid in the trabecular meshwork-derived cellular environment. Exp. Eye Res. 2017, 155, 91–98. [Google Scholar] [CrossRef]
- Bendixen, E.; Danielsen, M.; Hollung, K.; Gianazza, E.; Miller, I. Farm animal proteomics. J. Proteom. 2011, 74, 282–293. [Google Scholar] [CrossRef]
- Shevchenko, A.; Tomas, H.; Havlis, J.; Olsen, J.V.; Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006, 1, 2856–2860. [Google Scholar] [CrossRef]
- Bell, K.; Wilding, C.; Funke, S.; Pfeiffer, N.; Grus, F.H. Protective effect of 14-3-3 antibodies on stressed neuroretinal cells via the mitochondrial apoptosis pathway. BMC Ophthalmol. 2015, 15, 64. [Google Scholar] [CrossRef] [PubMed]
- Bell, K.; Funke, S.; Pfeiffer, N.; Grus, F.H. Serum and antibodies of glaucoma patients lead to changes in the proteome, especially cell regulatory proteins, in retinal cells. PLoS ONE 2012, 7, e46910. [Google Scholar] [CrossRef]
- Schmelter, C.; Funke, S.; Treml, J.; Beschnitt, A.; Perumal, N.; Manicam, C.; Pfeiffer, N.; Grus, F.H. Comparison of two solid-phase extraction (spe) methods for the identification and quantification of porcine retinal protein markers by lc-ms/ms. Int. J. Mol. Sci. 2018, 19, 3847. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. String v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Oliveros, J.C. An Interactive Tool for Comparing Lists with Venn’s Diagrams. Available online: http://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 22 May 2019).
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).