Overexpression of a Multiprotein Bridging Factor 1 Gene DgMBF1 Improves the Salinity Tolerance of Chrysanthemum
Abstract
1. Introduction
2. Results
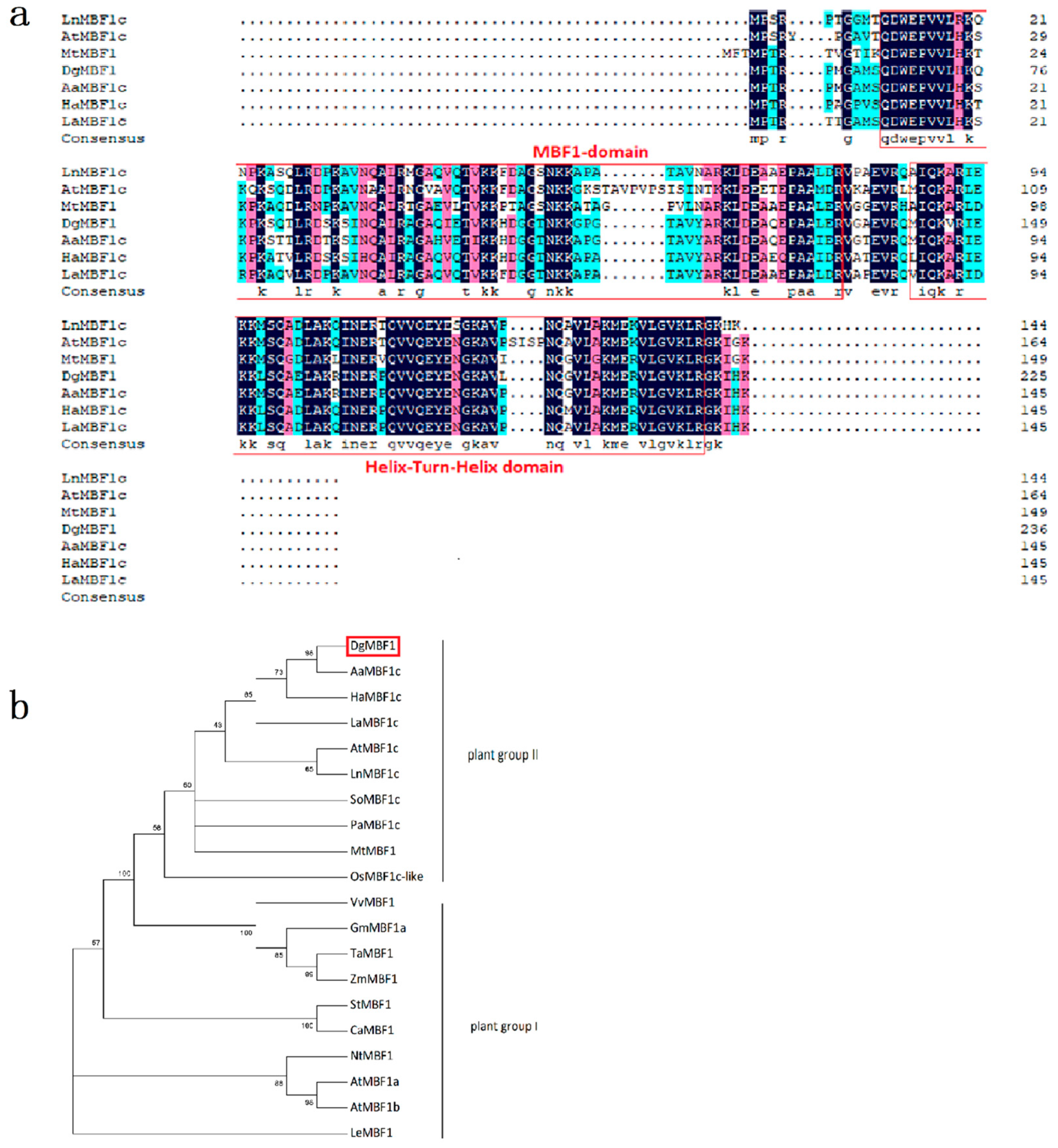
2.1. DgMBF1 Clone and Sequence Analysis
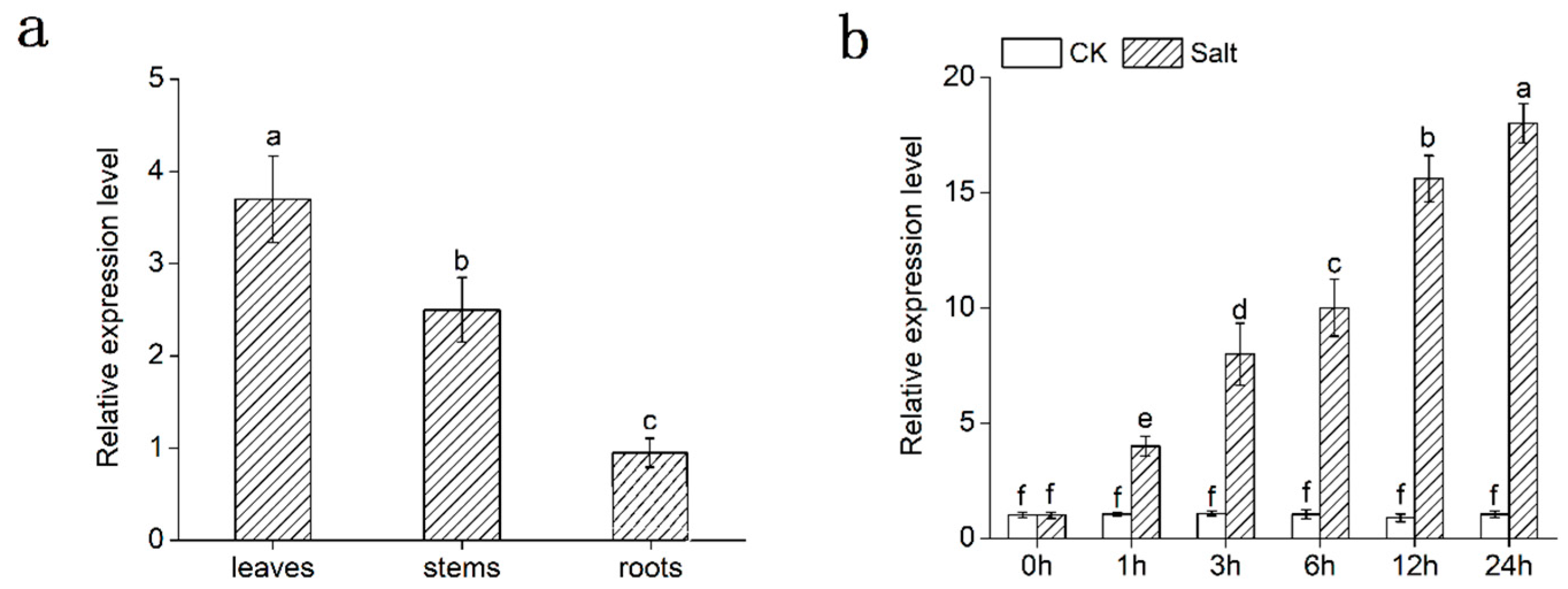
2.2. Expression Analysis of DgMBF1
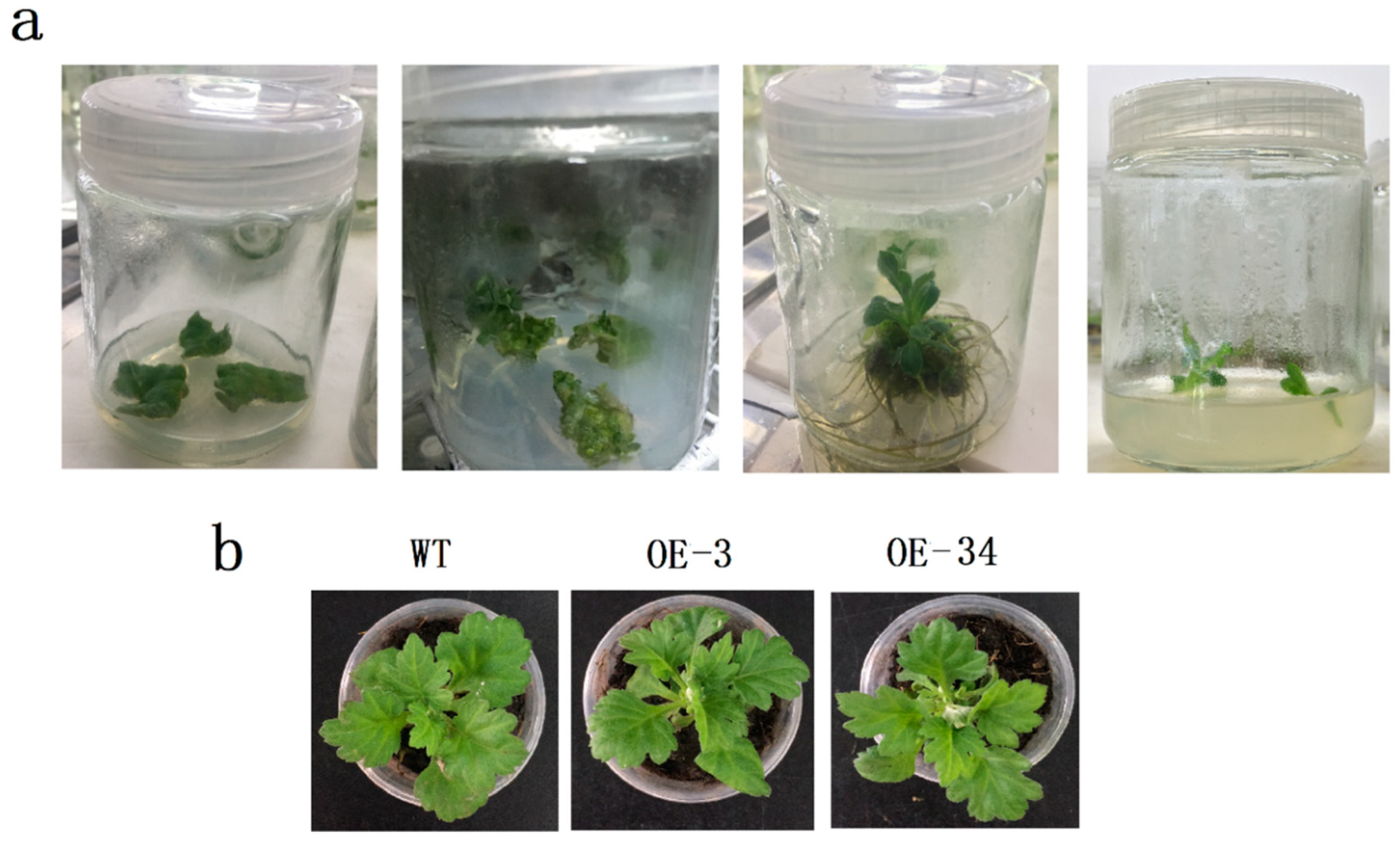
2.3. Observation of Callus and Phenotype
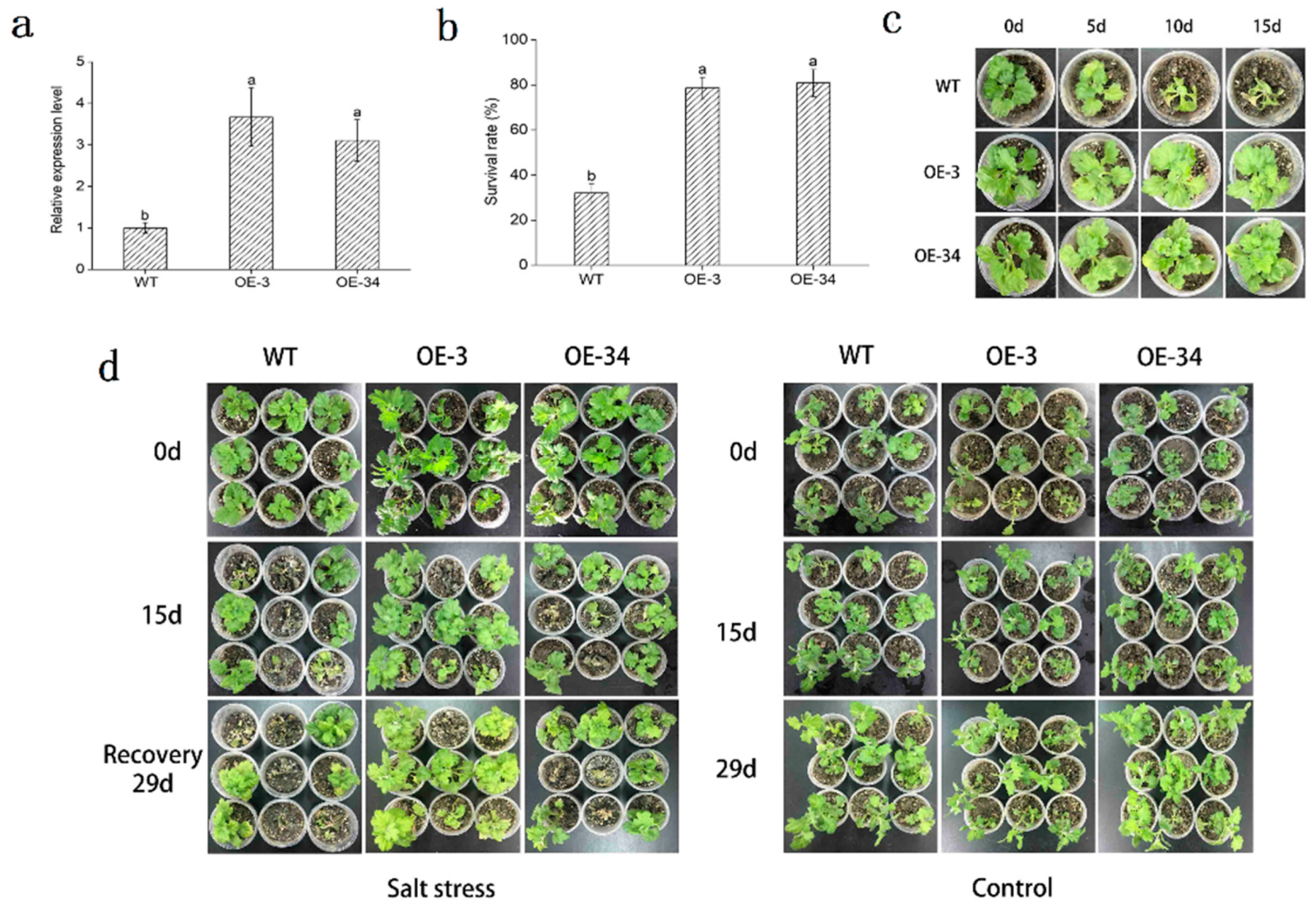
2.4. Overexpression of DgMBF1 in Chrysanthemum Enhanced the Salt Tolerance
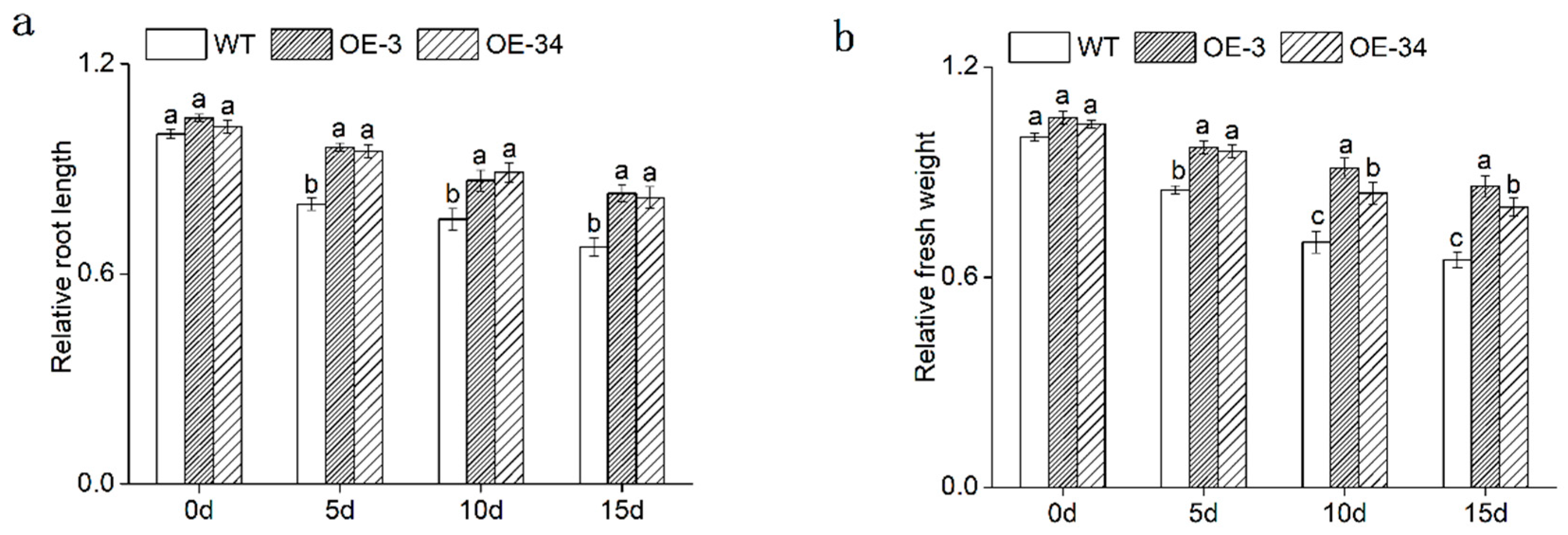
2.5. Influence of Salt Stress on the Growth and Development of Chrysanthemum
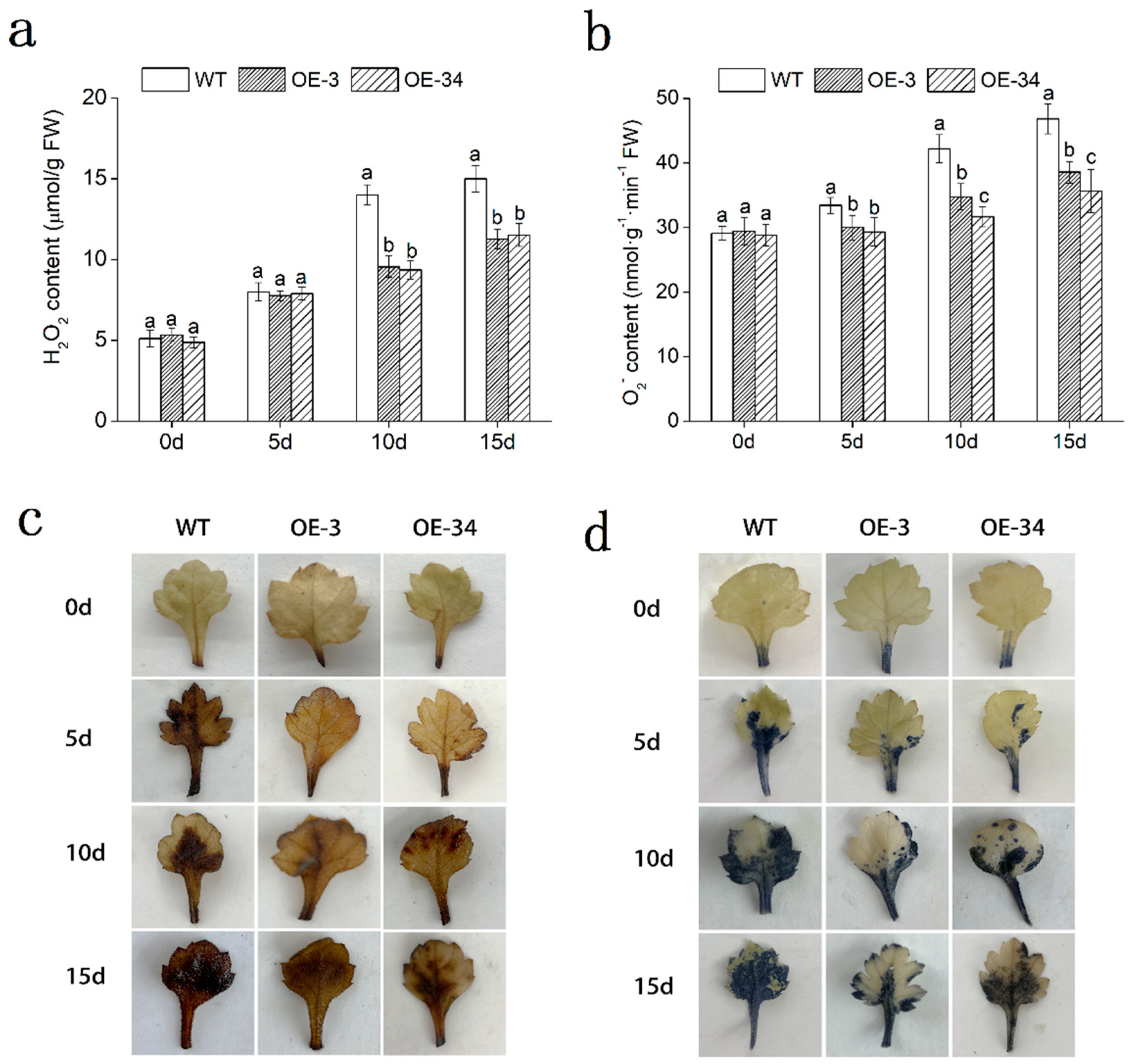
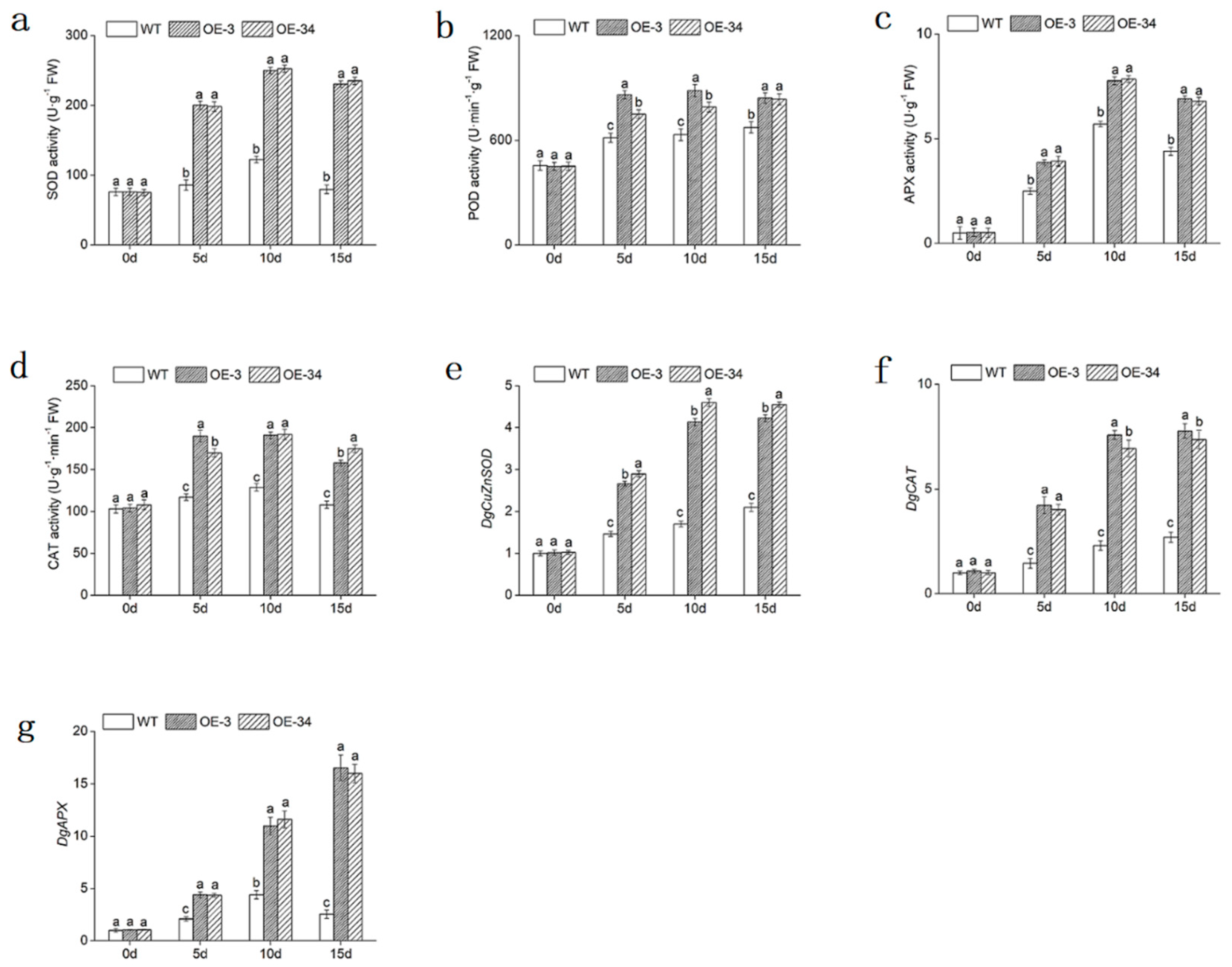
2.6. Overexpression of DgMBF1 Enhanced Oxidation Tolerance under Salt Stress
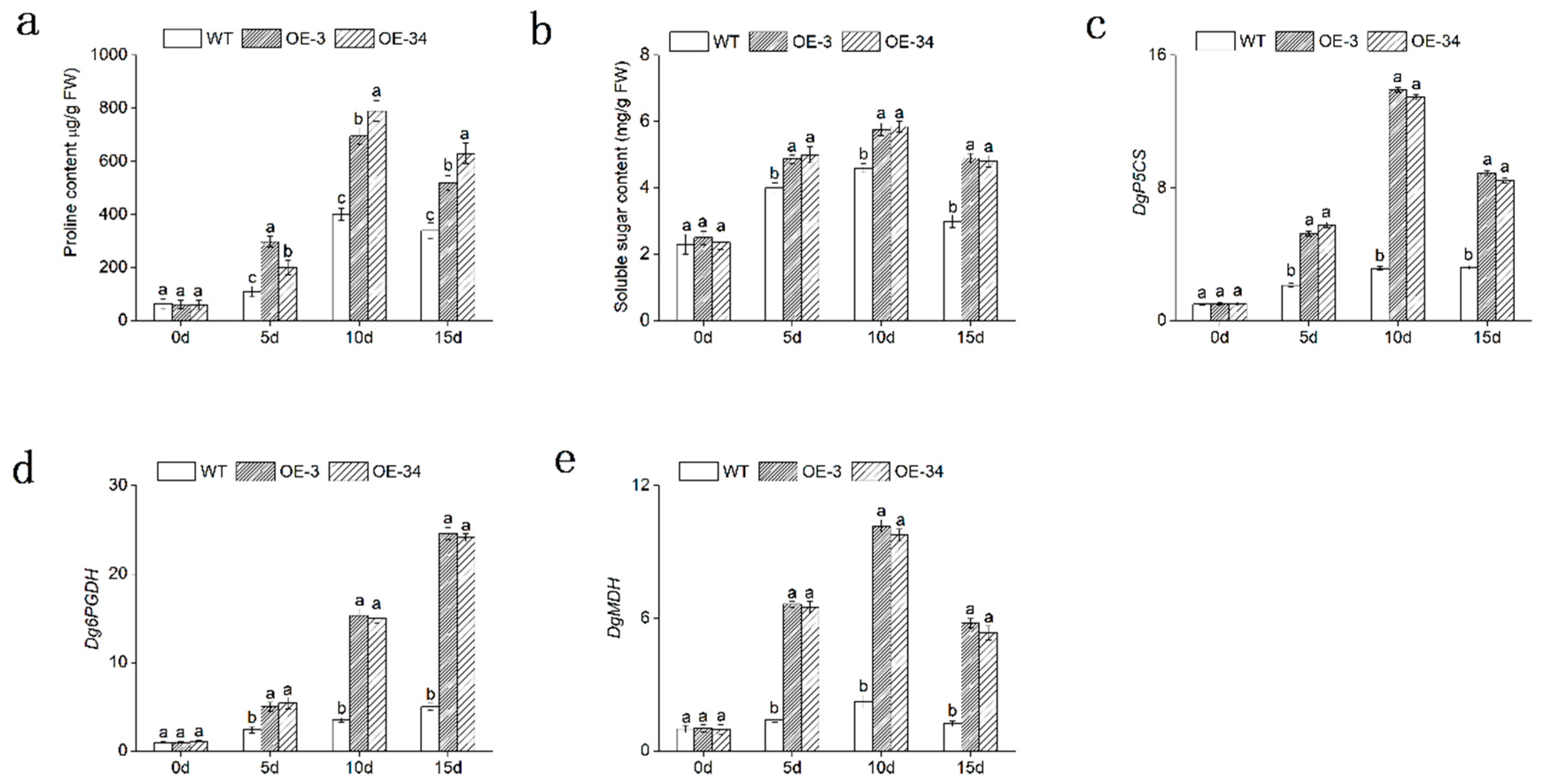
2.7. Overexpression of DgMBF1 Promoted the Accumulation of Osmotic Substances under Salt Stress
2.8. Overexpression of DgMBF1 Enhanced the K+/Na+ Selectivity under Salt Stress
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Stress Treatment
4.2. DgMBF1 Clone and Sequence Analyses
4.3. Sequence Alignment and Phylogenetic Analysis
4.4. Expression Vector Constructs and Chrysanthemum Transformations
4.5. Gene Expression Levels Analysis
4.6. Determination of Salt Tolerance
4.7. Determination of Physiological Indexes of Chrysanthemum under Salt Stress
4.8. Expression of Stress-Related Genes
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| APX | superoxide Anion |
| CaMV | cauliflower mosaic virus |
| CAT | catalase |
| DAB | diaminobenzidine |
| HTH | helix-turn-helix |
| H2O2 | hydrogen peroxide |
| MBF1 | multiprotein bridging factor 1 |
| NBT | nitro blue tetrazolium |
| O2− | superoxide anion |
| OE | overexpressed |
| ORF | open reading frame |
| PCR | polymerase chain reaction |
| POD | peroxidase |
| qRT-PCR | quantitative real-time PCR |
| ROS | reactive oxygen species |
| SOD | superoxide |
| SS | soluble sugar |
| WT | wild type |
References
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef]
- Zhao, Q.; He, L.; Wang, B.; Liu, Q.; Pan, Y.; Zhang, F.; Jiang, B.; Zhang, L.; Liu, G.; Jia, Y. Transcriptome Comparative Analysis of Salt Stress Responsiveness in Chrysanthemum (Dendranthema grandiflorum) Roots by Illumina and Single-Molecule Real-Time-Based RNA Sequencing. DNA Cell Biol. 2018, 37, 1–15. [Google Scholar] [CrossRef]
- Kaleem, F.; Shabir, G.; Aslam, K.; Rasul, S.; Manzoor, H.; Shah, M.; Khan, A. An overview of the genetics of plant response to salt stress: present status and the way forward. Appl. Biochem. Biotech. 2018, 9, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Tester, M. Mechanisms of Salinity Tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Deinlein, U.; Stephan, A.; Horie, T.; Luo, W.; Xu, G.; Schroeder, J. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014, 19, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Apel, K.; Hirt, H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef]
- Tripathy, B.; Ralf, O. Reactive oxygen species generation and signaling in plants. Plant Signal. Behav. 2012, 7, 1621–1633. [Google Scholar] [CrossRef]
- Prabucki, A.; Serek, M.; Andersen, A.S. Influence of salt stress on stock plant growth and cutting performance of Chrysanthemum morifolium Ramat. J. Hortic. Sci. Biotech. 1999, 74, 132–134. [Google Scholar] [CrossRef]
- Lee, M.; Iersel, M. Sodium chloride effects on growth, morphology, and physiology of chrysanthemum (Chrysanthemum X morifolium). HortScience 2008, 43, 1888–1891. [Google Scholar] [CrossRef]
- Lindemose, S.; O’Shea, C.; Jensen, M.; Skriver, K. Structure, function and networks of transcription factors involved in abiotic stress responses. Int. J. Mol. Sci. 2013, 14, 5842–5878. [Google Scholar] [CrossRef]
- Nakashima, K.; Ito, Y.; Yamaguchi-Shinozaki, K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 2009, 149, 88–95. [Google Scholar] [CrossRef]
- Wang, K.; Wu, Y.; Tian, X.; Bai, Z.; Liang, Q.; Liu, Q.; Pan, Y.; Zhang, L.; Jiang, B. Overexpression of DgWRKY4 enhances salt tolerance in chrysanthemum seedlings. Front. Plant Sci. 2017, 8, 1592–1605. [Google Scholar] [CrossRef] [PubMed]
- Alavilli, H.; Lee, H.; Park, M.; Lee, B. Antarctic moss multiprotein bridging factor 1c overexpression in Arabidopsis resulted in enhanced tolerance to salt stress. Front. Plant Sci. 2017, 8, 1206–1220. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Lim, G.; Kim, E.; Ko, C.; Yang, K.; Jeong, J.; Lee, M.; Kim, C. Abiotic and biotic stress tolerance in Arabidopsis overexpressing the multiprotein bridging factor 1a (MBF1a) transcriptional co-activator gene. Biochem. Biophys. Res. Commun. 2017, 354, 440–446. [Google Scholar] [CrossRef] [PubMed]
- Mauro, M.; Iglesias, M.; Arce, D.; Valle, E.; Arnold, B.; Tsuda, K.; Yamazaki, K.; Casalongué, C.; Godoy, A. MBF1s regulate ABA-dependent germination of Arabidopsis seeds. Plant Signal. Behav. 2012, 7, 188–192. [Google Scholar] [CrossRef]
- Brendel, C.; Gelman, L.; Auwerx, J. Multiprotein bridging factor-1 (MBF-1) is a cofactor for nuclear receptors that regulate lipid metabolism. Mol. Endocrinol. 2002, 16, 1367–1377. [Google Scholar] [CrossRef]
- Takemaru, K.; Harashima, S.; Ueda, H.; Hirose, S. Yeast co-activator MBF1 mediates GCN4-dependent transcriptional activation. Mol. Cell Biol. 1998, 18, 4971–4976. [Google Scholar] [CrossRef]
- Fan, G.; Zhang, K.; Huang, H.; Zhang, H.; Zhao, A.; Chen, L.; Chen, R.; Li, G.; Wang, Z.; Lu, G. Multiprotein-bridging factor 1 regulates vegetative growth, osmotic stress, and virulence in magnaporthe oryzae. Curr. Genet. 2017, 63, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, K.; Tsuji, T.; Hirose, S.; Yamazaki, K. Three Arabidopsis MBF1 homologs with distinct expression profiles play roles as transcriptional co-activator. Plant Cell Physiol. 2004, 45, 225–231. [Google Scholar] [CrossRef]
- Tsuda, K.; Yamazaki, K. Structure and expression analysis of three subtypes of Arabidopsis MBF1 genes. Biochim. Biophys. Acta 2004, 1680, 1–10. [Google Scholar] [CrossRef]
- Rizhsky, L.; Liang, H.; Mittler, R. The combined effect of drought stress and heat shock on gene expression in tobacco. Plant Physiol. 2002, 130, 1143–1151. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Rizhsky, L.; Liang, H.; Shuman, J.; Mittler, R. Enhanced tolerance to environmental stress in transgenic plants expressing the transcriptional co-activator multiprotein bridging factor 1c. Plant Physiol. 2005, 139, 1313–1322. [Google Scholar] [CrossRef]
- Guo, W.; Chen, R.; Du, X.; Zhang, Z.; Yin, Y.; Gong, Z.; Wang, G. Reduced tolerance to abiotic stress in transgenic Arabidopsis overexpressing a Capsicum annuum multiprotein bridging factor 1. BMC Plant Biol. 2014, 14, 138–150. [Google Scholar] [CrossRef]
- Arce, D.; Tonón, C.; Zanetti, M.; Godoy, A.; Hirose, S.; Casalongué, C. The potato transcriptional co-activator StMBF1 is up-regulated in response to oxidative stress and interacts with the TATA-box binding protein. J. Biochem. Mol. Biol. Biophys. 2006, 39, 355–360. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, T.; Wang, K.; Liang, Q.; Bai, Z.; Liu, Q.; Pan, Y.; Jiang, B.; Zhang, L. Comparative analysis of the chrysanthemum leaf transcript profiling in response to salt stress. PLoS ONE 2016, 11, e0159721. [Google Scholar] [CrossRef]
- An, J.; Song, A.; Guan, Z.; Jiang, J.; Chen, F.; Lou, W.; Fang, W.; Liu, Z.; Chen, S. The over-expression of Chrysanthemum crassum CcSOS1 improves the salinity tolerance of chrysanthemum. Mol. Biol. Rep. 2014, 41, 4155–4162. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Zhong, M.; Wu, Y.; Bai, Z.; Liang, Q.; Liu, Q.; Pan, Y.; Zhang, L.; Jiang, B.; Jia, Y.; et al. Overexpression of a chrysanthemum transcription factor gene DgNAC1, improves the salinity tolerance in chrysanthemum. Plant Cell Rep. 2017, 36, 1–11. [Google Scholar] [CrossRef]
- Zhao, Q.; Zhong, M.; He, L.; Wang, B.; Liu, Q.; Pan, Y.; Jiang, B.; Zhang, L. Overexpression of a chrysanthemum transcription factor gene DgNAC1, improves drought tolerance in chrysanthemum. Plant Cell Tissue Organ 2018, 135, 119–132. [Google Scholar] [CrossRef]
- He, L.; Wu, Y.; Zhao, Q.; Wang, B.; Liu, Q.; Zhang, L. Chrysanthemum DgWRKY2 gene enhances tolerance to salt stress in transgenic chrysanthemum. Int. J. Mol. Sci. 2018, 19, 2062. [Google Scholar] [CrossRef]
- Liang, Q.; Wu, Y.; Wang, K.; Bai, Z.; Liu, Q.; Pan, Y.; Zhang, L.; Jiang, B. Chrysanthemum WRKY gene DgWRKY5 enhances tolerance to salt stress in transgenic chrysanthemum. Sci. Rep. 2017, 7, 4799–4808. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Sejima, H.; Tam, R.; Schlauch, K.; Mittler, R. Identification of the MBF1 heat-response regulon of Arabidopsis thaliana. Plant J. 2011, 66, 844–851. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, G.; Dong, Y.; Guo, J.; Huang, L.; Kang, Z. Cloning and characterization of a MBF1 transcriptional co-activator factor in wheat induced by stripe rust pathogen. Acta Agron. Sin. 2009, 35, 11–17. [Google Scholar] [CrossRef]
- Qin, D.; Wang, F.; Geng, X.; Zhang, L.; Yao, Y.; Ni, Z.; Peng, H.; Sun, Q. Overexpression of heat stress-responsive TaMBF1c, a wheat (Triticum aestivum L.) multiprotein bridging factor, confers heat tolerance in both yeast and rice. Plant Mol. Biol. 2015, 87, 31–45. [Google Scholar] [CrossRef]
- Yan, Q.; Hou, H.; Singer, S.; Yan, X.; Guo, R.; Wang, X. Grape VvMBF1 gene improves drought stress tolerance in transgenic Arabidopsis thaliana. Plant Cell Tissue Organ 2014, 118, 571–582. [Google Scholar] [CrossRef]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Wu, L.; Yu, Z. Effect of salt and drought stress on antioxidant enzymes activities and SOD isoenzymes of liquorice (Glycyrrhiza uralensis fisch). Plant Growth Regul. 2006, 49, 157–165. [Google Scholar] [CrossRef]
- Negi, N.; Shrivastava, D.; Sharma, V.; Sarin, N. Overexpression of CuZnSOD from Arachis hypogaea alleviates salinity and drought stress in tobacco. Plant Cell Rep. 2015, 34, 1109–1126. [Google Scholar] [CrossRef]
- Luo, X.; Wu, J.; Li, Y.; Nan, Z.; Xing, G.; Wang, Y.; Zhang, A.; Wang, Z.; Xia, G.; Tian, Y. Synergistic effects of GhSOD1 and GhCAT1 overexpression in cotton chloroplasts on enhancing tolerance to methyl viologen and salt stresses. PLoS ONE 2013, 8, e54002. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, J.; Li, J.; Li, H.; Zhang, G. The functional and regulatory mechanisms of the Thellungiella Salsuginea ascorbate peroxidase 6 (TsAPX6) in response to salinity and water deficit stresses. PLoS ONE 2016, 11, e0154042. [Google Scholar] [CrossRef]
- Yang, Y.; Shah, J.; Klessig, D. Signal perception and transduction in plant defense responses. Genes Dev. 1997, 11, 1621–1639. [Google Scholar] [CrossRef]
- Watanabe, S.; Kojima, K.; Ide, Y.; Sasaki, S. Effects of saline and osmotic stress on proline and sugar accumulation in Populus euphratica in vitro. Plant Cell Tissue Organ 2000, 63, 199–206. [Google Scholar] [CrossRef]
- Yamchi, A.; Rastgar, J.; Mousavi, A.; Karkhane, A.; Renu. Proline accumulation in transgenic tobacco as a result of expression of Arabidopsis 1-pyrroline-5-carboxylate synthetase (P5CS) during osmotic stress. J. Plant Biochem. Biot. 2007, 16, 9–15. [Google Scholar] [CrossRef]
- Vendruscolo, E.; Schuster, I.; Pileggi, M.; Scapim, C.; Molinari, H.; Marur, C.; Vieira, L. Stress-induced synthesis of proline confers tolerance to water deficit in transgenic wheat. J. Plant Physiol. 2007, 164, 1367–1376. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Shriram, V.; Kavi, K.; Jawali, N.; Shitole, M. Enhanced proline accumulation and salt stress tolerance of transgenic Indica rice by over-expressing P5CSF129A gene. Plant Biotechnol. Rep. 2010, 4, 37–48. [Google Scholar] [CrossRef]
- Hmida-Sayari, A.; Gargouri-Bouzid, R.; Bidani, A.; Jaoua, L.; Savouré, A.; Jaoua, S. Overexpression of Δ1-pyrroline-5-carboxylate synthetase increases proline production and confers salt tolerance in transgenic potato plants. Plant Sci. 2005, 169, 746–752. [Google Scholar] [CrossRef]
- Maathuis, F.; Amtmann, A. K+ nutrition and Na+ toxicity: the basis of cellular K+/Na+ ratios. Ann. Bot. 1999, 84, 123–133. [Google Scholar] [CrossRef]
- Rus, A.; Estan, M.; Gisbert, C.; Garcia-Sogo, B.; Serrano, R.; Caro, M. Expressing the yeast HAL1 gene in tomato increases fruit yield and enhances K+/Na+ selectivity under salt stress. Plant Cell Environ. 2010, 24, 875–880. [Google Scholar] [CrossRef]
- Yue, Y.; Zhang, M.; Zhang, J.; Duan, L.; Li, Z. SOS1, gene overexpression increased salt tolerance in transgenic tobacco by maintaining a higher K+/Na+ ratio. J. Plant Physiol. 2012, 169, 255–261. [Google Scholar] [CrossRef]
- Rodriguez-Rosales, M.; Galvez, F.J.; Huertas, R.; Aranda, M.N.; Baghour, M.; Cagnac, O.; Venema, K. Plant NHX cation/proton antiporters. Plant Signal Behav. 2009, 4, 265–276. [Google Scholar] [CrossRef]
- Galvez, F.J.; Baghour, M.; Hao, G.; Cagnac, O.; Rodríguez-Rosales, M.; Venema, K. Expression of LeNHX isoforms in response to salt stress in salt sensitive and salt tolerant tomato species. Plant Physiol. Bioch. 2012, 51, 109–115. [Google Scholar] [CrossRef]
- Yang, Q.; Chen, Z.; Zhou, X.; Yin, H.; Li, X.; Xin, X.; Hong, X.; Zhu, J.K.; Gong, Z. Overexpression of SOS (Salt Overly Sensitive) genes increases salt tolerance in transgenic Arabidopsis. Mol. Plant 2009, 2, 22–31. [Google Scholar] [CrossRef]
- Fuchs, I.; Stolzle, S.; Ivashikina, N.; Hedrich, R. Rice K+uptake channel OsAKT1 is sensitive to salt stress. Planta 2005, 221, 212–221. [Google Scholar] [CrossRef] [PubMed]
- Ardie, S.W.; Liu, S.; Takano, T. Expression of the AKT1-type K+ channel gene from Puccinellia tenuiflora, PutAKT1, enhances salt tolerance in Arabidopsis. Plant Cell Rep. 2010, 29, 865–874. [Google Scholar] [CrossRef]
- Shen, Y.; Shen, L.; Shen, Z.; Jing, W.; Ge, H.; Zhao, J.; Zhang, W. The potassium transporter OsHAK21 functions in the maintenance of ion homeostasis and tolerance to salt stress in rice. Plant Cell Environ. 2016, 38, 2766–2779. [Google Scholar] [CrossRef]
- Horie, T.; Sugawara, M.; Okada, T.; Taira, K.; Kaothien-Nakayama, P.; Katsuhara, M.; Shinmyo, A.; Nakayama, H. Rice sodium-insensitive potassium transporter, OsHAK5, confers increased salt tolerance in tobacco BY2 cells. J. Biosci. Bioeng. 2011, 111, 346–356. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Golldack, D.; Zhao, C.; Bohnert, H.J. The expression of HAK type K+ transporters is regulated in response to salinity stress in common ice plant. Plant Physiol. 2002, 129, 1482–1493. [Google Scholar] [CrossRef] [PubMed]
- An, G.; Watson, B.; Chiang, C. Transformation of tobacco, tomato, potato, and Arabidopsis thaliana using a binary Ti vector system. Plant Physiol. 1986, 81, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Chen, F.; Chen, S. Establishment of regeneration and transformation system of ground cover chrysanthemum Yuhuaxunzhang. J. Nanjing Agric. Univ. 2009, 32, 40–46. [Google Scholar] [CrossRef]
- Livak, K.; Schmittgen, T. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Chen, Y.; Jiang, J.; Chen, S.; Chen, F.; Guan, Z.; Fang, W. The constitutive expression of Chrysanthemum dichrum ICE1 in Chrysanthemum grandiflorum improves the level of low temperature, salinity and drought tolerance. Plant Cell Rep. 2012, 31, 1747–1758. [Google Scholar] [CrossRef]

| Forward Primers | Reverse Primers | |
|---|---|---|
| Primers used for cloning of DgMBF1 | ||
| DgMBF1 | TCCAGACCCTCAACTCCTA | ACAAACTCGACACAATACAAAG |
| DNA detection | GAGTCAAAGATTCAAATAGAGGACCT | ACAAACTCGACACAATACAAAG |
| Primers used for qRT-PCR | ||
| DgMBF1 | TGCCGACAAGACCAATGGG | TGGACAACTTGCGGCCTTT |
| EF1α | TTTTGGTATCTGGTCCTGGAG | CCATTCAAGCGACAGACTCA |
| DgCuZnSOD | CCATTGTTGACAAGCAGATTCCACTCA | ATCATCAGGATCAGCATGGACGACTAC |
| DgCAT | TACAAGCAACGCCCTTCAA | GACCTCTGTTCCCAACAGTCA |
| DgAPX | GTTGGCTGGTGTTGTTGCT | GATGGTCGTTTCCCTTAGTTG |
| DgP5CS | TTGGAGCAGAGGTTGGAAT | GCAGGTCTTTGTGGGTGTAG |
| Dg6PGDH | CGAGGTACTTCGTTCTCCGG | CTCCCTTCTTCCCCGGTAGA |
| DgMDH | GGTTGCCCCAGATGATCACA | GTTGGTCATCCAGATCGCCA |
| DgNHX | TGGTGGTAAAAGCTCGCACA | TCATTAACAACGCCCTCCCC |
| DgSOS | AGCTTCGACAAAGGGATGGG | GCTTTCTCGTCGGCTACCTT |
| DgAKT | ATTGCAGCACTTTCAGCAGC | ACTTGCCAAAGGTCCAACCA |
| DgHAK | TGGAACTTGCCATGGCCAA | GGCTTCCAACAACTGCAGC |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Q.; He, L.; Wang, B.; Liu, Q.; Pan, Y.; Zhang, F.; Jiang, B.; Zhang, L. Overexpression of a Multiprotein Bridging Factor 1 Gene DgMBF1 Improves the Salinity Tolerance of Chrysanthemum. Int. J. Mol. Sci. 2019, 20, 2453. https://doi.org/10.3390/ijms20102453
Zhao Q, He L, Wang B, Liu Q, Pan Y, Zhang F, Jiang B, Zhang L. Overexpression of a Multiprotein Bridging Factor 1 Gene DgMBF1 Improves the Salinity Tolerance of Chrysanthemum. International Journal of Molecular Sciences. 2019; 20(10):2453. https://doi.org/10.3390/ijms20102453
Chicago/Turabian StyleZhao, Qian, Ling He, Bei Wang, Qinglin Liu, Yuanzhi Pan, Fan Zhang, Beibei Jiang, and Lei Zhang. 2019. "Overexpression of a Multiprotein Bridging Factor 1 Gene DgMBF1 Improves the Salinity Tolerance of Chrysanthemum" International Journal of Molecular Sciences 20, no. 10: 2453. https://doi.org/10.3390/ijms20102453
APA StyleZhao, Q., He, L., Wang, B., Liu, Q., Pan, Y., Zhang, F., Jiang, B., & Zhang, L. (2019). Overexpression of a Multiprotein Bridging Factor 1 Gene DgMBF1 Improves the Salinity Tolerance of Chrysanthemum. International Journal of Molecular Sciences, 20(10), 2453. https://doi.org/10.3390/ijms20102453




