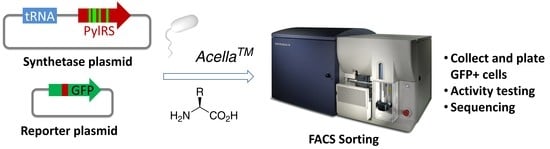
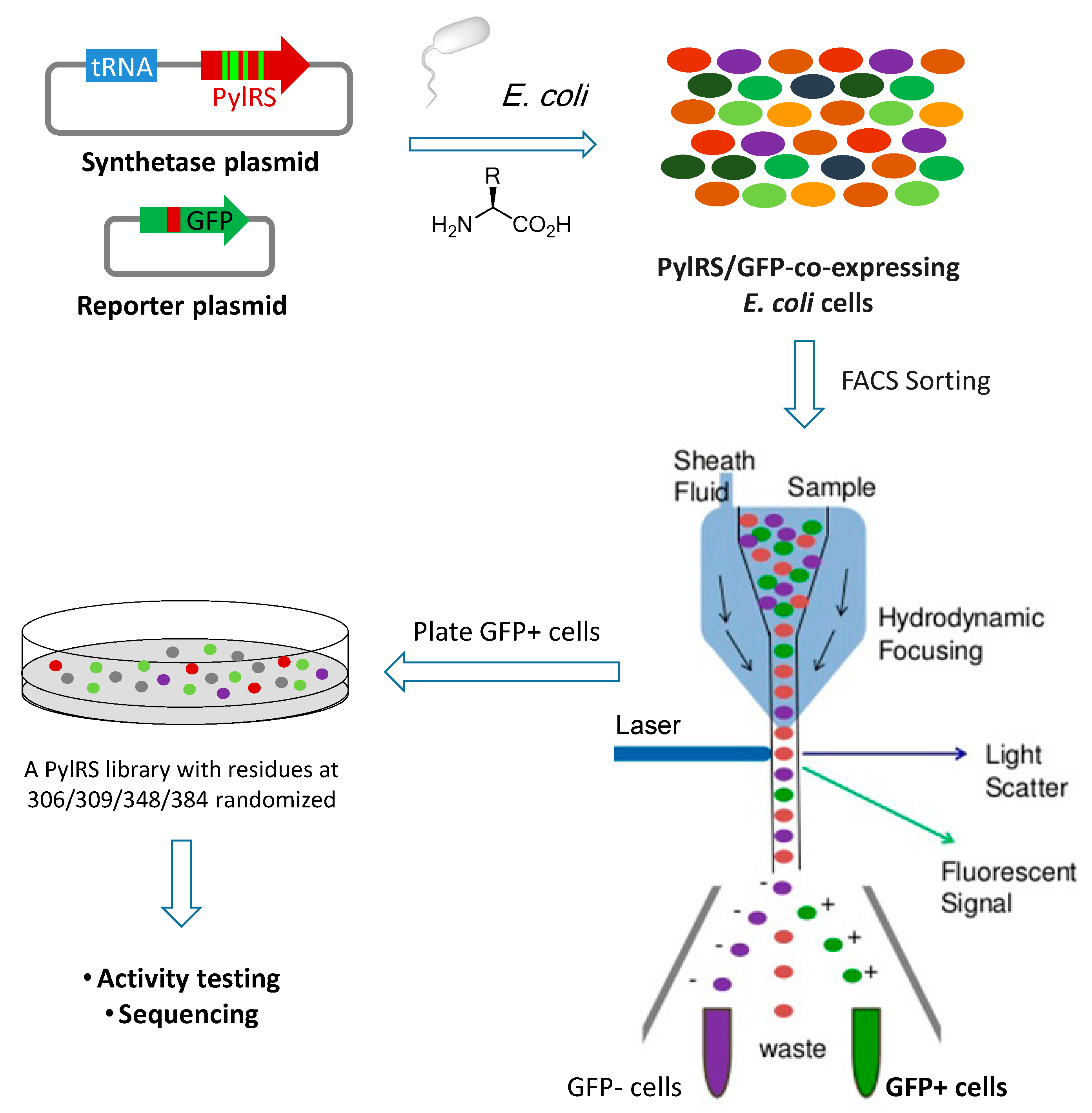
Rapid Identification of Functional Pyrrolysyl-tRNA Synthetases via Fluorescence-Activated Cell Sorting
Abstract
1. Introduction
2. Results
2.1. Construction of the PylRS Library
2.2. Assessment of PylRS Library Activity via FACS Analysis
2.3. Identification of Functional PylRS Variants via FACS Sorting and Verification of Their Activity
3. Discussion
4. Materials and Methods
4.1. Construction of the PylRS Library Using Gibson Assembly
4.2. Assessment of the PylRS Library Quality
4.3. FACS Analysis of AcellaTM Cells Co-Expressing the PylRS Library and sfGFP
4.4. FACS Sorting of Acella™ Cells Co-Expressing the PylRS Library and sfGFP
4.5. Verification of Biological Activity of the Selected PylRS Variants
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BocK | Nε-tert-butoxycarbonyl-L-lysine |
| E. coli | Escherichia coli |
| FACS | fluorescence-activated cell sorting |
| GFP | green fluorescent protein |
| IPTG | isopropyl β-d-1-thiogalactopyranoside |
| LB medium | Luria–Bertani medium |
| PBS | phosphate buffered saline |
| PylRS | pyrrolysyl-tRNA synthetase |
| sfGFP | super folder green fluorescent protein |
| SphK | Nε-spiro[2.3]hex-1-enylmethoxycarbonyl-L-lysine |
References
- Srinivasan, G.; James, C.M.; Krzycki, J.A. Pyrrolysine encoded by UAG in Archaea: Charging of a UAG-decoding specialized tRNA. Science 2002, 296, 1459–1462. [Google Scholar] [CrossRef] [PubMed]
- Blight, S.K.; Larue, R.C.; Mahapatra, A.; Longstaff, D.G.; Chang, E.; Zhao, G.; Kang, P.T.; Green-Church, K.B.; Chan, M.K.; Krzycki, J.A. Direct charging of tRNA(CUA) with pyrrolysine in vitro and in vivo. Nature 2004, 431, 333–335. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.C.; Schultz, P.G. Adding new chemistries to the genetic code. Annu. Rev. Biochem. 2010, 79, 413–444. [Google Scholar] [CrossRef] [PubMed]
- Wan, W.; Tharp, J.M.; Liu, W.R. Pyrrolysyl-tRNA synthetase: An ordinary enzyme but an outstanding genetic code expansion tool. Biochim. Biophys. Acta Proteins Proteom. 2014, 1844, 1059–1070. [Google Scholar] [CrossRef] [PubMed]
- Neuman, H.; Peak-Chew, S.Y.; Chin, J.W. Genetically encoding N(epsilon)-acetyllysine in recombinant proteins. Nat. Chem. Biol. 2008, 4, 232–234. [Google Scholar] [CrossRef] [PubMed]
- Bryson, D.I.; Fan, C.; Guo, L.T.; Miller, C.; Söll, D.; Liu, D.R. Continuous directed evolution of aminoacyl-tRNA synthetases. Nat. Chem. Biol. 2017, 13, 1253–1260. [Google Scholar] [CrossRef] [PubMed]
- Santoro, S.W.; Wang, L.; Herberich, B.; King, D.S.; Schultz, P.G. An efficient system for the evolution of aminoacyl-tRNA synthetase specificity. Nat. Biotechnol. 2002, 20, 1044–1048. [Google Scholar] [CrossRef]
- Kuhn, S.M.; Rubini, M.; Fuhrmann, M.; Theobald, I.; Skerra, A. Engineering of an orthogonal aminoacyl-tRNA synthetase for efficient incorporation of the non-natural amino acid O-methyl-L-tyrosine using fluorescence-based bacterial cell sorting. J. Mol. Biol. 2010, 404, 70–87. [Google Scholar] [CrossRef]
- Cooley, R.B.; Feldman, J.L.; Driggers, C.M.; Bundy, T.A.; Stokes, A.L.; Karplus, P.A.; Mehl, R.A. Structural basis of improved second-generation 3-nitro-tyrosine tRNA synthetases. Biochemistry 2014, 53, 1916–1924. [Google Scholar] [CrossRef]
- Link, A.J.; Vink, M.K.S.; Agard, N.J.; Prescher, J.A.; Bertozzi, C.R.; Tirrell, D.A. Discovery of aminoacyl-tRNA synthetase activity through cell-surface display of noncanonical amino acids. Proc. Natl. Acad. Sci. USA 2006, 103, 10180–10185. [Google Scholar] [CrossRef]
- Gibson, D.G.; Young, L.; Chuang, R.Y.; Venter, J.C.; Hutchison, C.A., 3rd; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef] [PubMed]
- Mukai, T.; Kobayashi, T.; Hino, N.; Yanagisawa, T.; Sakamoto, K.; Yokoyama, S. Adding l-lysine derivatives to the genetic code of mammalian cells with engineered pyrrolysyl-tRNA synthetases. Biochem. Biophys. Res. Commun. 2008, 371, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, T.; Ishii, R.; Fukunaga, R.; Kobayashi, T.; Sakamoto, K.; Yokoyama, S. Multistep engineering of pyrrolysyl-tRNA synthetase to genetically encode N(epsilon)-(o-azidobenzyloxycarbonyl) lysine for site-specific protein modification. Chem. Biol. 2008, 15, 1187–1197. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.P.; Garcia Alai, M.M.; Kapadnis, P.B.; Neumann, H.; Chin, J.W. Genetically encoding N(epsilon)-methyl-L-lysine in recombinant histones. J. Am. Chem. Soc. 2009, 131, 14194–14195. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Lin, Q. Design of spiro[2.3]hex-1-ene, a genetically encodable double-strained alkene for superfast photoclick chemistry. J. Am. Chem. Soc. 2014, 136, 4153–4156. [Google Scholar] [CrossRef]
- Nguyen, D.P.; Lusic, H.; Neumann, H.; Kapadnis, P.B.; Deiters, A.; Chin, J.W. Genetic encoding and labeling of aliphatic azides and alkynes in recombinant proteins via a pyrrolysyl-tRNA Synthetase/tRNA(CUA) pair and click chemistry. J. Am. Chem. Soc. 2009, 131, 8720–8721. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.M.; Yang, M.Y.; Huang, Y.C.; Song, X.D.; Liu, L.; Chen, P.R. Genetically encoded alkenyl–pyrrolysine analogues for thiol–ene reaction mediated site-specific protein labeling. Chem. Sci. 2012, 3, 2766–2770. [Google Scholar] [CrossRef]
- Li, N.; Ramil, C.P.; Lim, R.K.; Lin, Q. A Genetically Encoded Alkyne Directs Palladium-Mediated Protein Labeling on Live Mammalian Cell Surface. ACS Chem. Biol. 2015, 10, 379–384. [Google Scholar] [CrossRef]
- Odoi, K.A.; Huang, Y.; Rezenom, Y.H.; Liu, W.R. Nonsense and sense suppression abilities of original and derivative Methanosarcina mazei pyrrolysyl-tRNA synthetase-tRNA(Pyl) pairs in the Escherichia coli BL21(DE3) cell strain. PLoS ONE 2013, 8, e57035. [Google Scholar] [CrossRef]
- Lacey, V.K.; Louie, G.V.; Noel, J.P.; Wang, L. Expanding the library and substrate diversity of the pyrrolysyl-tRNA synthetase to incorporate unnatural amino acids containing conjugated rings. ChemBioChem 2013, 14, 2100–2105. [Google Scholar] [CrossRef]
- An, P.; Lewandowski, T.M.; Erbay, T.G.; Liu, P.; Lin, Q. Sterically shielded, stabilized nitrile imine for rapid bioorthogonal protein labeling in live cells. J. Am. Chem. Soc. 2018, 140, 4860–4868. [Google Scholar] [CrossRef] [PubMed]
- An, P.; Wu, H.Y.; Lewandowski, T.M.; Lin, Q. Hydrophilic azaspiroalkenes as robust bioorthogonal reporters. Chem. Commun. 2018, 54, 14005–14008. [Google Scholar] [CrossRef] [PubMed]



© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, A.E.; Lin, Q. Rapid Identification of Functional Pyrrolysyl-tRNA Synthetases via Fluorescence-Activated Cell Sorting. Int. J. Mol. Sci. 2019, 20, 29. https://doi.org/10.3390/ijms20010029
Lin AE, Lin Q. Rapid Identification of Functional Pyrrolysyl-tRNA Synthetases via Fluorescence-Activated Cell Sorting. International Journal of Molecular Sciences. 2019; 20(1):29. https://doi.org/10.3390/ijms20010029
Chicago/Turabian StyleLin, Andrew E., and Qing Lin. 2019. "Rapid Identification of Functional Pyrrolysyl-tRNA Synthetases via Fluorescence-Activated Cell Sorting" International Journal of Molecular Sciences 20, no. 1: 29. https://doi.org/10.3390/ijms20010029
APA StyleLin, A. E., & Lin, Q. (2019). Rapid Identification of Functional Pyrrolysyl-tRNA Synthetases via Fluorescence-Activated Cell Sorting. International Journal of Molecular Sciences, 20(1), 29. https://doi.org/10.3390/ijms20010029