OsASN1 Plays a Critical Role in Asparagine-Dependent Rice Development
Abstract
1. Introduction
2. Results
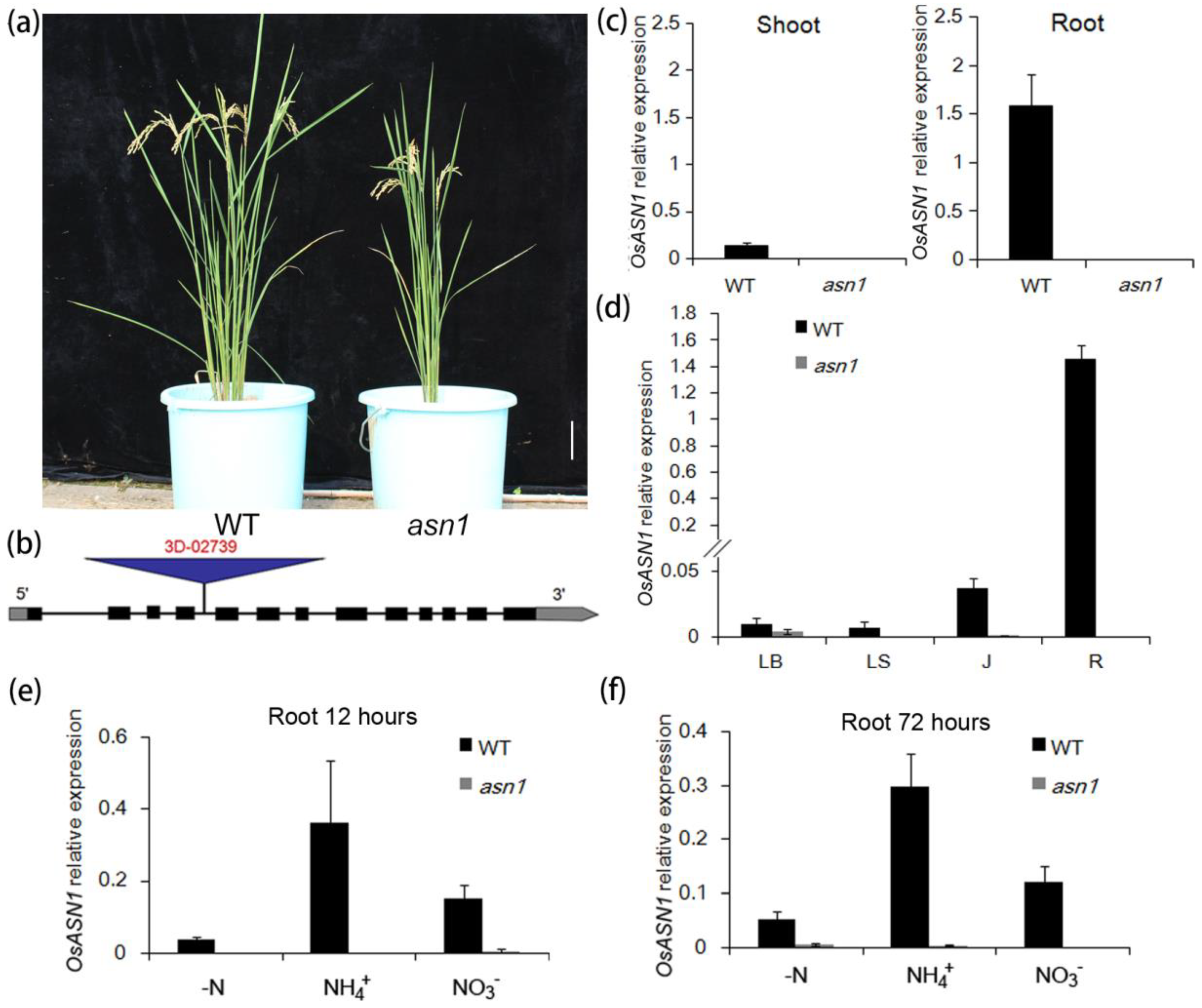
2.1. Screening of a New T-DNA Insertion Mutant with Growth Defects
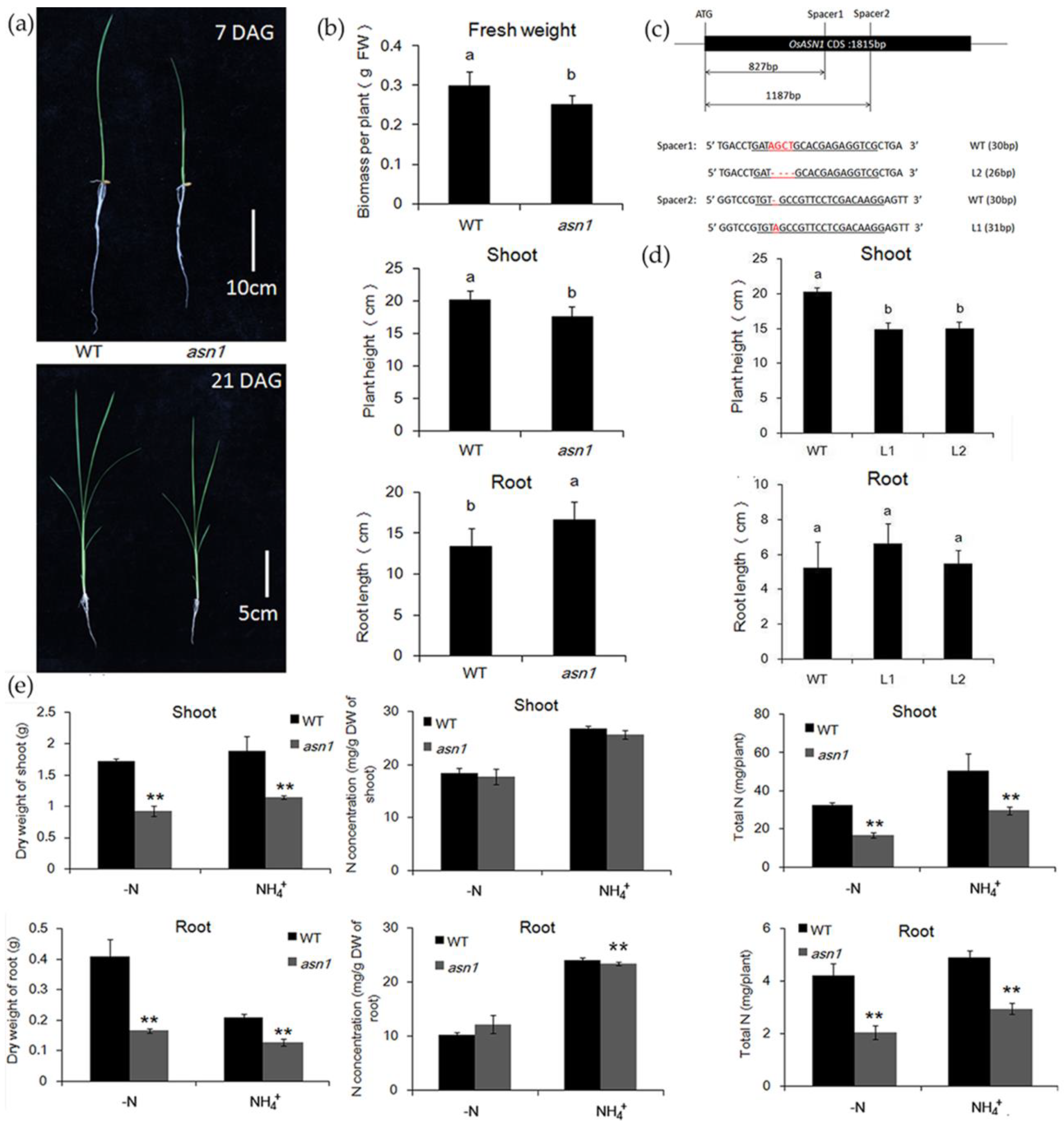
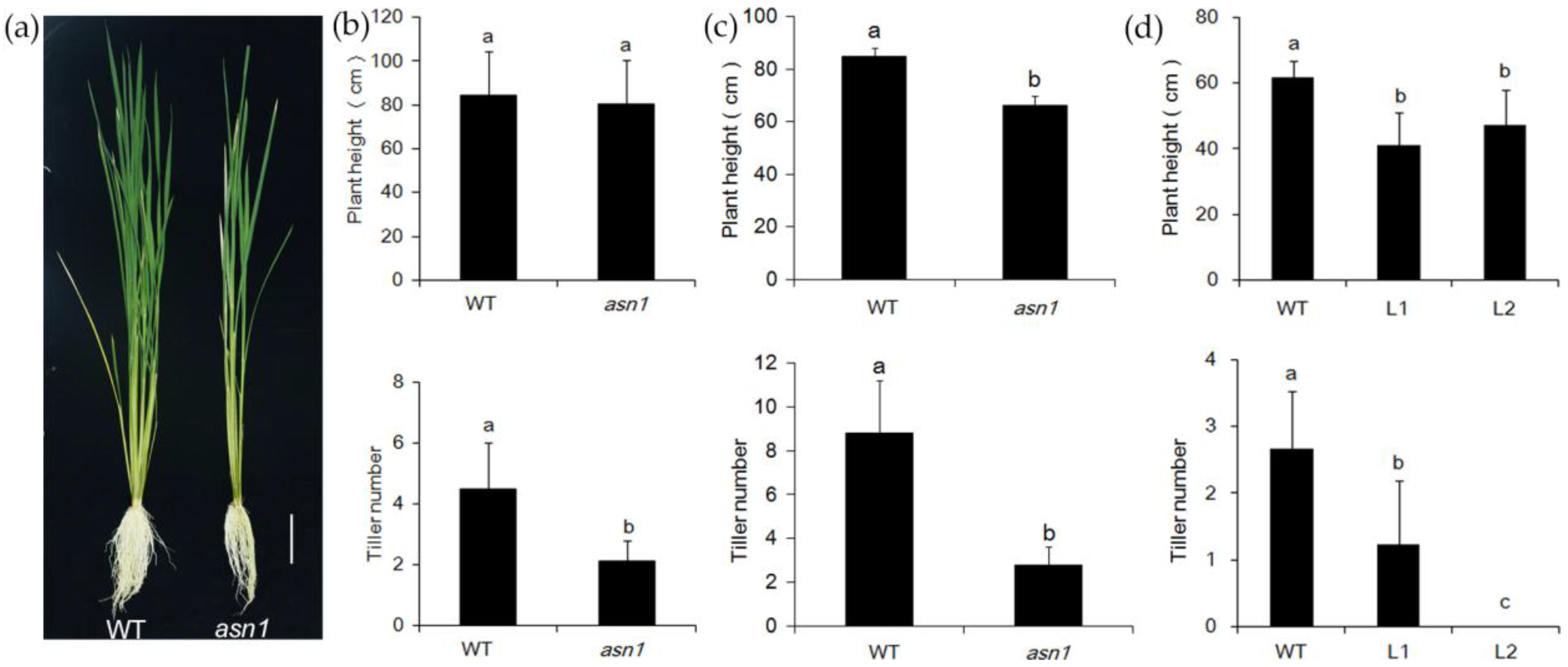
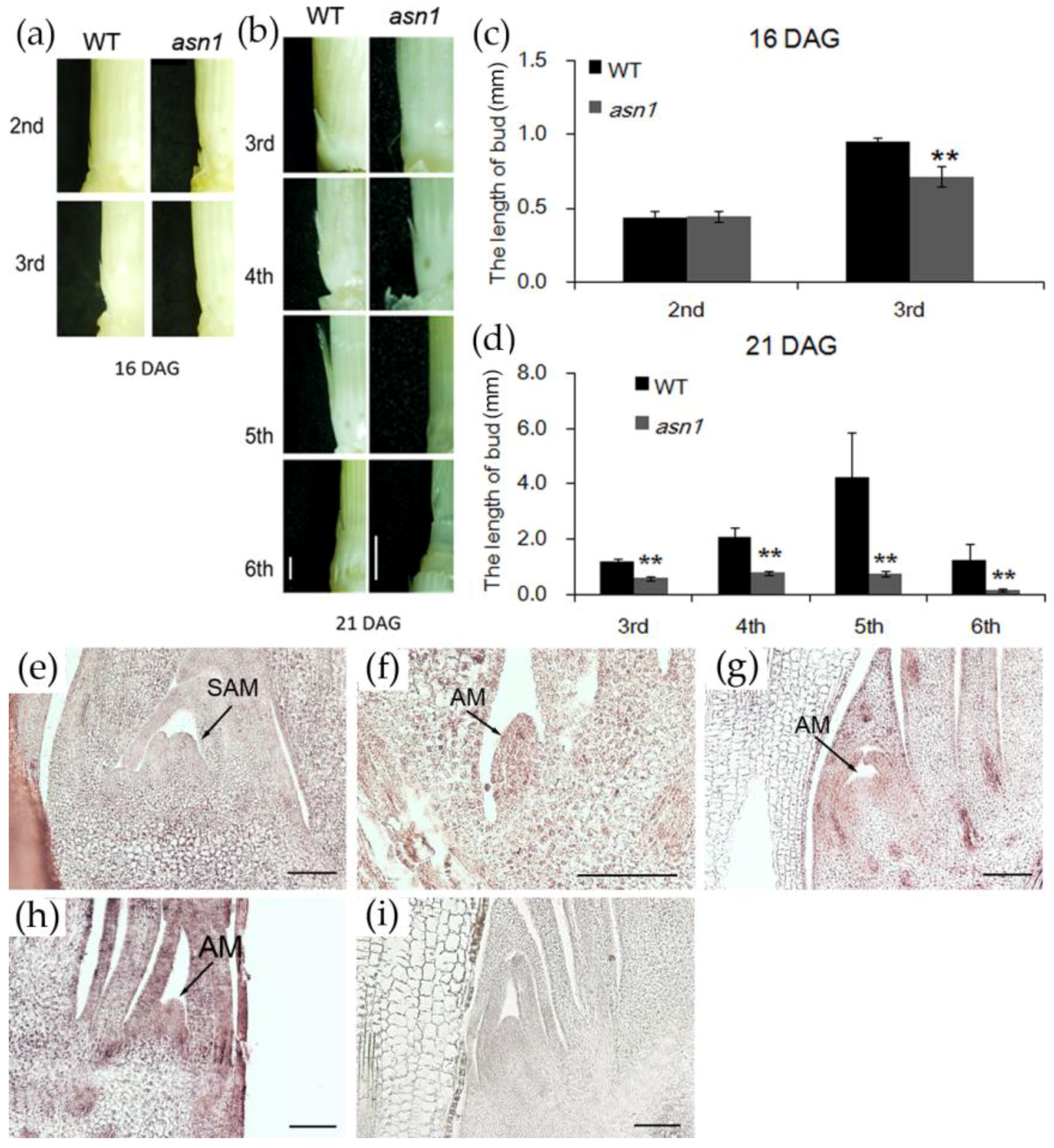
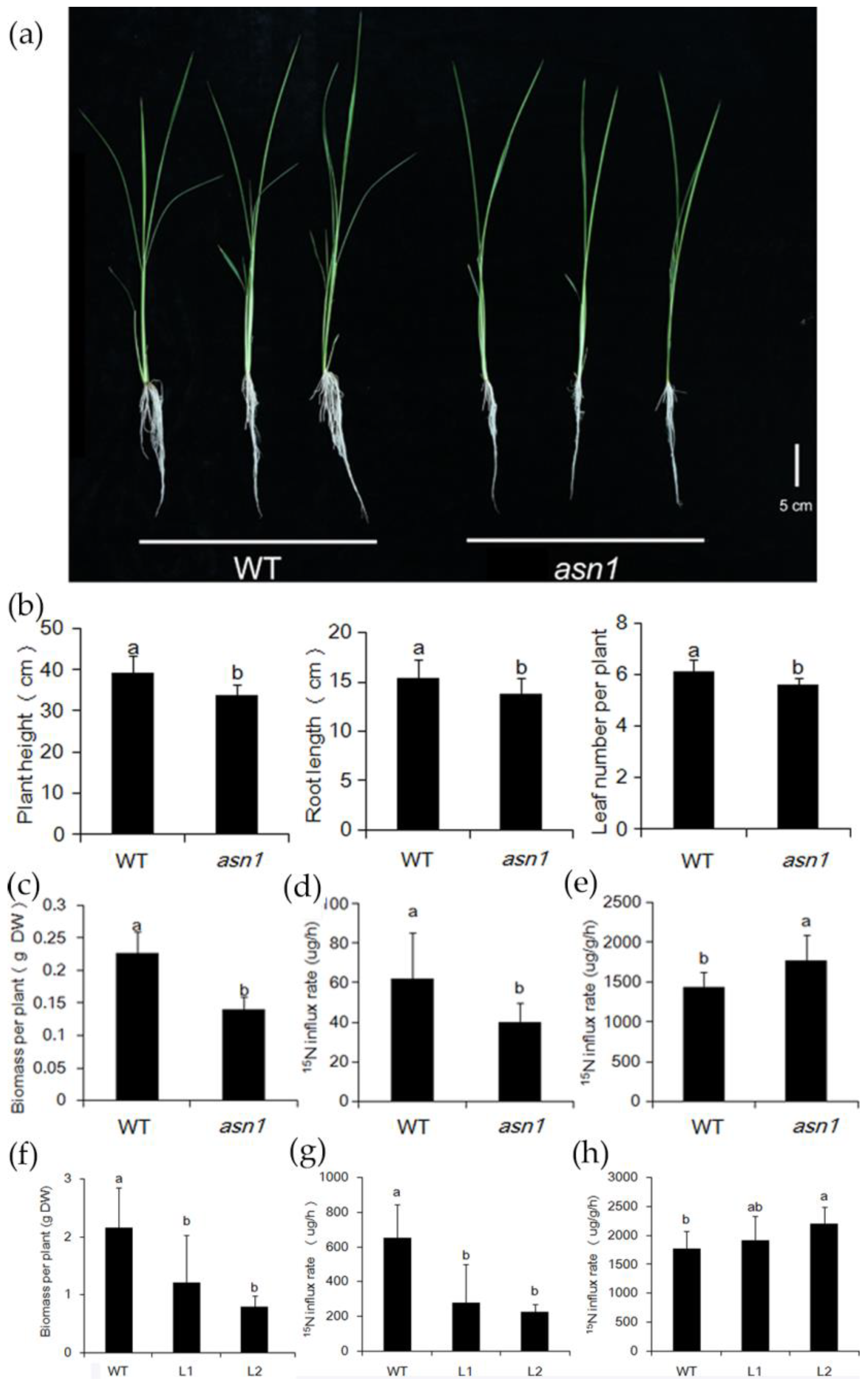
2.2. asn1 Shows Growth Defects at Both Early and Late Stages
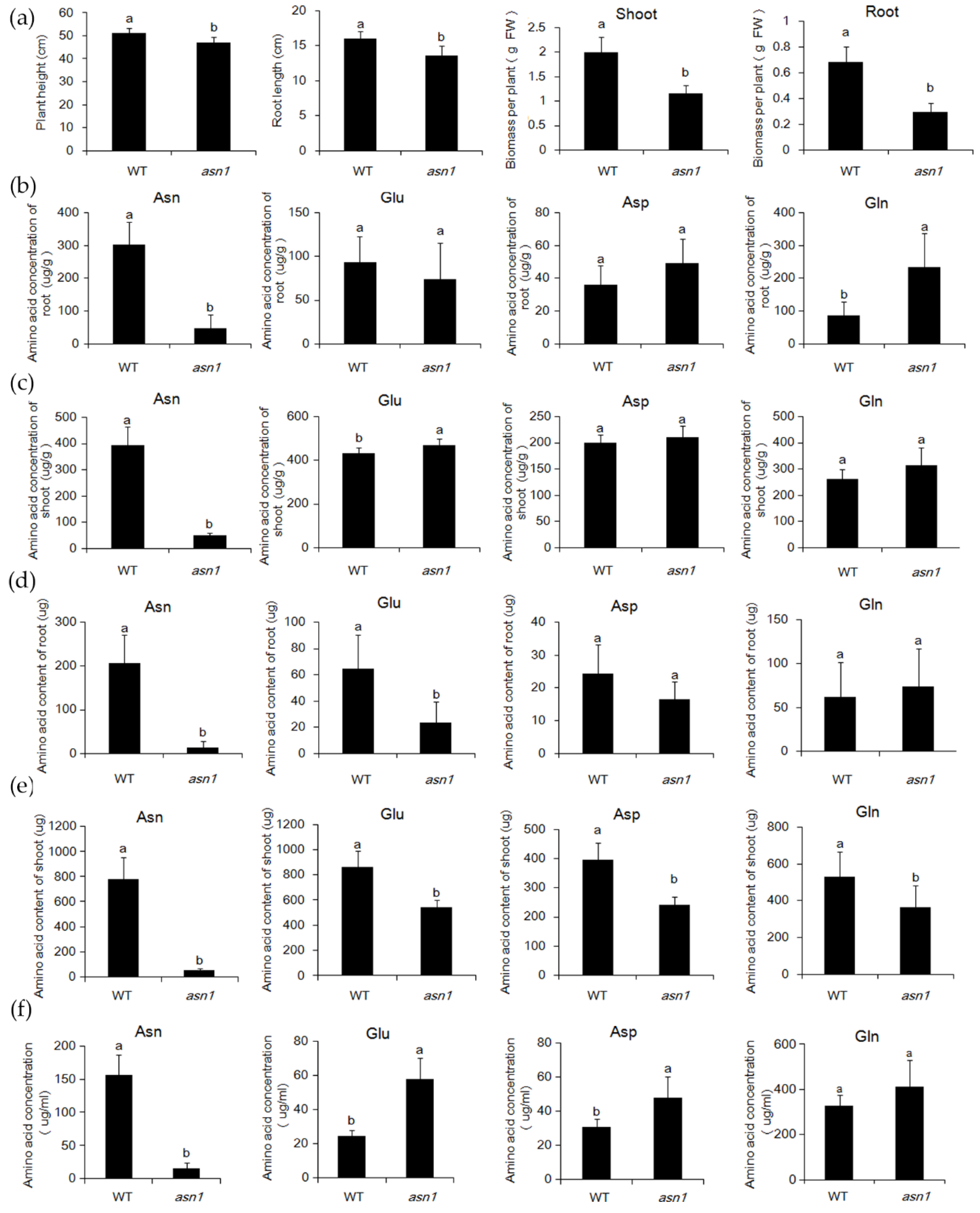
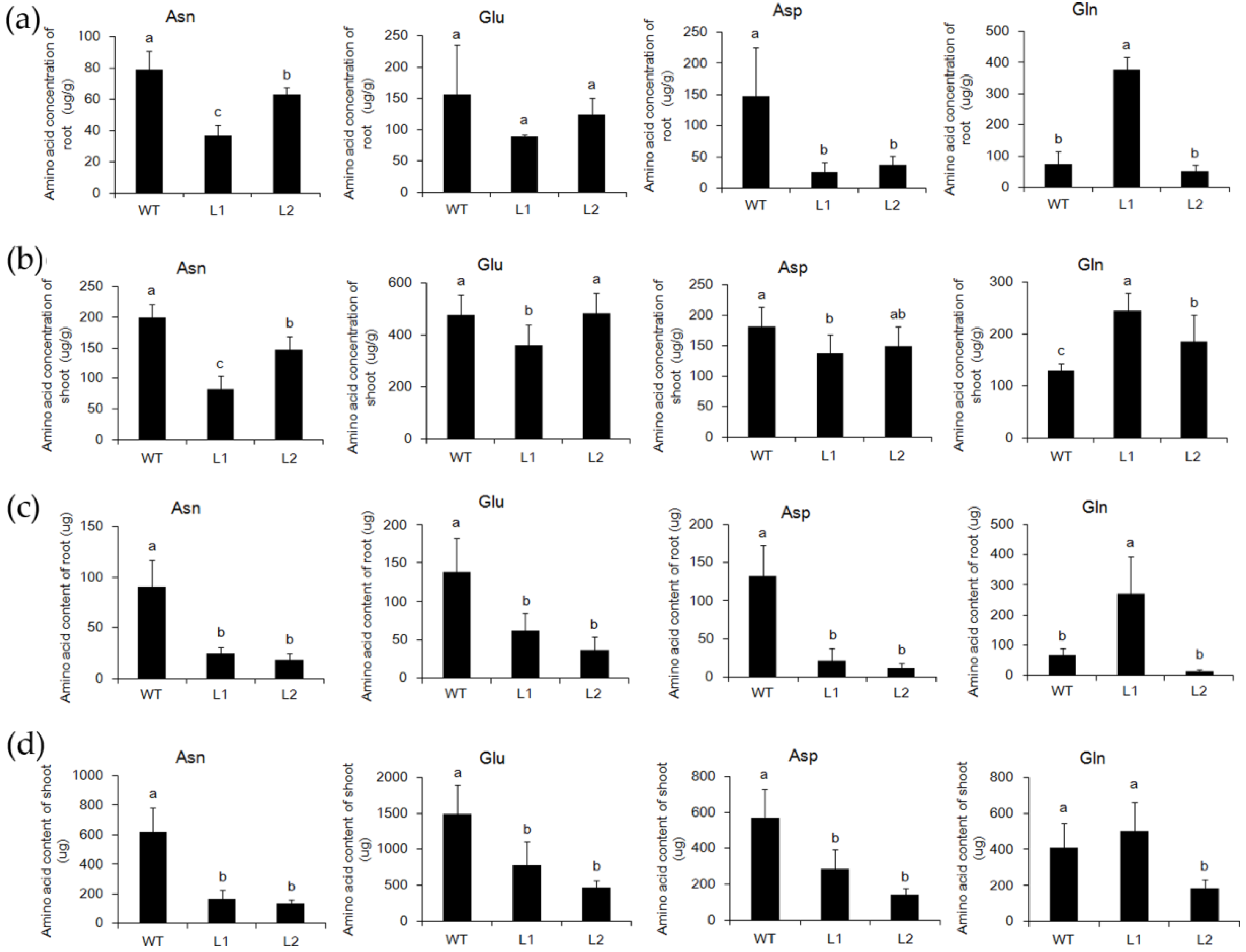
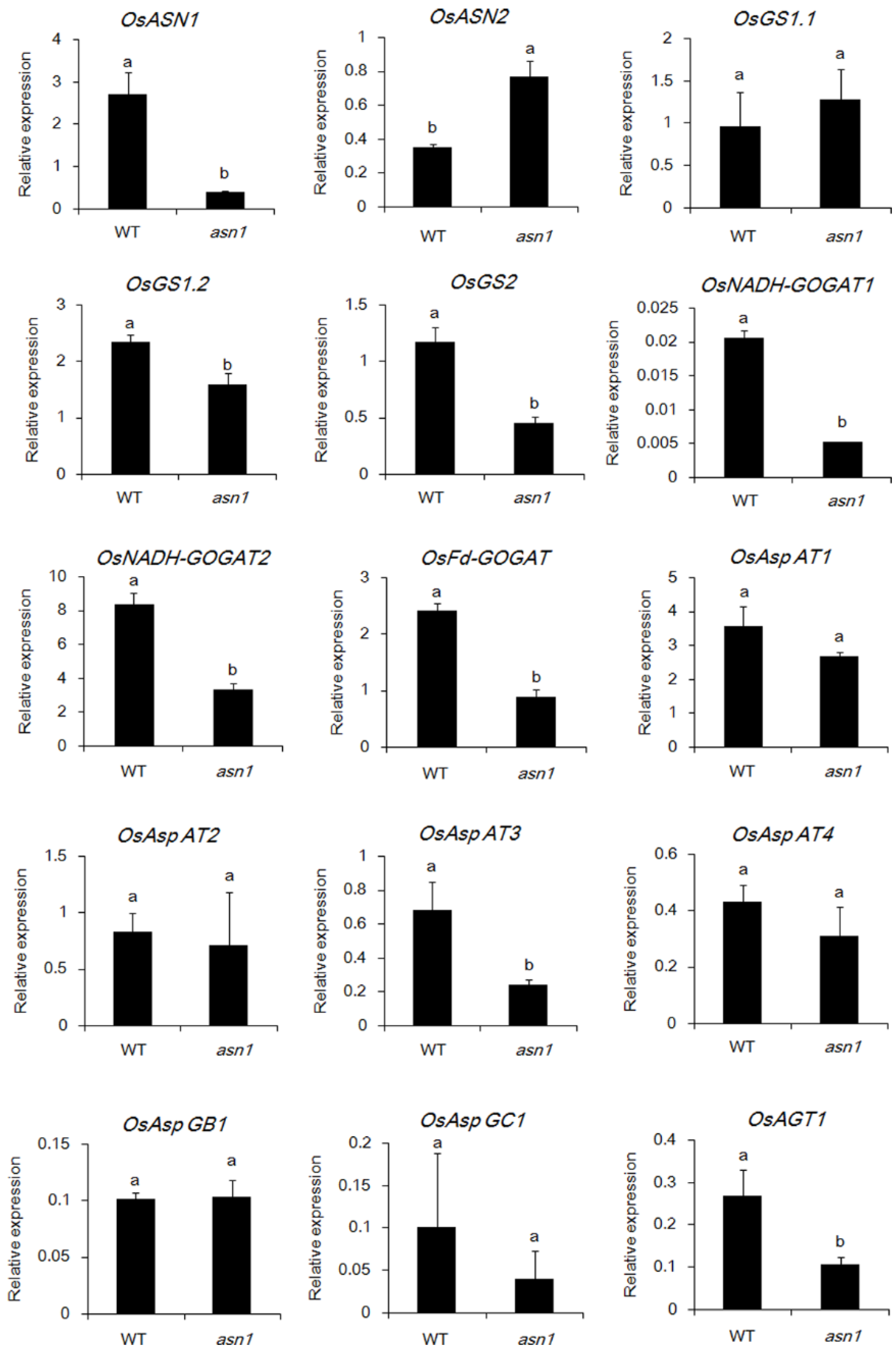
2.3. The Asparagine Metabolism Pathway Is Strongly Influenced in asn1 Mutant
2.4. OsASN1 Influences the Tiller Phenotype
2.5. ASN1 Does Not Influence the N Absorption
2.6. ASN1 Metabolism Is Significantly Influenced Which Caused the Phenotype Difference
3. Discussion
3.1. OsASN1 Influence the Development of Plants
3.2. OsASN1 Is a Gene with Multiple Functions
3.3. Lack of Asparagine Suppressed the Cell Division
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Generation of OsASN1 CRSPR/CAS9 Mutants
4.3. RNA Extraction and Quantitative PCR Analysis
4.4. Analysis of Total N Concentration
4.5. Analysis of Amino Acids Concentration
4.6. Determination of the 15N-NH4+ Influx Rate
4.7. RNA In Situ Hybridization
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Xu, G.H.; Fan, X.R.; Miller, A.J. Plant Nitrogen Assimilation and Use Efficiency. Annu. Rev. Plant Biol. 2012, 63, 153–182. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, H.; Chino, M. Chemical-Composition of Phloem Sap from the Uppermost Internode of the Rice Plant. Plant Cell Physiol. 1990, 31, 247–251. [Google Scholar]
- Lea, P.J.; Sodek, L.; Parry, M.A.J.; Shewry, R.; Halford, N.G. Asparagine in plants. Ann. Appl. Biol. 2007, 150, 1–26. [Google Scholar] [CrossRef]
- Ireland, R.J.; John Lea, P. The enzymes of glutamine, glutamate, asparagine and aspartate metabolism. In Plant Amino Acids: Biochemistry and Biotechnology; Marcel Dekker Inc.: New York, NY, USA, 1999; pp. 49–109. [Google Scholar]
- Yamaya, T.; Kusano, M. Evidence supporting distinct functions of three cytosolic glutamine synthetases and two NADH-glutamate synthases in rice. J. Exp. Bot. 2014, 65, 5519–5525. [Google Scholar] [CrossRef] [PubMed]
- McNally, S.F.; Hirel, B.; Gadal, P.; Mann, A.F.; Stewart, G.R. Glutamine Synthetases of Higher Plants: Evidence for a Specific Isoform Content Related to their Possible Physiological Role and their Compartmentation within the Leaf. Plant Physiol. 1983, 72, 22–25. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.; Lee, J.; Kichey, T.; Gerentes, D.; Zivy, M.; Tatout, C.; Dubois, F.; Balliau, T.; Valot, B.; Davanture, M.; et al. Two cytosolic glutamine synthetase isoforms of maize are specifically involved in the control of grain production. Plant Cell 2006, 18, 3252–3274. [Google Scholar] [CrossRef]
- Ishiyama, K.; Inoue, E.; Watanabe-Takahashi, A.; Obara, M.; Yamaya, T.; Takahashi, H. Kinetic properties and ammonium-dependent regulation of cytosolic isoenzymes of glutamine synthetase in Arabidopsis. J. Biol. Chem. 2004, 279, 16598–16605. [Google Scholar] [CrossRef]
- Bernard, S.M.; Moller, A.L.B.; Dionisio, G.; Kichey, T.; Jahn, T.P.; Dubois, F.; Baudo, M.; Lopes, M.S.; Terce-Laforgue, T.; Foyer, C.H.; et al. Gene expression, cellular localisation and function of glutamine synthetase isozymes in wheat (Triticum aestivum L.). Plant Mol. Biol. 2008, 67, 89–105. [Google Scholar] [CrossRef]
- Lam, H.M.; Coschigano, K.T.; Oliveira, I.C.; MeloOliveira, R.; Coruzzi, G.M. The molecular-genetics of nitrogen assimilation into amino acids in higher plants. Annu. Rev. Plant Phys. 1996, 47, 569–593. [Google Scholar] [CrossRef]
- Fukumorita, T.; Chino, M. Sugar, Amino Acid and Inorganic Contents in Rice Phloem Sap. Plant Cell Physiol. 1982, 23, 273–283. [Google Scholar] [CrossRef]
- Funayama, K.; Kojima, S.; Tabuchi-Kobayashi, M.; Sawa, Y.; Nakayama, Y.; Hayakawa, T.; Yamaya, T. Cytosolic Glutamine Synthetase1;2 is Responsible for the Primary Assimilation of Ammonium in Rice Roots. Plant Cell Physiol. 2013, 54, 934–943. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, M.; Ishiyama, K.; Kojima, S.; Konishi, N.; Nakano, K.; Kanno, K.; Hayakawa, T.; Yamaya, T. Asparagine Synthetase1, but not Asparagine Synthetase2, is Responsible for the Biosynthesis of Asparagine Following the Supply of Ammonium to Rice Roots. Plant Cell Physiol. 2015, 56, 769–778. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, I.C.; Brenner, E.; Chiu, J.; Hsieh, M.H.; Kouranov, A.; Lam, H.M.; Shin, M.J.; Coruzzi, G. Metabolite and light regulation of metabolism in plants: Lessons from the study of a single biochemical pathway. Br. J. Med. Biol. Res. 2001, 34, 567–575. [Google Scholar] [CrossRef]
- Gaufichon, L.; Reisdorf-Cren, M.; Rothstein, S.J.; Chardon, F.; Suzuki, A. Biological functions of asparagine synthetase in plants. Plant Sci. 2010, 179, 141–153. [Google Scholar] [CrossRef]
- Igarashi, D.; Ishizaki, T.; Totsuka, K.; Ohsumi, C. ASN2 is a key enzyme in asparagine biosynthesis under ammonium sufficient conditions. Plant Biotechnol. 2009, 26, 153–159. [Google Scholar] [CrossRef]
- Gaufichon, L.; Masclaux-Daubresse, C.; Tcherkez, G.; Reisdorf-Cren, M.; Sakakibara, Y.; Hase, T.; Clement, G.; Avice, J.C.; Grandjean, O.; Marmagne, A.; et al. Arabidopsis thaliana ASN2 encoding asparagine synthetase is involved in the control of nitrogen assimilation and export during vegetative growth. Plant Cell Environ. 2013, 36, 328–342. [Google Scholar] [CrossRef] [PubMed]
- Nakano, K.; Suzuki, T.; Hayakawa, T.; Yamaya, T. Organ and cellular localization of asparagine synthetase in rice plants. Plant Cell Physiol. 2000, 41, 874–880. [Google Scholar] [CrossRef]
- Kawachi, T.; Sueyoshi, K.; Nakajima, A.; Yamagata, H.; Sugimoto, T.; Oji, Y. Expression of asparagine synthetase in rice (Oryza sativa) roots in response to nitrogen. Physiol. Plant. 2002, 114, 41–46. [Google Scholar] [CrossRef]
- Luo, L.; Pan, S.; Liu, X.H.; Wang, H.X.; Xu, G.H. Nitrogen deficiency inhibits cell division-determined elongation, but not initiation, of rice tiller buds. Isr. J. Plant Sci. 2017, 64, 32–40. [Google Scholar] [CrossRef]
- Umehara, M.; Hanada, A.; Magome, H.; Takeda-Kamiya, N.; Yamaguchi, S. Contribution of Strigolactones to the Inhibition of Tiller Bud Outgrowth under Phosphate Deficiency in Rice. Plant Cell Physiol. 2010, 51, 1118–1126. [Google Scholar] [CrossRef]
- Huang, W.T.; Bai, G.X.; Wang, J.; Zhu, W.; Zeng, Q.S.; Lu, K.; Sun, S.Y.; Fang, Z.M. Two Splicing Variants of OsNPF7.7 Regulate Shoot Branching and Nitrogen Utilization Efficiency in Rice. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.M.; Bai, G.X.; Huang, W.T.; Wang, Z.X.; Wang, X.L.; Zhang, M.Y. The Rice Peptide Transporter OsNPF7.3 is Induced by Organic Nitrogen, and Contributes to Nitrogen Allocation and Grain Yield. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lu, K.; Nie, H.P.; Zeng, Q.S.; Wu, B.W.; Qian, J.J.; Fang, Z.M. Rice nitrate transporter OsNPF7.2 positively regulates tiller number and grain yield. Rice 2018, 11. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Wu, B.W.; Wang, J.; Zhu, W.; Nie, H.P.; Qian, J.J.; Huang, W.T.; Fang, Z.M. Blocking amino acid transporter OsAAP3 improves grain yield by promoting outgrowth buds and increasing tiller number in rice. Plant Biotechnol. J. 2018, 16, 1710–1722. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, M.; Ishiyama, K.; Kojima, S.; Kojima, M.; Sakakibara, H.; Yamaya, T.; Hayakawa, T. Lack of Cytosolic Glutamine Synthetase1;2 Activity Reduces Nitrogen-Dependent Biosynthesis of Cytokinin Required for Axillary Bud Outgrowth in Rice Seedlings. Plant Cell Physiol. 2017, 58, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Gaufichon, L.; Rothstein, S.J.; Suzuki, A. Asparagine Metabolic Pathways in Arabidopsis. Plant Cell Physiol. 2016, 57, 675–689. [Google Scholar] [CrossRef] [PubMed]
- Wong, H.K.; Chan, H.K.; Coruzzi, G.M.; Lam, H.M. Correlation of ASN2 gene expression with ammonium metabolism in Arabidopsis. Plant Physiol. 2004, 134, 332–338. [Google Scholar] [CrossRef]
- Lam, H.M.; Wong, P.; Chan, H.K.; Yam, K.M.; Chen, L.; Chow, C.M.; Coruzzi, G.M. Overexpression of the ASN1 gene enhances nitrogen status in seeds of Arabidopsis. Plant Physiol. 2003, 132, 926–935. [Google Scholar] [CrossRef]
- Gaufichon, L.; Marmagne, A.; Belcram, K.; Yoneyama, T.; Sakakibara, Y.; Hase, T.; Grandjean, O.; Clement, G.; Citerne, S.; Boutet-Mercey, S.; et al. ASN1-encoded asparagine synthetase in floral organs contributes to nitrogen filling in Arabidopsis seeds. Plant J. 2017, 91, 371–393. [Google Scholar] [CrossRef]
- Ohashi, M.; Ishiyama, K.; Kojima, S.; Konishi, N.; Sasaki, K.; Miyao, M.; Hayakawa, T.; Yamaya, T. Outgrowth of Rice Tillers Requires Availability of Glutamine in the Basal Portions of Shoots. Rice 2018, 11. [Google Scholar] [CrossRef]
- Krall, A.S.; Xu, S.L.; Graeber, T.G.; Braas, D.; Christofk, H.R. Asparagine promotes cancer cell proliferation through use as an amino acid exchange factor. Nat. Commun. 2016, 7, 11457. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhou, F.S.; Du, W.H.; Dou, J.F.; Xu, Y.; Gao, W.W.; Chen, G.; Zuo, X.B.; Sun, L.D.; Zhang, X.J.; et al. Knockdown of asparagine synthetase by RNAi suppresses cell growth in human melanoma cells and epidermoid carcinoma cells. Biotechnol. Appl. Biochem. 2016, 63, 328–333. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.X.; Wang, X.Y.; Wang, L.; Zheng, J.; Wang, J.; Wang, B.M. Knockdown of asparagine synthetase (ASNS) suppresses cell proliferation and inhibits tumor growth in gastric cancer cells. Scand. J. Gastroenterol. 2016, 51, 1220–1226. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Lv, F.Z.; Zhu, X.X.; Wu, Y.; Shen, X.Y. Loss of asparagine synthetase suppresses the growth of human lung cancer cells by arresting cell cycle at G0/G1 phase. Cancer Gene Ther. 2016, 23, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Miao, J.; Guo, D.S.; Zhang, J.Z.; Huang, Q.P.; Qin, G.J.; Zhang, X.; Wan, J.M.; Gu, H.Y.; Qu, L.J. Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 2013, 23, 1233–1236. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.F.; Ren, H.Y.; Gu, M.; Zhao, J.N.; Sun, S.B.; Zhang, X.; Chen, J.Y.; Wu, P.; Xu, G.H. The Phosphate Transporter Gene OsPht1;8 Is Involved in Phosphate Homeostasis in Rice. Plant 2011, 156, 1164–1175. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, L.; Qin, R.; Liu, T.; Yu, M.; Yang, T.; Xu, G. OsASN1 Plays a Critical Role in Asparagine-Dependent Rice Development. Int. J. Mol. Sci. 2019, 20, 130. https://doi.org/10.3390/ijms20010130
Luo L, Qin R, Liu T, Yu M, Yang T, Xu G. OsASN1 Plays a Critical Role in Asparagine-Dependent Rice Development. International Journal of Molecular Sciences. 2019; 20(1):130. https://doi.org/10.3390/ijms20010130
Chicago/Turabian StyleLuo, Le, Ruyi Qin, Tao Liu, Ming Yu, Tingwen Yang, and Guohua Xu. 2019. "OsASN1 Plays a Critical Role in Asparagine-Dependent Rice Development" International Journal of Molecular Sciences 20, no. 1: 130. https://doi.org/10.3390/ijms20010130
APA StyleLuo, L., Qin, R., Liu, T., Yu, M., Yang, T., & Xu, G. (2019). OsASN1 Plays a Critical Role in Asparagine-Dependent Rice Development. International Journal of Molecular Sciences, 20(1), 130. https://doi.org/10.3390/ijms20010130



