Comparative Metabolic Phenotyping of Tomato (Solanum lycopersicum) for the Identification of Metabolic Signatures in Cultivars Differing in Resistance to Ralstonia solanacearum
Abstract
1. Introduction
2. Results
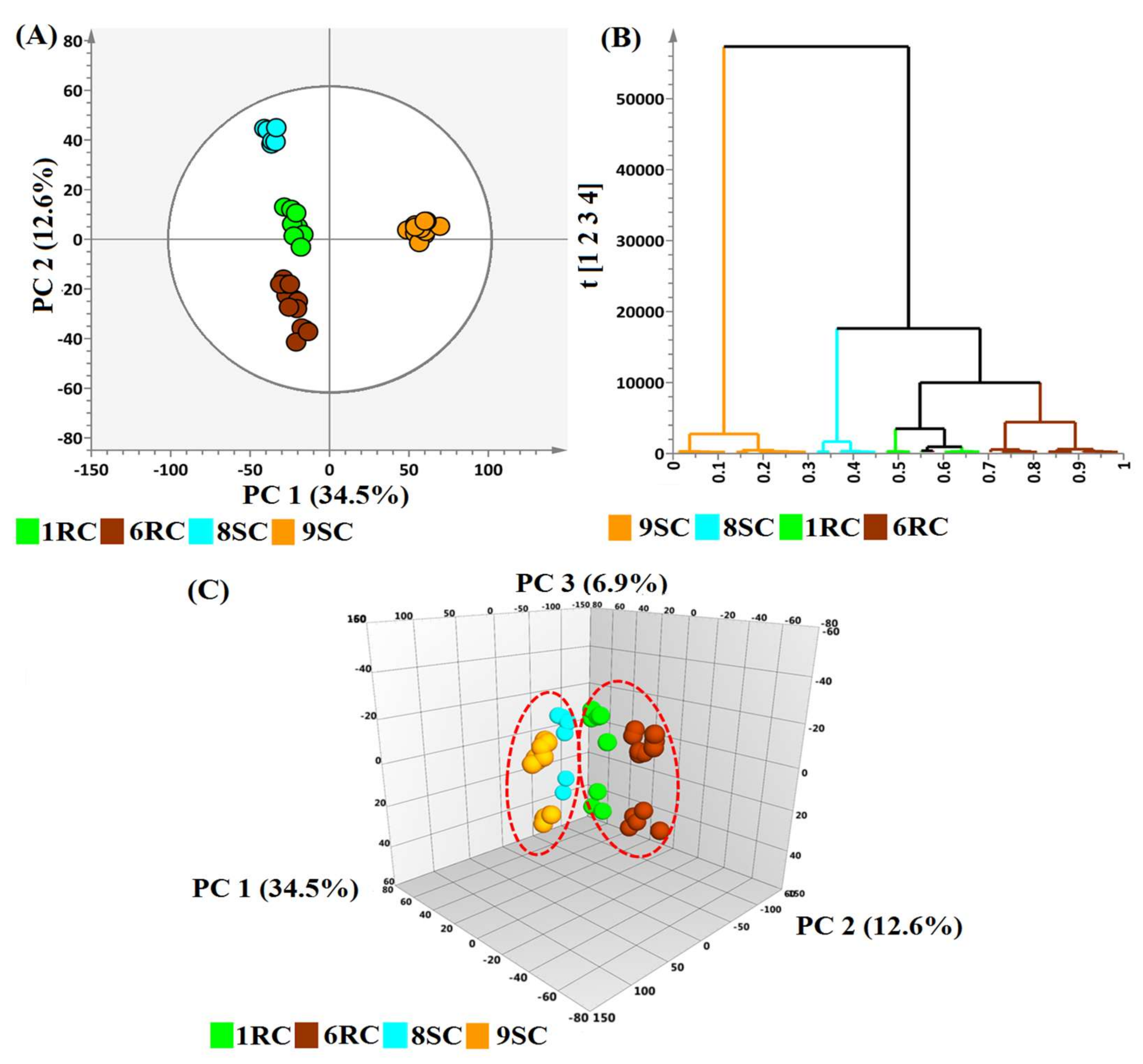
2.1. Differential Metabolic Profiles as Described by Chemometric Models
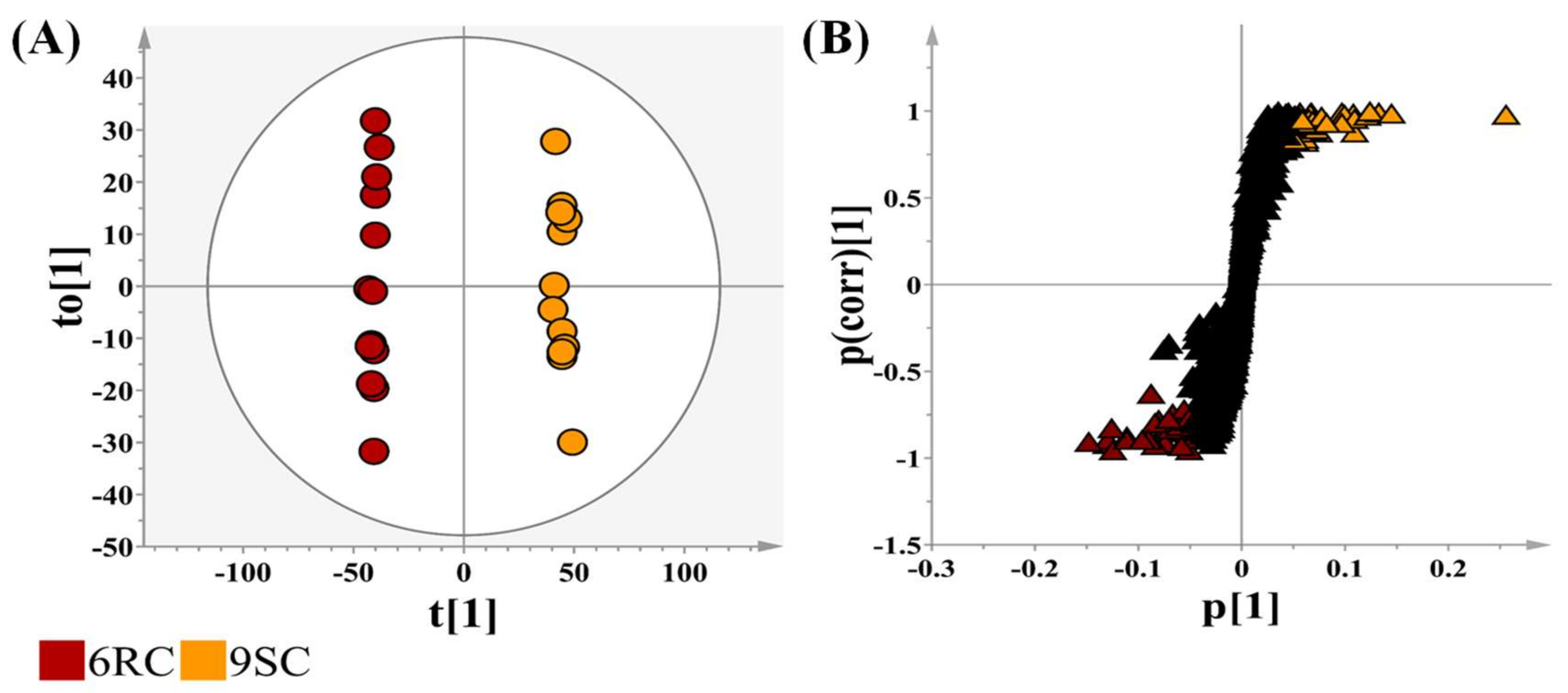
2.2. Metabolic Signatures Differentiating the Four Tomato Cultivars
3. Discussion
4. Materials and Methods
4.1. Plant Cultivation
4.2. Metabolite Extraction and Sample Preparation
4.3. Ultra-High-Performance Liquid Chromatography (UHPLC) Analyses
4.4. Quadrupole Time-of-Flight Mass Spectrometry (q-TOF-MS) Analyses
4.5. Data Analyses
4.6. Metabolite Annotation and Qualitative Comparison
4.7. Semi-Quantitative Comparison
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Grandillo, S.; Zamir, D.; Tanksley, S.D. Genetic improvement of processing tomatoes: A 20 years perspective. Euphytica 1999, 110, 85–97. [Google Scholar] [CrossRef]
- Beckles, D.M. Factors affecting the postharvest soluble solids and sugar content of tomato (Solanum lycopersicum L.) fruit. Postharvest Biol. Technol. 2012, 63, 129–140. [Google Scholar] [CrossRef]
- FAOSTAT. Value of Agricultural Production. Available online: http://www.fao.org/faostat/en/#data/QV (accessed on 29 May 2017).
- Gómez-Romero, M.; Segura-Carretero, A.; Fernández-Gutiérrez, A. Metabolite profiling and quantification of phenolic compounds in methanol extracts of tomato fruit. Phytochemistry 2010, 71, 1848–1864. [Google Scholar] [CrossRef] [PubMed]
- Lima, P.G.P.; Vianello, F.; Corrêa, C.R.; Arnoux, R.; Campos, D.S.; Borguini, M.G. Polyphenols in Fruits and Vegetables and Its Effect on Human Health. Food Nutr. Sci. 2014, 5, 1065–1082. [Google Scholar] [CrossRef]
- Raiola, A.; Rigano, M.I.M.; Calafiore, R.; Frusciante, L.; Barone, A. Enhancing the health-promoting effects of tomato fruit for biofortified food. Mediat. Inflamm. 2014, 2014, 139873. [Google Scholar] [CrossRef] [PubMed]
- Arab, L.; Steck, S. Lycopene and cardiovascular disease. Am. J. Clin. Nutr. 2000, 71, 1691S–1695S. [Google Scholar] [CrossRef] [PubMed]
- Bauchet, G.; Causse, M. Genetic diversity in tomato (Solanum lycopersicum) and its wild relatives. In Genetic Diversity in Plants; Caliskan, M., Ed.; InTech: Shanghai, China, 2012; pp. 134–162. [Google Scholar]
- Zamir, D. Improving plant breeding with exotic genetic libraries. Nat. Rev. Genet. 2001, 2, 983–989. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Lindhout, P. Domestication and breeding of tomatoes: What have we gained and what can we gain in the future? Ann. Bot. 2007, 100, 1085–1094. [Google Scholar] [CrossRef] [PubMed]
- Klee, H.J.; Tieman, D.M. Genetic challenges of flavor improvement in tomato. Trends Genet. 2013, 29, 257–262. [Google Scholar] [CrossRef] [PubMed]
- Perez-de-Castro, A.M.; Vilanova, S.; Canizares, J.; Pascual, L.; Blanca, J.M.; Diez, M.J.; Prohens, J.; Pico, B. Application of genomic tools in plant breeding. Curr. Genom. 2012, 13, 179–195. [Google Scholar] [CrossRef] [PubMed]
- Tugizimana, F.; Piater, L.; Dubery, I. Plant metabolomics: A new frontier in phytochemical analysis. S. Afr. J. Sci. 2013, 109, 1–11. [Google Scholar] [CrossRef]
- Kumar, R.; Bohra, A.; Pandey, A.K.; Pandey, M.K.; Kumar, A. Metabolomics for plant improvement: Status and prospects. Front. Plant Sci. 2017, 8, 01302. [Google Scholar] [CrossRef] [PubMed]
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Sumner, L.W.; Mendes, P.; Dixon, R.A. Plant metabolomics: Large-scale phytochemistry in the functional genomics era. Phytochemistry 2003, 62, 817–836. [Google Scholar] [CrossRef]
- Verpoorte, R.; Choi, Y.H.; Mustafa, N.R.; Kim, H.K. Metabolomics: Back to basics. Phytochem. Rev. 2008, 7, 525–537. [Google Scholar] [CrossRef]
- Finnegan, T.; Steenkamp, P.A.; Piater, L.A.; Dubery, I.A. The lipopolysaccharide-induced metabolome signature in Arabidopsis thaliana reveals dynamic reprogramming of phytoalexin and phytoanticipin pathways. PLoS ONE 2016, 11, e0163572. [Google Scholar] [CrossRef] [PubMed]
- Mhlongo, M.I.; Piater, L.A.; Madala, N.E.; Steenkamp, P.A.; Dubery, I.A. Phenylpropanoid defences in Nicotiana tabacum cells: Overlapping metabolomes indicate common aspects to priming responses induced by lipopolysaccharides, chitosan and flagellin-22. PLoS ONE 2016, 11, e0151350. [Google Scholar] [CrossRef] [PubMed]
- Mhlongo, M.I.; Steenkamp, P.A.; Piater, L.A.; Madala, N.E.; Dubery, I.A. Profiling of altered metabolomics states in Nicotiana tabacum cells induced by priming agents. Front. Plant Sci. 2016, 7, 1527. [Google Scholar] [CrossRef] [PubMed]
- Harrigan, G.G.; Martino-Catt, S.; Glenn, K.C. Metabolomics, metabolic diversity and genetic variation in crops. Metabolomics 2007, 3, 259–272. [Google Scholar] [CrossRef]
- Liu, X.; Locasale, J.W. Metabolomics: A primer. Trends Biochem. Sci. 2017, 42, 274–284. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.; Hinzman, A.A.; Kang, E.L.; Szczesniak, R.D.; Lu, L.J. Computational and statistical analysis of metabolomics data. Metabolomics 2015, 11, 1492–1513. [Google Scholar] [CrossRef]
- Wiklund, S.; Johansson, E.; Sjöström, L.; Mellerowicz, E.J.; Edlund, U.; Shockcor, J.P.; Gottfries, J.; Moritz, T.; Trygg, J. Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Anal. Chem. 2007, 80, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, L.; Trygg, J.; Wold, S. CV-ANOVA for significance testing of PLS and OPLS® models. J. Chemom. 2008, 22, 594–600. [Google Scholar] [CrossRef]
- Westerhuis, J.A.; Hoefsloot, H.C.J.; Smit, S.; Vis, D.J.; Smilde, A.K.; Velzen, E.J.J.; Duijnhoven, J.P.M.; Dorsten, F.A. Assessment of PLSDA cross validation. Metabolomics 2008, 4, 81–89. [Google Scholar] [CrossRef]
- Triba, M.N.; Le Moyec, L.; Amathieu, R.; Goossens, C.; Bouchemal, N.; Nahon, P.; Rutledge, D.N.; Savarin, P. PLS/OPLS models in metabolomics: Impact of permutation of dataset rows on the K-fold cross-validation quality parameters. Mol. Biosyst. 2014, 11, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Mhlongo, M.I.; Piater, L.A.; Steenkamp, P.A.; Madala, N.E.; Dubery, I.A. Priming agents of plant defence stimulate the accumulation of mono- and di-acylated quinic acids in cultured tobacco cells. Physiol. Mol. Plant Pathol. 2014, 88, 61–66. [Google Scholar] [CrossRef]
- Tugizimana, F.; Ncube, E.N.; Steenkamp, P.A.; Dubery, I.A. Metabolomics-derived insights into the manipulation of terpenoid synthesis in Centella asiatica cells by methyl jasmonate. Plant Biotechnol. Rep. 2015, 9, 125–136. [Google Scholar] [CrossRef]
- Nebbioso, A.; De Martino, A.; Eltlbany, N.; Smalla, K.; Piccolo, A. Phytochemical profiling of tomato roots following treatments with different microbial inoculants as revealed by IT-TOF mass spectrometry. Chem. Biol. Technol. Agric. 2016, 3, 12. [Google Scholar] [CrossRef]
- Itkin, M.; Rogachev, I.; Alkan, N.; Rosenberg, T.; Malitsky, S.; Masini, L.; Meir, S.; Iijima, Y.; Aoki, K.; de Vos, R.; et al. Glycoalkaloid metabolism is required for steroidal alkaloid glycosylation and prevention of phytotoxicity in tomato. Plant Cell 2011, 23, 4507–4525. [Google Scholar] [CrossRef] [PubMed]
- Liang, N.; Kitts, D.D. Role of chlorogenic acids in controlling oxidative and inflammatory stress conditions. Nutrients 2015, 8, 16. [Google Scholar] [CrossRef] [PubMed]
- Miedes, E.; Vanholme, R.; Boerjan, W.; Molina, A. The role of the secondary cell wall in plant resistance to pathogens. Front. Plant Sci. 2014, 5, 358. [Google Scholar] [CrossRef] [PubMed]
- Kulbat, K. The role of phenolic compounds in plant stress responses. Food Sci. Biotechnol. 2016, 80, 97–108. [Google Scholar]
- Mazid, M.; Khan, T.A.; Mohammad, F. Role of secondary metabolites in defense mechanisms of plants. Biol. Med. 2011, 3, 232–249. [Google Scholar]
- Baker, C.J.; Mock, N.M.; Smith, J.M.; Aver’yanov, A.A. The dynamics of apoplast phenolics in tobacco leaves following inoculation with bacteria. Front. Plant Sci. 2015, 6, 649. [Google Scholar] [CrossRef] [PubMed]
- Hammerschmidt, R. Chlorogenic acid: A versatile defense compound. Physiol. Mol. Plant Pathol. 2014, 88, 3–6. [Google Scholar] [CrossRef]
- Wojciechowska, E.; Weinert, C.H.; Egert, B.; Trierweiler, B.; Schmidt-Heydt, M.; Horneburg, B.; Graeff-Hönninger, S.; Kulling, S.E.; Geisen, R. Chlorogenic acid, a metabolite identified by untargeted metabolome analysis in resistant tomatoes, inhibits the colonization by Alternaria alternata by inhibiting alternariol biosynthesis. Eur. J. Plant Pathol. 2014, 139, 735–747. [Google Scholar] [CrossRef]
- Ncube, E.N.; Steenkamp, P.A.; Madala, N.E.; Dubery, I.A. Stimulatory effects of acibenzolar-S-methyl on chlorogenic acids biosynthesis in Centella asiatica cells. Front. Plant Sci. 2016, 7, 1469. [Google Scholar] [CrossRef] [PubMed]
- Bollina, V.; Kumaraswamy, G.K.; Kushalappa, A.C.; Choo, T.M.; Dion, Y.; Rioux, S.; Faubert, D.; Hamzehzarghani, H. Mass spectrometry-based metabolomics application to identify quantitative resistance-related metabolites in barley against Fusarium head blight. Mol. Plant Pathol. 2010, 11, 769–782. [Google Scholar] [CrossRef] [PubMed]
- Vicente, O.; Boscaiu, M. Flavonoids: Antioxidant compounds for plant defence… and for a healthy human diet. Not. Bot. Horti Agrobot. Cluj-Napoca 2017, 46, 14–21. [Google Scholar] [CrossRef]
- Natella, F.; Nardini, M.; Di Felice, M.; Scaccini, C. Benzoic and cinnamic acid derivatives as antioxidants: Structure-activity relation. J. Agric. Food Chem. 1999, 47, 1453–1459. [Google Scholar] [CrossRef] [PubMed]
- Senaratna, T.; Merritt, D.; Dixon, K.; Bunn, E.; Touchell, D.; Sivasithamparam, K. Benzoic acid may act as the functional group in salicylic acid and derivatives in the induction of multiple stress tolerance in plants. Plant Growth Regul. 2003, 39, 77–81. [Google Scholar] [CrossRef]
- Huang, X.-X.; Zhu, G.-O.; Liu, Q.; Chen, L.; Li, Y.-J.; Hou, B.-K. Modulation of plant salicylic acid-associated immune responses via glycosylation of dihydroxybenzoic acids. Plant Physiol. 2018, 176, 3103–3119. [Google Scholar] [CrossRef] [PubMed]
- Mehdy, M. Active Oxygen Species in Plant Defense against Pathogens. Plant Physiol. 1994, 105, 468–472. [Google Scholar] [CrossRef]
- Okazaki, Y.; Saito, K. Roles of lipids as signaling molecules and mitigators during stress response in plants. Plant J. 2014, 79, 584–596. [Google Scholar] [CrossRef] [PubMed]
- Kachroo, A.; Kachroo, P. Fatty acid-derived signals in plant defense. Annu. Rev. Phytopathol. 2009, 47, 153–176. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Xiao, S. Lipids in salicylic acid-mediated defense in plants: Focusing on the roles of phosphatidic acid and phosphatidylinositol 4-phosphate. Front. Plant Sci. 2015, 6, 387. [Google Scholar] [CrossRef] [PubMed]
- Shimada, T.L.; Takano, Y.; Shimada, T.; Fujiwara, M.; Fukao, Y.; Mori, M.; Okazaki, Y.; Saito, K.; Sasaki, R.; Aoki, K.; et al. Leaf oil body functions as a subcellular factory for the production of a phytoalexin in Arabidopsis. Plant Physiol. 2014, 164, 105–118. [Google Scholar] [CrossRef] [PubMed]
- Skórzynska-Polit, E. Lipid peroxidation in plant cells, its physiological role and changes under heavy metal stress. Acta Soc. Bot. Pol. 2007, 76, 49–54. [Google Scholar] [CrossRef]
- Yahara, S.; Uda, N.; Yoshio, E.; Yae, E. Steroidal alkaloid glycosides from tomato (Lycopersicon esculentum). J. Nat. Prod. 2004, 67, 500–502. [Google Scholar] [CrossRef] [PubMed]
- Cataldi, T.R.I.; Lelario, F.; Bufo, S.A. Analysis of tomato glycoalkaloids by liquid chromatography coupled with electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2005, 19, 3103–3110. [Google Scholar] [CrossRef] [PubMed]
- Cárdenas, P.D.; Sonawane, P.D.; Heinig, U.; Bocobza, S.E.; Burdman, S.; Aharoni, A. The bitter side of the nightshades: Genomics drives discovery in Solanaceae steroidal alkaloid metabolism. Phytochemistry 2015, 113, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Distl, M.; Wink, M. Identification and quantification of steroidal alkaloids from wild tuber-bearing solanum species by HPLC and LC-ESI-MS. Potato Res. 2009, 52, 79–104. [Google Scholar] [CrossRef]
- Friedman, M. Tomato glycoalkaloids: Role in the plant and in the diet. J. Agric. Food Chem. 2002, 50, 5751–5780. [Google Scholar] [CrossRef] [PubMed]
- Simons, V.; Morrissey, J.P.; Latijnhouwers, M.; Csukai, M.; Cleaver, A.; Yarrow, C.; Osbourn, A. Dual effects of plant steroidal alkaloids on Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 2006, 50, 2732–2740. [Google Scholar] [CrossRef] [PubMed]
- Moco, S.; Capanoglu, E.; Tikunov, Y.; Bino, R.J.; Boyacioglu, D.; Hall, R.D.; Vervoort, J.; De Vos, R.C.H. Tissue specialization at the metabolite level is perceived during the development of tomato fruit. J. Exp. Bot. 2007, 58, 4131–4146. [Google Scholar] [CrossRef] [PubMed]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Inititative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [PubMed]
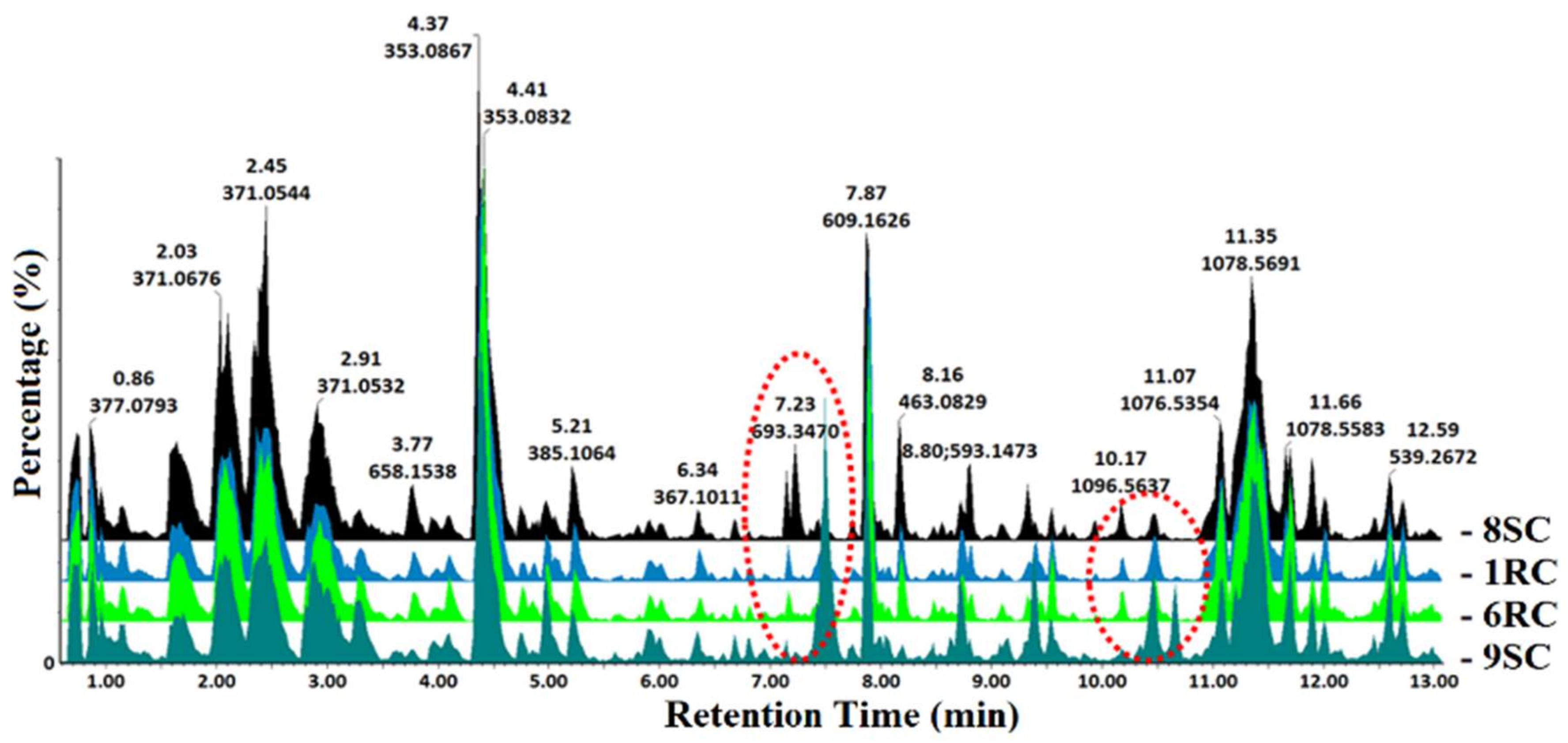
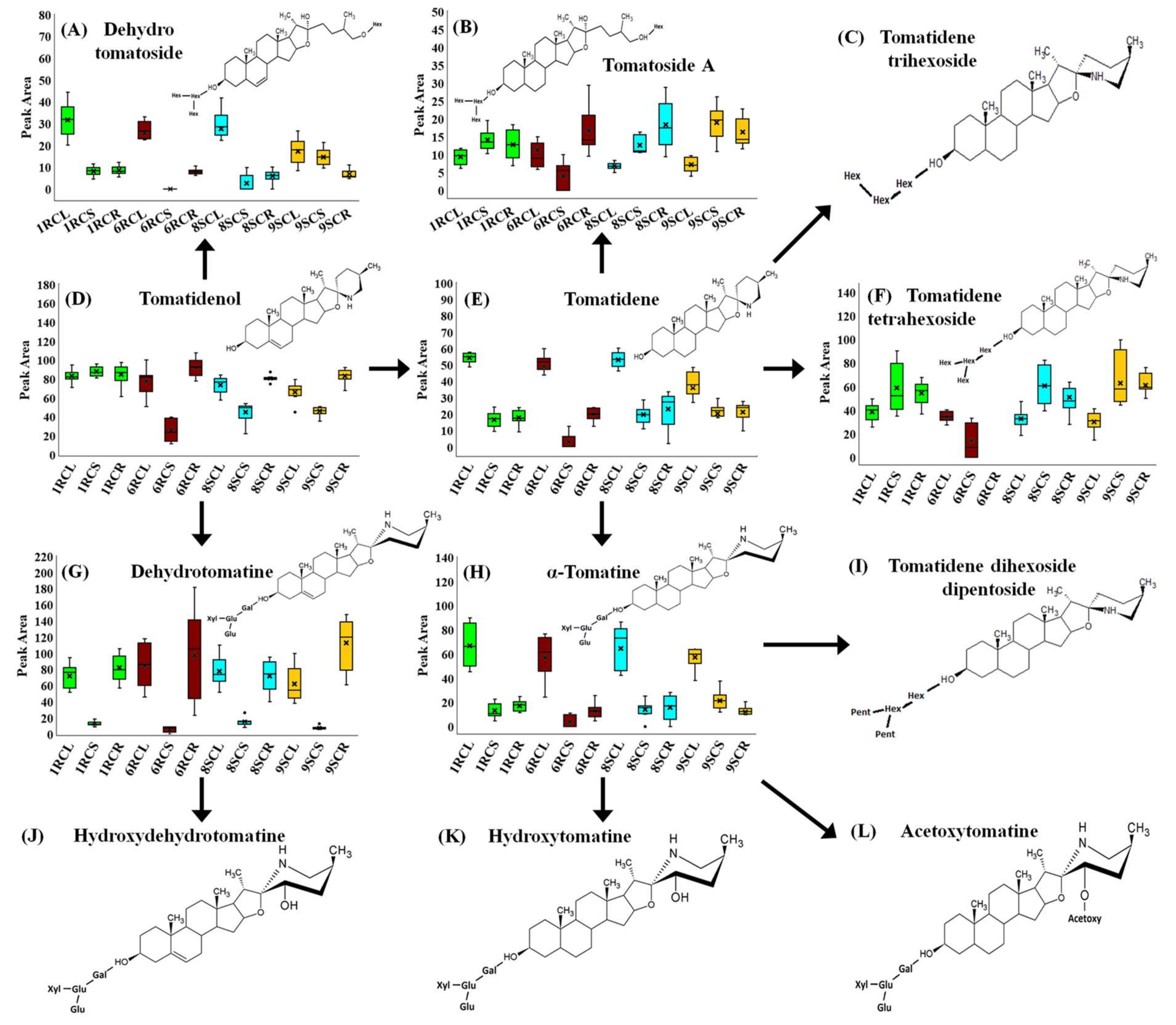

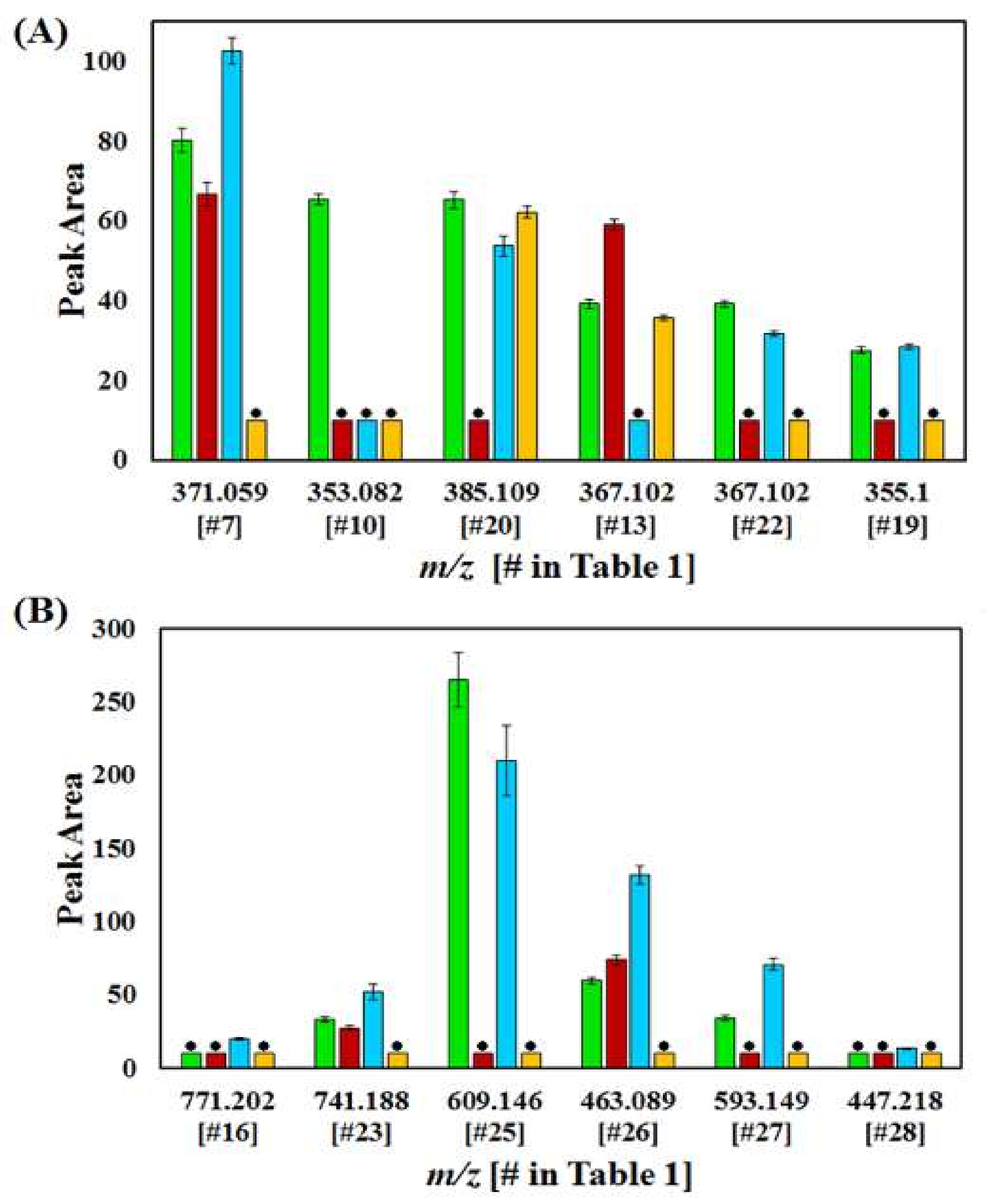
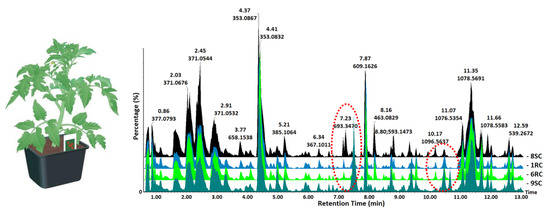
| # | Rt (min) | Compound Name | Mass (m/z) | 1RC | 6RC | 8SC | 9SC | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L | S | R | L | S | R | L | S | R | L | S | R | ||||
| Amino Acids | [M − H]− | ||||||||||||||
| 1 | 0.85 | Glutamic acid | 146.043 | o | |||||||||||
| 5 | 1.17 | Pyroglutamic acid | 128.032 | o | o | o | o | ||||||||
| 6 | 1.38 | Leucine/Isoleucine | 130.084 | o | o | o | o | o | |||||||
| 8 | 1.90 | Phenylalanine | 164.070 | o | o | o | |||||||||
| 24 | 7.51 | Acetyl tryptophan | 245.088 | o | o | ||||||||||
| Organic Acids | [M − H]− | ||||||||||||||
| 3 | 0.97 | Malic acid | 133.011 | o | o | o | o | o | o | ||||||
| 4 | 0.98 | Fumaric acid | 115.000 | o | o | o | o | ||||||||
| 15 | 4.77 | Citrate pentoside | 323.131 | o | o | ||||||||||
| 18 | 5.07 | Ascorbic acid | 175.036 | o | o | o | |||||||||
| Cinnamic Acid Derivatives | [M − H]− | ||||||||||||||
| 2 | 1.41 | Caffeoylglycoside | 341.105 | o | o | o | o | o | |||||||
| 7 | 1.64 | Caffeoylglucaric acid isomer 1 | 371.059 | o | o | o | |||||||||
| 9 | 2.07 | Caffeoylglucaric acid isomer 2 | 371.054 | o | o | ||||||||||
| 10 | 2.83 | Caffeoylquinic acid isomer 1 | 353.082 | o | o | o | |||||||||
| 12 | 3.98 | Sinapoylglycoside isomer 1 | 385.072 | o | |||||||||||
| 13 | 4.10 | Feruloylquinic acid isomer 1 | 367.102 | o | o | o | o | ||||||||
| 14 | 4.41 | Caffeoylquinic acid isomer 2 | 353.083 | o | o | o | o | o | o | o | o | ||||
| 19 | 5.07 | Feruloylglycoside | 355.100 | o | o | o | o | o | o | o | o | ||||
| 20 | 5.21 | Sinapoylglycoside isomer 2 | 385.109 | o | o | o | o | o | o | o | o | ||||
| 21 | 5.92 | Feruloylglucaric acid | 385.184 | o | o | ||||||||||
| 22 | 6.36 | Feruloylquinic acid isomer 2 | 367.102 | o | o | o | o | ||||||||
| Hydroxybenzoic Acid Derivatives | [M − H]− | ||||||||||||||
| 11 | 3.31 | 2,5-Dihydroxybenzoic acid pentose | 285.055 | o | o | o | o | o | |||||||
| 17 | 4.99 | Benzyl alcohol hexose pentoside | 401.142 | o | o | o | o | o | o | o | |||||
| Flavonoids | [M − H]− | ||||||||||||||
| 16 | 4.96 | Quercetin dihexose deoxyhexoside | 771.202 | o | |||||||||||
| 23 | 7.16 | Quercetin hexose deoxyhexose pentoside | 741.188 | o | o | o | o | o | |||||||
| 25 | 7.90 | Quercetin rutinoside | 609.146 | o | o | o | o | o | o | o | |||||
| 26 | 8.17 | Quercetin glycoside | 463.089 | o | o | o | o | o | |||||||
| 27 | 8.83 | Kaempferol rutinoside | 593.149 | o | o | o | o | o | |||||||
| 28 | 9.12 | Kaempferol glycoside | 447.218 | o | |||||||||||
| Steroidal Glycoalkaloids | [M + H]+ | ||||||||||||||
| 29 | 10.18 | Hydroxytomatine | 1050.548 | o | o | ||||||||||
| 30 | 11.07 | Dehydrotomatine isomer 1 | 1032.540 | o | o | o | o | o | o | o | o | o | |||
| 31 | 11.31 | Lycoperoside G/Lycoperoside F/Esculeoside A | 1136.560 | o | o | o | |||||||||
| 32 | 11.43 | Dehydrotomatoside | 1079.560 | o | o | o | o | o | o | o | |||||
| 33 | 11.43 | Tomatidene tetrahexoside | 1062.560 | o | o | o | |||||||||
| 34 | 11.44 | Dehydrotomatine isomer 2 | 1032.550 | o | o | o | |||||||||
| 35 | 11.45 | Tomatoside A | 1081.570 | o | |||||||||||
| 36 | 11.66 | α-Tomatine | 1034.543 | o | o | o | o | o | |||||||
| Lipids | [M − H]− | ||||||||||||||
| 37 | 13.92 | Hydroxyoctadecanedioic acid | 329.230 | o | |||||||||||
| 38 | 14.00 | Trihydroxyoctadecadienoic acid | 327.220 | o | |||||||||||
| 39 | 14.35 | 13-Amino-13-oxo-tridecanoic acid | 242.170 | o | |||||||||||
| 40 | 15.00 | Hydroxyoctadecanedioic acid | 329.230 | o | |||||||||||
| 41 | 16.20 | Hydroxyoctadecanedioic acid | 329.230 | o | o | ||||||||||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeiss, D.R.; Mhlongo, M.I.; Tugizimana, F.; Steenkamp, P.A.; Dubery, I.A. Comparative Metabolic Phenotyping of Tomato (Solanum lycopersicum) for the Identification of Metabolic Signatures in Cultivars Differing in Resistance to Ralstonia solanacearum. Int. J. Mol. Sci. 2018, 19, 2558. https://doi.org/10.3390/ijms19092558
Zeiss DR, Mhlongo MI, Tugizimana F, Steenkamp PA, Dubery IA. Comparative Metabolic Phenotyping of Tomato (Solanum lycopersicum) for the Identification of Metabolic Signatures in Cultivars Differing in Resistance to Ralstonia solanacearum. International Journal of Molecular Sciences. 2018; 19(9):2558. https://doi.org/10.3390/ijms19092558
Chicago/Turabian StyleZeiss, Dylan R., Msizi I. Mhlongo, Fidele Tugizimana, Paul A. Steenkamp, and Ian A. Dubery. 2018. "Comparative Metabolic Phenotyping of Tomato (Solanum lycopersicum) for the Identification of Metabolic Signatures in Cultivars Differing in Resistance to Ralstonia solanacearum" International Journal of Molecular Sciences 19, no. 9: 2558. https://doi.org/10.3390/ijms19092558
APA StyleZeiss, D. R., Mhlongo, M. I., Tugizimana, F., Steenkamp, P. A., & Dubery, I. A. (2018). Comparative Metabolic Phenotyping of Tomato (Solanum lycopersicum) for the Identification of Metabolic Signatures in Cultivars Differing in Resistance to Ralstonia solanacearum. International Journal of Molecular Sciences, 19(9), 2558. https://doi.org/10.3390/ijms19092558