rs495139 in the TYMS-ENOSF1 Region and Risk of Ovarian Carcinoma of Mucinous Histology
Abstract
1. Introduction
2. Results
2.1. Association Testing
2.2. Expression Quantitative Trait Locus (eQTL) Analysis
3. Discussion
4. Materials and Methods
4.1. Study Subjects and Genotyping
4.2. In Silico and eQTL Analysis
4.3. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| dTMP | Deoxythymidine monophosphate |
| dUMP | Deoxyuridine monophosphate |
| ENOSF1 | Enolase superfamily member 1 |
| eQTL | Expression Quantitative Trait Locus |
| GTEx | Genotype Tissue Expression project |
| iCOGS | international Collaborative Oncology Gene-environment Study |
| MOC | Mucinous ovarian carcinoma |
| mRNA | Messenger ribonucleic acid |
| OCAC | Ovarian Cancer Association Consortium |
| SNP | Single nucleotide polymorphism |
| TYMS | Thymidylate synthase |
| UTR | Untranslated region |
References
- Kelemen, L.E.; Kobel, M. Mucinous carcinomas of the ovary and colorectum: Different organ, same dilemma. Lancet Oncol. 2011, 12, 1071–1080. [Google Scholar] [CrossRef]
- Risch, H.A.; Marrett, L.D.; Jain, M.; Howe, G.R. Differences in risk factors for epithelial ovarian cancer by histologic type. Results of a case-control study. Am. J. Epidemiol. 1996, 144, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Rossing, M.A.; Cushing-Haugen, K.L.; Wicklund, K.G.; Weiss, N.S. Cigarette smoking and risk of epithelial ovarian cancer. Cancer Causes Control 2008, 19, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Faber, M.T.; Kjaer, S.K.; Dehlendorff, C.; Chang-Claude, J.; Andersen, K.K.; Hogdall, E.; Webb, P.M.; Jordan, S.J.; Rossing, M.A.; Doherty, J.A.; et al. Cigarette smoking and risk of ovarian cancer: A pooled analysis of 21 case-control studies. Cancer Causes Control 2013, 24, 989–1004. [Google Scholar] [CrossRef] [PubMed]
- Olsen, C.M.; Nagle, C.M.; Whiteman, D.C.; Ness, R.; Pearce, C.L.; Pike, M.C.; Rossing, M.A.; Terry, K.L.; Wu, A.H.; Australian Cancer, S.; et al. Obesity and risk of ovarian cancer subtypes: Evidence from the Ovarian Cancer Association Consortium. Endocr. Relat. Cancer 2013, 20, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Kelemen, L.E.; Lawrenson, K.; Tyrer, J.; Li, Q.; Lee, J.M.; Seo, J.H.; Phelan, C.M.; Beesley, J.; Chen, X.; Spindler, T.J.; et al. Genome-wide significant risk associations for mucinous ovarian carcinoma. Nat. Genet. 2015, 47, 888–897. [Google Scholar] [CrossRef] [PubMed]
- Phelan, C.M.; Kuchenbaecker, K.B.; Tyrer, J.P.; Kar, S.P.; Lawrenson, K.; Winham, S.J.; Dennis, J.; Pirie, A.; Riggan, M.J.; Chornokur, G.; et al. Identification of 12 new susceptibility loci for different histotypes of epithelial ovarian cancer. Nat. Genet. 2017, 49, 680–691. [Google Scholar] [CrossRef] [PubMed]
- Blount, B.C.; Mack, M.M.; Wehr, C.M.; MacGregor, J.T.; Hiatt, R.A.; Wang, G.; Wickramasinghe, S.N.; Everson, R.B.; Ames, B. Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: Implications for cancer and neuronal damage. Proc. Natl. Acad. Sci. USA 1997, 94, 3290–3295. [Google Scholar] [CrossRef] [PubMed]
- Kelemen, L.E.; Sellers, T.A.; Schildkraut, J.M.; Cunningham, J.M.; Vierkant, R.A.; Pankratz, V.S.; Fredericksen, Z.S.; Gadre, M.K.; Rider, D.N.; Liebow, M.; et al. Genetic variation in the one-carbon transfer pathway and ovarian cancer risk. Cancer Res. 2008, 68, 2498–2506. [Google Scholar] [CrossRef] [PubMed]
- Kelemen, L.E.; Goodman, M.T.; McGuire, V.; Rossing, M.A.; Webb, P.M.; Kobel, M.; Anton-Culver, H.; Beesley, J.; Berchuck, A.; Brar, S.; et al. Genetic variation in TYMS in the one-carbon transfer pathway is associated with ovarian carcinoma types in the Ovarian Cancer Association Consortium. Cancer Epidemiol. Biomark. Prev. 2010, 19, 1822–1830. [Google Scholar] [CrossRef] [PubMed]
- Dolnick, B.J. Cloning and characterization of a naturally occurring antisense RNA to human thymidylate synthase mRNA. Nucleic Acids Res. 1993, 21, 1747–1752. [Google Scholar] [CrossRef] [PubMed]
- Dolnick, B.J.; Black, A.R.; Winkler, P.M.; Schindler, K.; Hsueh, C.T. rTS gene expression is associated with altered cell sensitivity to thymidylate synthase inhibitors. Adv. Enzym. Regul. 1996, 36, 165–180. [Google Scholar] [CrossRef]
- Chu, J.; Dolnick, B.J. Natural antisense (rTSalpha) RNA induces site-specific cleavage of thymidylate synthase mRNA. Biochim. Biophys. Acta 2002, 1587, 183–193. [Google Scholar] [CrossRef]
- Vogelstein, B.; Fearon, E.R.; Hamilton, S.R.; Kern, S.E.; Preisinger, A.C.; Leppert, M.; Nakamura, Y.; White, R.; Smits, A.M.; Bos, J.L. Genetic alterations during colorectal-tumor development. N. Engl. J. Med. 1988, 319, 525–532. [Google Scholar] [CrossRef] [PubMed]
- Marquez, R.T.; Baggerly, K.A.; Patterson, A.P.; Liu, J.; Broaddus, R.; Frumovitz, M.; Atkinson, E.N.; Smith, D.I.; Hartmann, L.; Fishman, D.; et al. Patterns of gene expression in different histotypes of epithelial ovarian cancer correlate with those in normal fallopian tube, endometrium, and colon. Clin. Cancer Res. 2005, 11, 6116–6126. [Google Scholar] [CrossRef] [PubMed]
- Heinzelmann-Schwarz, V.A.; Gardiner-Garden, M.; Henshall, S.M.; Scurry, J.P.; Scolyer, R.A.; Smith, A.N.; Bali, A.; Vanden Bergh, P.; Baron-Hay, S.; Scott, C.; et al. A distinct molecular profile associated with mucinous epithelial ovarian cancer. Br. J. Cancer 2006, 94, 904–913. [Google Scholar] [CrossRef] [PubMed]
- Auersperg, N.; Wong, A.S.; Choi, K.C.; Kang, S.K.; Leung, P.C. Ovarian surface epithelium: Biology, endocrinology, and pathology. Endocr. Rev. 2001, 22, 255–288. [Google Scholar] [CrossRef] [PubMed]
- Tavassoulu, F.A.; Devilee, P. World Health Organization Classification of Tumors. Pathology and Genetics of Tumors of the Breast and Female Genital Organs; IARC Press: Lyon, France, 2003. [Google Scholar]
- Wang, K.K.; Sampliner, R.E. Updated guidelines 2008 for the diagnosis, surveillance and therapy of Barrett’s esophagus. Am. J. Gastroenterol. 2008, 103, 788–797. [Google Scholar] [CrossRef] [PubMed]
- Pharoah, P.D.; Tsai, Y.Y.; Ramus, S.J.; Phelan, C.M.; Goode, E.L.; Lawrenson, K.; Buckley, M.; Fridley, B.L.; Tyrer, J.P.; Shen, H.; et al. GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nat. Genet. 2013, 45, 362–370e2. [Google Scholar] [CrossRef] [PubMed]
- Sakoda, L.C.; Jorgenson, E.; Witte, J.S. Turning of COGS moves forward findings for hormonally mediated cancers. Nat. Genet. 2013, 45, 345–348. [Google Scholar] [CrossRef] [PubMed]
- GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 2015, 348, 648–660. [Google Scholar] [CrossRef] [PubMed]
- Konecny, G.E.; Wang, C.; Hamidi, H.; Winterhoff, B.; Kalli, K.R.; Dering, J.; Ginther, C.; Chen, H.W.; Dowdy, S.; Cliby, W.; et al. Prognostic and therapeutic relevance of molecular subtypes in high-grade serous ovarian cancer. J. Natl. Cancer Inst. 2014, 106. [Google Scholar] [CrossRef] [PubMed]
- Higgins, J.P.; Thompson, S.G. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002, 21, 1539–1558. [Google Scholar] [CrossRef] [PubMed]
- Hosmer, D.W.; Lemeshow, S.L. Applied Logistic Regression, 2nd ed.; John Wiley and Sons, Inc.: New York, NY, USA, 2000. [Google Scholar]
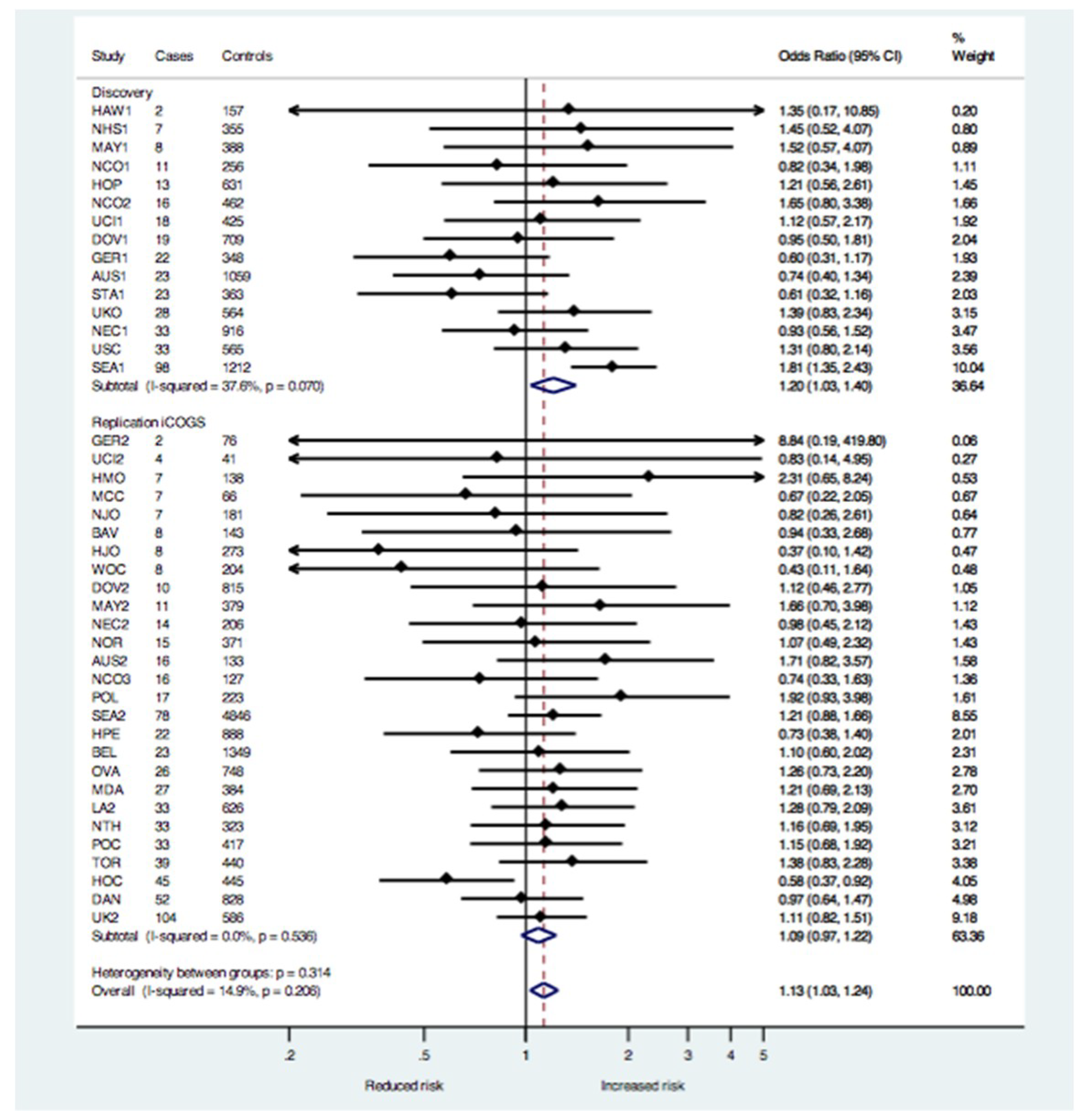
| Study Sample and Tumor Histology | Analysis Method | Cases, N | Controls, N | OR | 95% CI | p-Value |
|---|---|---|---|---|---|---|
| iCOGS sample | ||||||
| Mucinous invasive only | Meta-analysis | 665 2 | 15,256 2 | 1.09 | 0.97–1.22 | 0.16 |
| Discovery + iCOGS samples | ||||||
| Mucinous invasive only | Meta-analysis | 1019 2 | 23,666 2 | 1.13 | 1.03–1.24 | 0.01 |
| All invasive tumors 3 | Pooled | 15,000 | 24,351 | 1.00 | 0.97–1.03 | 0.84 |
| Mucinous invasive only | Pooled | 1021 2 | 24,351 | 1.12 | 1.02–1.22 | 0.02 |
| Mucinous borderline | Pooled | 621 | 24,351 | 0.97 | 0.86–1.09 | 0.59 |
| Mucinous invasive and borderline combined | Pooled | 1642 | 24,351 | 1.06 | 0.99–1.14 | 0.11 |
| Serous invasive | Pooled | 8889 | 24,351 | 1.01 | 0.98–1.05 | 0.53 |
| Endometrioid invasive | Pooled | 2164 | 24,351 | 0.97 | 0.91–1.04 | 0.40 |
| Clear cell invasive | Pooled | 1046 | 24,351 | 0.93 | 0.85–1.02 | 0.11 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kelemen, L.E.; Earp, M.; Fridley, B.L.; Chenevix-Trench, G.; On behalf of Australian Ovarian Cancer Study Group; Fasching, P.A.; Beckmann, M.W.; Ekici, A.B.; Hein, A.; Lambrechts, D.; et al. rs495139 in the TYMS-ENOSF1 Region and Risk of Ovarian Carcinoma of Mucinous Histology. Int. J. Mol. Sci. 2018, 19, 2473. https://doi.org/10.3390/ijms19092473
Kelemen LE, Earp M, Fridley BL, Chenevix-Trench G, On behalf of Australian Ovarian Cancer Study Group, Fasching PA, Beckmann MW, Ekici AB, Hein A, Lambrechts D, et al. rs495139 in the TYMS-ENOSF1 Region and Risk of Ovarian Carcinoma of Mucinous Histology. International Journal of Molecular Sciences. 2018; 19(9):2473. https://doi.org/10.3390/ijms19092473
Chicago/Turabian StyleKelemen, Linda E., Madalene Earp, Brooke L. Fridley, Georgia Chenevix-Trench, On behalf of Australian Ovarian Cancer Study Group, Peter A. Fasching, Matthias W. Beckmann, Arif B. Ekici, Alexander Hein, Diether Lambrechts, and et al. 2018. "rs495139 in the TYMS-ENOSF1 Region and Risk of Ovarian Carcinoma of Mucinous Histology" International Journal of Molecular Sciences 19, no. 9: 2473. https://doi.org/10.3390/ijms19092473
APA StyleKelemen, L. E., Earp, M., Fridley, B. L., Chenevix-Trench, G., On behalf of Australian Ovarian Cancer Study Group, Fasching, P. A., Beckmann, M. W., Ekici, A. B., Hein, A., Lambrechts, D., Lambrechts, S., Van Nieuwenhuysen, E., Vergote, I., Rossing, M. A., Doherty, J. A., Chang-Claude, J., Behrens, S., Moysich, K. B., Cannioto, R., ... Ovarian Cancer Association Consortium. (2018). rs495139 in the TYMS-ENOSF1 Region and Risk of Ovarian Carcinoma of Mucinous Histology. International Journal of Molecular Sciences, 19(9), 2473. https://doi.org/10.3390/ijms19092473












