Comparative Proteomic Analysis of Two Ralstonia solanacearum Isolates Differing in Aggressiveness
Abstract
1. Introduction
2. Results
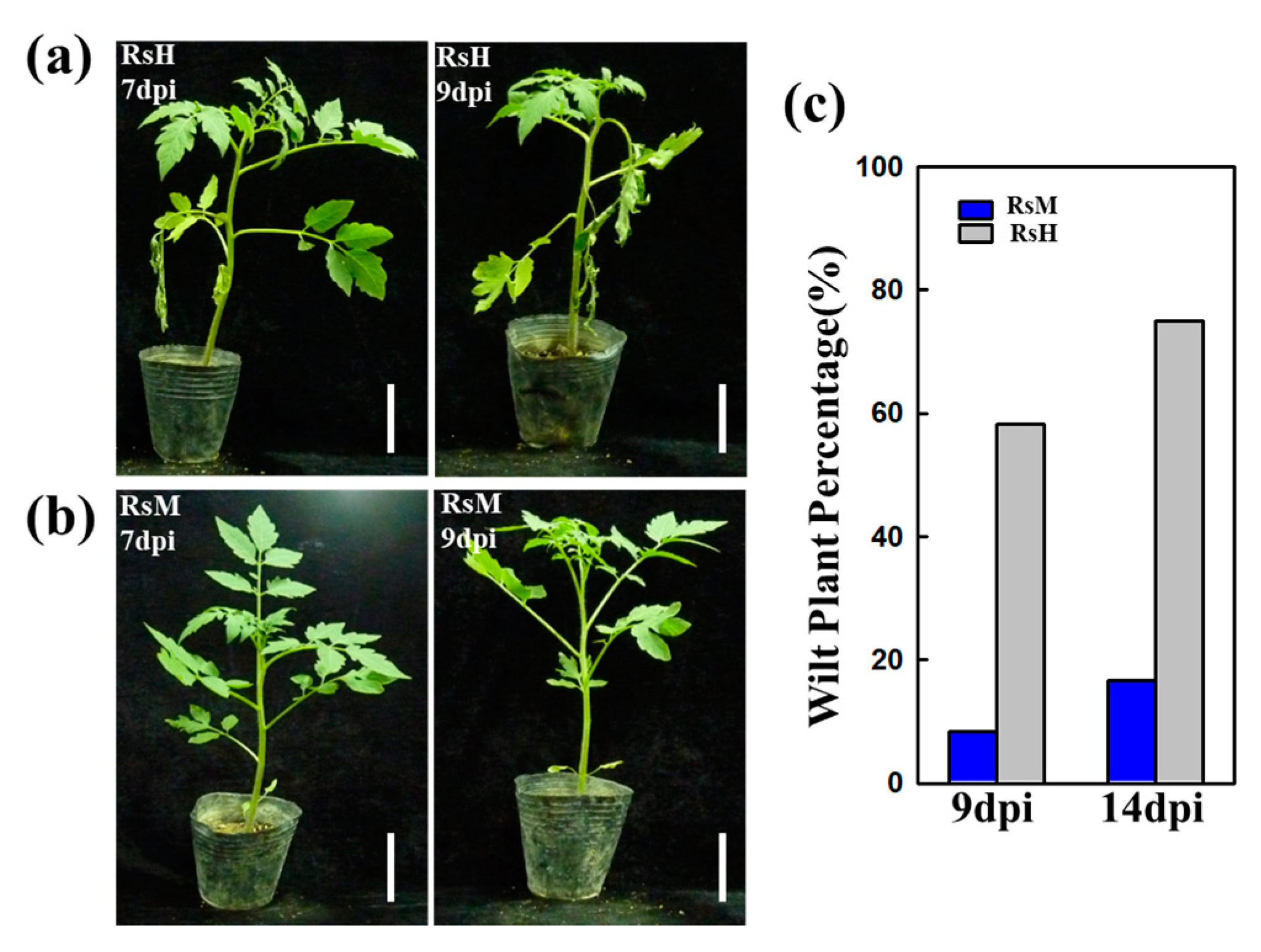
2.1. Evaluation of RsM and RsH Infection in the Hawaii 7996 Cultivar
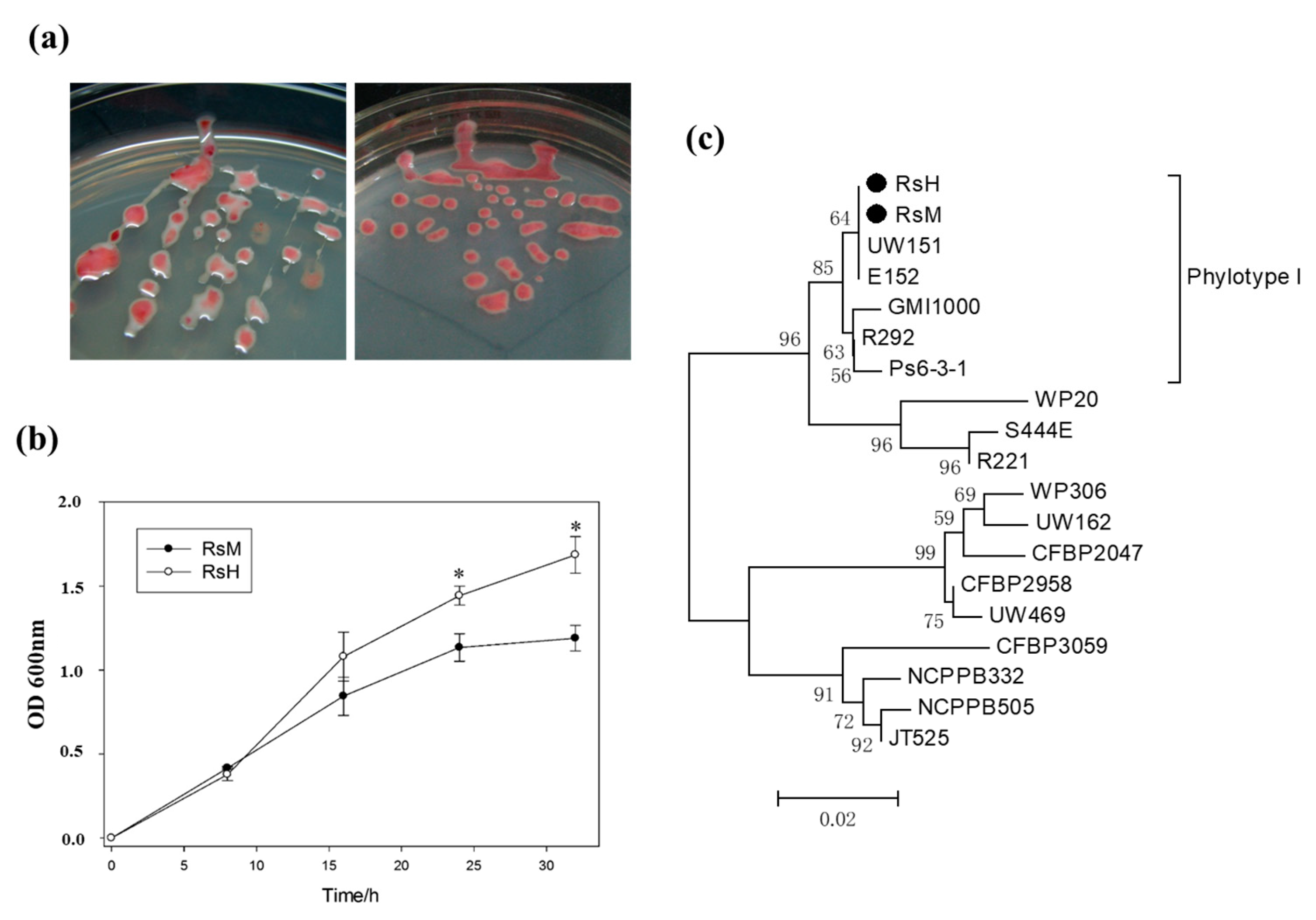
2.2. Characteristics of Two R. solanacearum Strains: RsM and RsH
2.3. Phylotype Identification of RsM and RsH
2.4. Proteome Analysis of the Two R. solanacearum Strains RsM and RsH
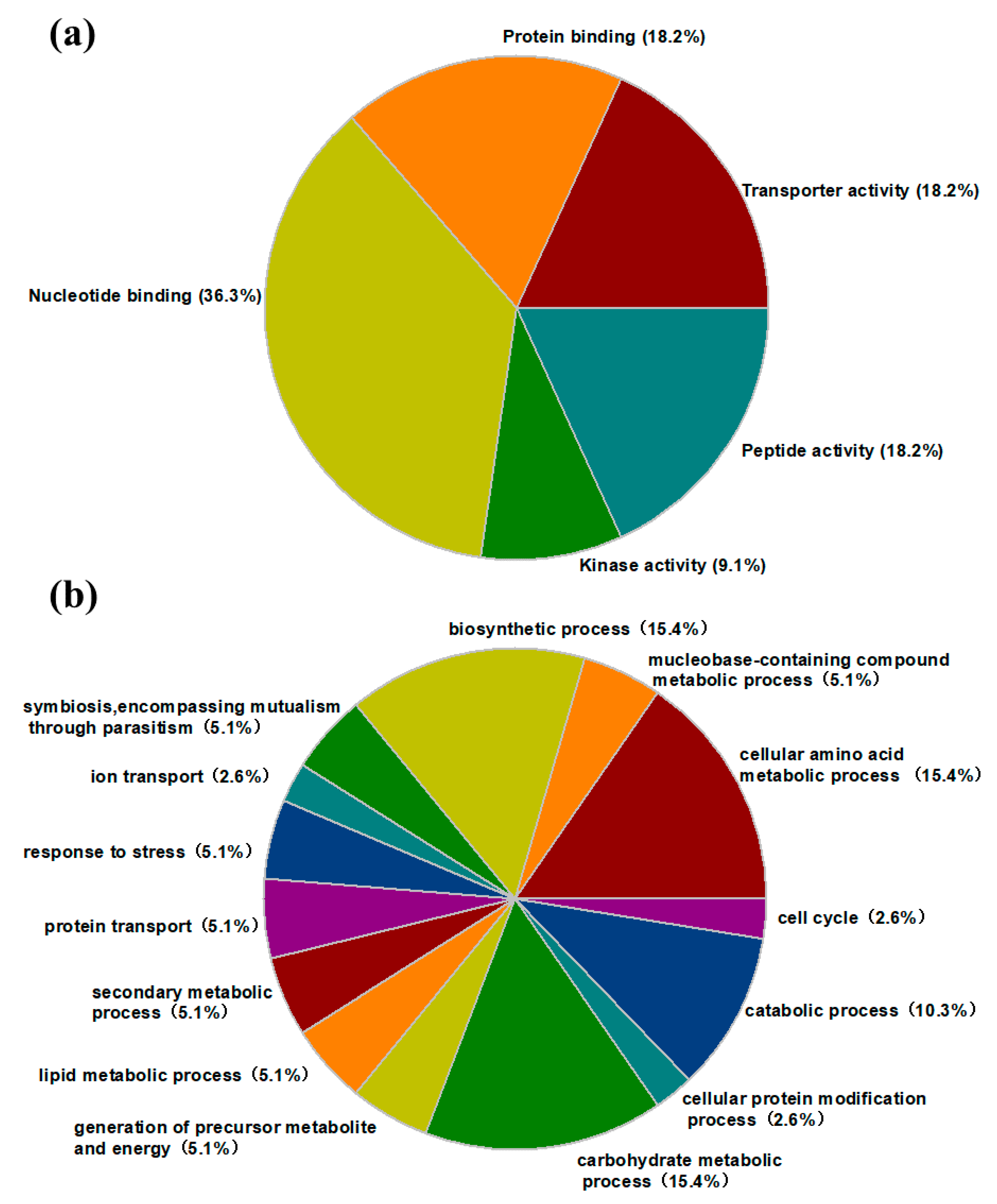
2.5. Protein Identification and Gene Ontology (GO) Annotation
2.6. Abundance Patterns of the Differential Proteins in RsM and RsH in CPZ and Min Medium
3. Discussion
3.1. RsM and RsH Exhibited Different Virulence
3.2. EPS as a Potential Factor Accounting for R. solanacearum Virulence
3.3. Isocitrate Lyase (ICL) is More Abundant in RsH
3.4. Membrane Proteins Possibly Play a Positive Role in Wilt Susceptibility in Tomatoes
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
4.2. RsM and RsH Infection
4.3. Phylotype Identification
4.4. Protein Extraction from RsM and RsH
4.6. Gel Imaging and Data Analysis
4.7. MALDI-TOF/MS-MS Identification of Differential Protein Spots
4.8. Differential Proteins Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hikichi, Y.; Yoshimochi, T.; Tsujimoto, S.; Shinohara, R.; Nakaho, K.; Kanda, A.; Kiba, A.; Ohnishi, K. Global regulation of pathogenicity mechanism of Ralstonia solanacearum. Plant Biotechnol. 2007, 24, 149–154. [Google Scholar] [CrossRef]
- Janse, J.D.; Van den Beld, H.E.; Elphinstone, J.; Simpkins, S.; Tjou-Tam-Sin, N.N.A.; Van Vaerenbergh, J. Introduction to Europe of Ralstonia solanacearum biovar 2, race 3 in Pelargonium zonale cuttings. J. Plant Pathol. 2004, 86, 147–155. [Google Scholar]
- Huet, G. Breeding for resistances to Ralstonia solanacearum. Front. Plant Sci. 2014, 5, 715. [Google Scholar] [CrossRef] [PubMed]
- Mansfield, J.; Genin, S.; Magori, S.; Citovsky, V.; Sriariyanum, M.; Ronald, P.; Dow, M.; Verdier, V.; Beer, S.V.; Machado, M.A.; et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 614–629. [Google Scholar] [CrossRef] [PubMed]
- Buddenhagen, I.W.; Kelman, A. Biological and physiological aspects of bacterial wilt caused by Pseudomonas solanacearum. Annu. Rev. Phytopathol. 1964, 2, 203–230. [Google Scholar] [CrossRef]
- Elphinstone, J.G. The current bacterial wilt situation: A global overview. Bacterial wilt disease and the Ralstonia solanacearum species complex. APS Press 2005, 1, 9–28. [Google Scholar]
- Wang, J.F.; Ho, F.I.; Truong, H.T.H.; Huang, S.M.; Balatero, C.H.; Dittapongpitch, V.; Hidayati, N. Identification of major QTLs associated with stable resistance of tomato cultivar ‘Hawaii 7996’ to Ralstonia solanacearum. Euphytica 2013, 190, 241–252. [Google Scholar] [CrossRef]
- Thoquet, P.; Olivier, J.; Sperisen, C.; Rogowsky, P.; Prior, P.; Anais, G.; Manhin, B.; Bazin, B.; Nazer, R.; Grimsley, N. Polygenic resistance of tomato plants to bacterial wilt in the French West Indies. Mol. Plant Microbe Interact. 1996, 9, 837. [Google Scholar] [CrossRef]
- Wang, J.F.; Olivier, J.; Thoquet, P.; Mangin, B.; Sauviac, L.; Grimsley, N.H. Resistance of tomato line Hawaii7996 to Ralstonia solanacearum Pss4 in Taiwan is controlled mainly by a major strain-specific locus. Mol. Plant Microbe Interact. 2010, 13, 6–13. [Google Scholar] [CrossRef] [PubMed]
- Carmeille, A.; Caranta, C.; Dintinger, J.; Prior, P.; Luisetti, J.; Besse, P. Identification of QTLs for Ralstonia solanacearum race 3-phylotype II resistance in tomato. Theor. Appl. Genet. 2006, 113, 110–121. [Google Scholar] [CrossRef] [PubMed]
- Böhmer, M.; Colby, T.; Böhmer, C.; Bräutigam, A.; Schmidt, J.; Bölker, M. Proteomic analysis of dimorphic transition in the phytopathogenic fungus Ustilago maydis. Proteomics 2007, 7, 675–685. [Google Scholar] [CrossRef] [PubMed]
- González-Fernández, R.; Aloria, K.; Valero-Galván, J.; Redondo, I.; Arizmendi, J.M.; Jorrín-Novo, J.V. Proteomic analysis of mycelium and secretome of different Botrytis cinerea wild-type strains. J. Proteom. 2014, 97, 195–221. [Google Scholar] [CrossRef] [PubMed]
- Pasquali, M.; Serchi, T.; Cocco, E.; Leclercq, C.C.; Planchon, S.; Guignard, C.; Renaut, J.; Hoffmann, L. A Fusarium graminearum strain-comparative proteomic approach identifies regulatory changes triggered by agmatine. J. Proteom. 2016, 137, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Lonjon, F.; Turner, M.; Henry, C.; Rengel, D.; Lohou, D.; van de Kerkhove, Q.; Cazale, A.C.; Peeters, N.; Genin, S.; Vailleau, F. Comparative secretome analysis of Ralstonia solanacearum Type 3 secretion-associated mutants reveals a fine control of effector delivery, essential for bacterial pathogenicity. Mol. Cell. Proteom. 2015, 15, 598. [Google Scholar] [CrossRef] [PubMed]
- Galán, J.E.; Wolf-Watz, H. Protein delivery into eukaryotic cells by type III secretion machines. Nature 2006, 444, 567–573. [Google Scholar] [CrossRef] [PubMed]
- Hauser, A.R. The type III secretion system of Pseudomonas aeruginosa: Infection by injection. Nat. Rev. Microbiol. 2009, 7, 654–665. [Google Scholar] [CrossRef] [PubMed]
- Jha, G.; Rajeshwari, R.; Sonti, R.V. Bacterial type two secretion system secreted proteins: Double-edged swords for plant pathogens. Mol. Plant Microbe Interact. 2005, 18, 891–898. [Google Scholar] [CrossRef] [PubMed]
- Korotkov, K.V.; Sandkvist, M.; Hol, W.G. The type II secretion system: Biogenesis, molecular architecture and mechanism. Nat. Rev. Microbiol. 2012, 10, 336–351. [Google Scholar] [CrossRef] [PubMed]
- Nivaskumar, M.; Francetic, O. Type II secretion system: A magic beanstalk or a protein escalator. BBA Mol. Cell Res. 2014, 1843, 1568–1577. [Google Scholar] [CrossRef] [PubMed]
- Grimault, V.; Prior, P.; Anais, G. A monogenic dominant resistance of tomato to bacterial wilt in Hawaii 7996 is associated with plant colonization by Pseudomonas solanacearum. J. Phytopathol. 1995, 143, 349–352. [Google Scholar] [CrossRef]
- Villa, J.E.; Tsuchiya, K.; Horita, M.; Natural, M.; Opina, N.; Hyakumachi, M. Phylogenetic relationships of Ralstonia solanacearum species complex strains from Asia and other continents based on 16S rDNA, endoglucanase, and h pB gene sequences. J. Gen. Plant Pathol. 2005, 71, 39–46. [Google Scholar] [CrossRef]
- Fegan, M.; Prior, P. How Complex Is the Ralstonia solanacearum Species Comple; APS Press: St. Paul, MN, USA, 2005; pp. 449–461. [Google Scholar]
- Cook, D.; Barlow, E.; Sequeira, L. Genetic diversity of Pseudomonas solanacearum: Detection of restriction fragment length polymorphisms with DNA probes that specify virulence and the hypersensitive response. Mol. Plant Microbe Interact. 1989, 1, 113–121. [Google Scholar] [CrossRef]
- Taghavi, M.; Hayward, C.; Sly, L.I.; Fegan, M. Analysis of the phylogenetic relationships of strains of Burkholderia solanacearum, Pseudomonas syzygii, and the blood disease bacterium of banana based on 16S rRNA gene sequences. Int. J. Syst. Evol. Microbiol. 1996, 46, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Poussier, S.; Prior, P.; Luisetti, J.; Hayward, C.; Fegan, M. Partial sequencing of the hpB and endoglucanase genes confirms and expands the known diversity within the Ralstonia solanacearum species complex. Syst. Appl. Microbiol. 2000, 23, 479–486. [Google Scholar] [CrossRef]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- McLoon, A.L.; Guttenplan, S.B.; Kearns, D.B.; Kolter, R.; Losick, R. Tracing the domestication of a biofilm-forming bacterium. J. Bacteriol. 2011, 193, 2027–2034. [Google Scholar] [CrossRef] [PubMed]
- Orgambide, G.; Montrozier, H.; Servin, P.; Roussel, J.; Trigalet-Demery, D.; Trigalet, A. High heterogeneity of the exopolysaccharides of Pseudomonas solanacearum strain GMI 1000 and the complete structure of the major polysaccharide. J. Biol. Chem. 1991, 266, 8312–8321. [Google Scholar] [PubMed]
- McGarvey, J.A.; Denny, T.P.; Schell, M.A. Spatial-temporal and quantitative analysis of growth and EPS I production by Ralstonia solanacearum in resistant and susceptible tomato cultivars. Phytopathology 1999, 89, 1233–1239. [Google Scholar] [CrossRef] [PubMed]
- Milling, A.; Babujee, L.; Allen, C. Ralstonia solanacearum extracellular polysaccharide is a specific elicitor of defense responses in wilt-resistant tomato plants. PLoS ONE 2011, 6, e15853. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, M.C.; Fink, G.R. The glyoxylate cycle is required for fungal virulence. Nature 2001, 412, 83–86. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, M.A.; Lorenz, M.C. Mutations in alternative carbon utilization pathways in Candida albicans attenuate virulence and confer pleiotropic phenotypes. Eukaryot. Cell 2007, 6, 280–290. [Google Scholar] [CrossRef] [PubMed]
- Piekarska, K.; Hardy, G.; Mol, E.; van den Burg, J.; Strijbis, K.; van Roermund, C.; van den Berg, M.; Distel, B. The activity of the glyoxylate cycle in peroxisomes of Candida albicans depends on a functional β-oxidation pathway: Evidence for reduced metabolite transport across the peroxisomal membrane. Microbiology 2008, 154, 3061–3072. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Y.; Thornton, C.R.; Kershaw, M.J.; Debao, L.; Talbot, N.J. The glyoxylate cycle is required for temporal regulation of virulence by the plant pathogenic fungus Magnaporthe grisea. Mol. Microbiol. 2003, 47, 1601–1612. [Google Scholar] [CrossRef] [PubMed]
- Brock, M. Fungal metabolism in host niches. Curr. Opin. Microbiol. 2009, 12, 371–376. [Google Scholar] [CrossRef] [PubMed]
- Vereecke, D.; Cornelis, K.; Temmerman, W.; Jaziri, M.; Van Montagu, M.; Holsters, M.; Goethals, K. Chomosomal locus that affects pathogenicity of Rhodococcus fascians. J. Bacteriol. 2002, 184, 1112–1120. [Google Scholar] [CrossRef] [PubMed]
- Idnurm, A.; Howlett, B.J. Isocitrate lyase is essential for pathogenicity of the fungus Leptosphaeria maculans to canola (Brassica napus). Eukaryot. Cell 2002, 1, 719–724. [Google Scholar] [CrossRef] [PubMed]
- Bocsanczy, A.M.; Achenbach, U.C.; Mangravita-Novo, A.; Chow, M.; Norman, D.J. Proteomic comparison of Ralstonia solanacearum strains reveals temperature dependent virulence factors. BMC Genom. 2014, 15, 280. [Google Scholar] [CrossRef] [PubMed]
- Dorman, C.J.; Chatfield, S.T.E.V.E.N.; Higgins, C.F.; Hayward, C.L.I.F.F.; Dougan, G. Characterization of porin and ompR mutants of a virulent strain of Salmonella typhimurium: OmpR mutants are attenuated in vivo. Infect. Immun. 1989, 57, 2136–2140. [Google Scholar] [PubMed]
- Bernardini, M.L.; Sanna, M.G.; Fontaine, A.; Sansonetti, P.J. OmpC is involved in invasion of epithelial cells by Shigella flexneri. Infect. Immun. 1993, 61, 3625–3635. [Google Scholar] [PubMed]
- Provenzano, D.; Klose, K.E. Altered expression of the ToxR-regulated porins OmpU and OmpT diminishes Vibrio cholerae bile resistance, virulence factor expression, and intestinal colonization. Proc. Natl. Acad. Sci. USA 2000, 97, 10220–10224. [Google Scholar] [CrossRef] [PubMed]
- Achouak, W.; Heulin, T. Multiple facets of bacterial porins. FEMS Microbiol. Lett. 2001, 199, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Kawakatsu, T.; Taramino, G.; Itoh, J.I.; Allen, J.; Sato, Y.; Hong, S.K.; Yule, R.; Nagasawa, N.; Kojima, M.; Kusaba, M.; et al. PLASTOCH ON3/GOLIATH encodes a glutamate carboxypeptidase required for proper development in rice. Plant J. 2009, 58, 1028–1040. [Google Scholar] [CrossRef] [PubMed]
- Xue, Q.Y.; Chen, Y.; Li, S.M.; Chen, L.F.; Ding, G.C.; Guo, D.W.; Guo, J.H. Evaluation of the strains of Acinetobacter and Enterobacter as potential biocontrol agents against Ralstonia wilt of tomato. Biol. Control 2009, 48, 252–258. [Google Scholar] [CrossRef]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Scali, M.; Vignani, R.; Spadafora, A.; Sensi, E.; Mazzuca, S.; Cresti, M. Protein extraction for two-dimensional electrophoresis from olive leaf, a plant tissue containing high levels of interfering compounds. Electrophoresis 2003, 24, 2369–2375. [Google Scholar] [CrossRef] [PubMed]
- Boucher, C.A.; Barberis, P.A.; Demery, D.A. Transposon mutagenesis of Pseudomonas solanacearum: Isolation of Tn5-induced avirulent mutants. Microbiology 1985, 131, 2449–2457. [Google Scholar] [CrossRef]
- Wang, W.; Vignani, R.; Scali, M.; Cresti, M. A universal and rapid protocol for protein extraction from recalcitrant plant tissues for proteomic analysis. Electrophoresis 2006, 27, 2782–2786. [Google Scholar] [CrossRef] [PubMed]
- Valledor, L.; Jorrín, J. Back to the basics: Maximizing the information obtained by quantitative two dimensional gel electrophoresis analyses by an appropriate experimental design and statistical analyses. J. Proteom. 2011, 74, 1–18. [Google Scholar] [CrossRef] [PubMed]


| Spot Number | Accession | Protein Name | Score | Theoretical Mr(kDa)/pI | Observed Mr(kDa)/pI | Organism | Protein Sequence Coverage |
|---|---|---|---|---|---|---|---|
| cluster 1 | |||||||
| AA-1 | 17546429 | trigger factor | 419 | 50.093/5.17 | 51.79/5.05 | R. solanacearum GMI1000 | 17% |
| AA-4 | 469776741 | Heat shock protein 60 family chaperone GroEL | 172 | 57.231/5.21 | 56.79/5.14 | R. solanacearum FQY_4 | 6% |
| AA-10 | 17549499 | sugar kinase | 503 | 32.734/5.57 | 32.67/5.77 | R. solanacearum GMI1000 | 30% |
| AA-13 | 17547209 | isocitrate dehydrogenase | 376 | 45.827/5.65 | 45.45/5.98 | R. solanacearum GMI1000 | 14% |
| AA-15 | 17548395 | methyltransferase | 412 | 31.336/5.53 | 29.34/6.05 | R. solanacearum GMI1000 | 23% |
| AA-16 | 17544951 | hypothetical protein RSc0232 | 352 | 30.243/6.1 | 30.3/6.1 | R. solanacearum GMI1000 | 22% |
| AA-18 | 17546077 | isocitrate lyase | 115 | 49.094/6.06 | 49.87/6.2 | R. solanacearum GMI1000 | 6% |
| AA-19 | 17549306 | secretion ATP | 499 | 49.500/5.91 | 49.67/6.32 | R. solanacearum GMI1000 | 18% |
| AA-37 | 17546445 | hypothetical protein RSc1726 | 631 | 22.449/6.29 | 22.07/7.01 | R. solanacearum GMI1000 | 56% |
| cluster 2 | |||||||
| AA-21 | 17544951 | hypothetical protein RSc0232 | 848 | 30.243/6.1 | 29.75/6.27 | R. solanacearum GMI1000 | 43% |
| AA-24 | 17549238 | udp-n-acetylglucosamine 2-epimerase | 599 | 41.656/5.95 | 38.3/6.38 | R. solanacearum GMI1000 | 25% |
| AA-28 | 17548464 | hemin transport protein | 362 | 41.267/6.23 | 40.7/6.48 | R. solanacearum GMI1000 | 16% |
| AA-34 | 17546446 | hypothetical protein RSc1727 | 350 | 20.751/6.15 | 20.46/6.64 | R. solanacearum GMI1000 | 29% |
| AA-35 | 17545056 | polyphenol oxidase B | 454 | 54.474/7.31 | 50.52/7.04 | R. solanacearum GMI1000 | 14% |
| AA-39 | 17549780 | hypothetical protein RS02117 | 996 | 16.136/6.74 | 17.48/6.95 | R. solanacearum GMI1000 | 66% |
| AA-42 | 17545056 | polyphenol oxidase B | 152 | 54.474/7.31 | 50.52/7.3 | R. solanacearum GMI1000 | 7% |
| cluster 3 | |||||||
| AA-41 | 17547209 | isocitrate dehydrogenase | 231 | 45.827/5.65 | 46.39/5.6 | R. solanacearum GMI1000 | 11% |
| AB-4 | 17548639 | ornithine cyclodeamin | 896 | 37.988/5.53 | 37.65/5.66 | R. solanacearum GMI1000 | 36% |
| AB-7 | 17545135 | hypothetical protein RSc0416 | 168 | 20.629/5.48 | 24.81/5.72 | R. solanacearum GMI1000 | 12% |
| cluster 4 | |||||||
| CB-9 | 17549238 | udp-n-acetylglucosamine 2-epimerase | 599 | 41.656/5.95 | 38.4/6.28 | R. solanacearum GMI1000 | 27% |
| CB-14 | 17548494 | glutamate carboxypeptidase | 202 | 41.526/6.42 | 40.47/6.62 | R. solanacearum GMI1000 | 13% |
| CB-15 | 17547574 | porin | 305 | 41.269/8.46 | 38/6.6 | R. solanacearum GMI1000 | 10% |
| CB-17 | 4.7E+08 | putative diaminopimelate decarboxylase protein | 265 | 45.900/6.47 | 46.98/6.98 | R. solanacearum FQY_4 | 13% |
| CA-25 | 17545414 | porin | 138 | 51.791/8.54 | 49/7.2 | R. solanacearum GMI1000 | 8% |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, G.; Kong, J.; Cui, D.; Zhao, H.; Zhao, P.; Feng, S.; Zhao, Y.; Wang, W. Comparative Proteomic Analysis of Two Ralstonia solanacearum Isolates Differing in Aggressiveness. Int. J. Mol. Sci. 2018, 19, 2444. https://doi.org/10.3390/ijms19082444
Wang G, Kong J, Cui D, Zhao H, Zhao P, Feng S, Zhao Y, Wang W. Comparative Proteomic Analysis of Two Ralstonia solanacearum Isolates Differing in Aggressiveness. International Journal of Molecular Sciences. 2018; 19(8):2444. https://doi.org/10.3390/ijms19082444
Chicago/Turabian StyleWang, Guoping, Jie Kong, Dandan Cui, Hongbo Zhao, Puyan Zhao, Shujie Feng, Yahua Zhao, and Wenyi Wang. 2018. "Comparative Proteomic Analysis of Two Ralstonia solanacearum Isolates Differing in Aggressiveness" International Journal of Molecular Sciences 19, no. 8: 2444. https://doi.org/10.3390/ijms19082444
APA StyleWang, G., Kong, J., Cui, D., Zhao, H., Zhao, P., Feng, S., Zhao, Y., & Wang, W. (2018). Comparative Proteomic Analysis of Two Ralstonia solanacearum Isolates Differing in Aggressiveness. International Journal of Molecular Sciences, 19(8), 2444. https://doi.org/10.3390/ijms19082444




