Arabidopsis Heat Stress-Induced Proteins Are Enriched in Electrostatically Charged Amino Acids and Intrinsically Disordered Regions
Abstract
1. Introduction
2. Results
2.1. Proteins That Are Overexpressed at High Temperatures Are Enriched in Electrostatically Charged Amino Acids and Depleted in Polar and Hydrophobic Amino Acids
2.2. The Amino Acid Composition of Heat-Induced Proteins Is Not due to Covariation of Amino Acid Composition with GC Content, Gene Expression Levels, or Subcellular Location
2.3. Proteins That Are Overexpressed at High Temperatures Are Highly Disordered
3. Discussion
4. Materials and Methods
4.1. Plant Material, Growth Conditions, and Experimental Design
4.2. Microarray Analysis
4.3. Gene Overexpression/Repression Analysis
4.4. Protein and Gene Sequence Analysis
4.5. Prediction of Protein Intrinsic Disorder
4.6. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| IDR | Intrinsically Disordered Region |
| MS | Murashige and Skoog |
| TAIR | The Arabidopsis Information Resource |
| BLAST | Basic Local Alignment Search Tool |
References
- Karshikoff, A.; Ladenstein, R. Ion pairs and the thermotolerance of proteins from hyperthermophiles: A ‘traffic rule’ for hot roads. Trends Biochem. Sci. 2001, 26, 550–557. [Google Scholar] [CrossRef]
- Strop, P.; Mayo, S.L. Contribution of surface salt bridges to protein stability. Biochemistry 2000, 39, 1251–1255. [Google Scholar] [CrossRef] [PubMed]
- Perutz, M.; Raidt, H. Stereochemical basis of heat stability in bacterial ferredoxins and in haemoglobin A2. Nature 1975, 255, 256–259. [Google Scholar] [CrossRef] [PubMed]
- Argos, P.; Rossmann, M.G.; Grau, U.M.; Zuber, H.; Frank, G.; Tratschin, J.D. Thermal stability and protein structure. Biochemistry 1979, 18, 5698–5703. [Google Scholar] [CrossRef] [PubMed]
- Beeby, M.; D O’Connor, B.; Ryttersgaard, C.; Boutz, D.R.; Perry, L.J.; Yeates, T.O. The genomics of disulfide bonding and protein stabilization in thermophiles. PLoS Biol. 2005, 3, e309. [Google Scholar] [CrossRef] [PubMed]
- Farias, S.T.; Bonato, M. Preferred amino acids and thermostability. Genet. Mol. Res. 2003, 2, 383–393. [Google Scholar] [PubMed]
- Haney, P.J.; Badger, J.H.; Buldak, G.L.; Reich, C.I.; Woese, C.R.; Olsen, G.J. Thermal adaptation analyzed by comparison of protein sequences from mesophilic and extremely thermophilic Methanococcus species. Proc. Natl. Acad. Sci. USA 1999, 96, 3578–3583. [Google Scholar] [CrossRef] [PubMed]
- Kreil, D.P.; Ouzounis, C.A. Identification of thermophilic species by the amino acid compositions deduced from their genomes. Nucleic Acids Res. 2001, 29, 1608–1615. [Google Scholar] [CrossRef] [PubMed]
- Tekaia, F.; Yeramian, E.; Dujon, B. Amino acid composition of genomes, lifestyles of organisms, and evolutionary trends: A global picture with correspondence analysis. Gene 2002, 297, 51–60. [Google Scholar] [CrossRef]
- Zeldovich, K.B.; Berezovsky, I.N.; Shakhnovich, E.I. Protein and DNA sequence determinants of thermophilic adaptation. PLoS Comput. Biol. 2007, 3, e5. [Google Scholar] [CrossRef] [PubMed]
- Chakravarty, S.; Varadarajan, R. Elucidation of determinants of protein stability through genome sequence analysis. FEBS Lett. 2000, 470, 65–69. [Google Scholar] [CrossRef]
- Cambillau, C.; Claverie, J.-M. Structural and genomic correlates of hyperthermostability. J. Biol. Chem. 2000, 275, 32383–32386. [Google Scholar] [CrossRef] [PubMed]
- Burra, P.V.; Kalmar, L.; Tompa, P. Reduction in structural disorder and functional complexity in the thermal adaptation of prokaryotes. PLoS ONE 2010, 5, e12069. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, Y.; Cao, Z.; Li, Z.; Zhao, H.; Zhou, Y. The role of semidisorder in temperature adaptation of bacterial FlgM proteins. Biophys. J. 2013, 105, 2598–2605. [Google Scholar] [CrossRef] [PubMed]
- Vicedo, E.; Schlessinger, A.; Rost, B. Environmental pressure may change the composition protein disorder in prokaryotes. PLoS ONE 2015, 10, e0133990. [Google Scholar] [CrossRef] [PubMed]
- Galea, C.A.; High, A.A.; Obenauer, J.C.; Mishra, A.; Park, C.-G.; Punta, M.; Schlessinger, A.; Ma, J.; Rost, B.; Slaughter, C.A. Large-scale analysis of thermostable, mammalian proteins provides insights into the intrinsically disordered proteome. J. Proteome Res. 2008, 8, 211–226. [Google Scholar] [CrossRef] [PubMed]
- Tsvetkov, P.; Myers, N.; Moscovitz, O.; Sharon, M.; Prilusky, J.; Shaul, Y. Thermo-resistant intrinsically disordered proteins are efficient 20S proteasome substrates. Mol. Biosyst. 2012, 8, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Galea, C.A.; Nourse, A.; Wang, Y.; Sivakolundu, S.G.; Heller, W.T.; Kriwacki, R.W. Role of intrinsic flexibility in signal transduction mediated by the cell cycle regulator, p27Kip1. J. Mol. Biol. 2008, 376, 827–838. [Google Scholar] [CrossRef] [PubMed]
- Van Noort, V.; Bradatsch, B.; Arumugam, M.; Amlacher, S.; Bange, G.; Creevey, C.; Falk, S.; Mende, D.R.; Sinning, I.; Hurt, E. Consistent mutational paths predict eukaryotic thermostability. BMC Evol. Biol. 2013, 13, 7. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.-Z.; Lercher, M.J. Amino acid composition in endothermic vertebrates is biased in the same direction as in thermophilic prokaryotes. BMC Evol. Biol. 2010, 10, 263. [Google Scholar] [CrossRef] [PubMed]
- Windisch, H.S.; Lucassen, M.; Frickenhaus, S. Evolutionary force in confamiliar marine vertebrates of different temperature realms: Adaptive trends in zoarcid fish transcriptomes. BMC Genom. 2012, 13, 549. [Google Scholar] [CrossRef] [PubMed]
- Albanese, V.; Yam, A.Y.-W.; Baughman, J.; Parnot, C.; Frydman, J. Systems analyses reveal two chaperone networks with distinct functions in eukaryotic cells. Cell 2006, 124, 75–88. [Google Scholar] [CrossRef] [PubMed]
- Berry, J.; Bjorkman, O. Photosynthetic response and adaptation to temperature in higher plants. Annu. Rev. Plant Physiol. 1980, 31, 491–543. [Google Scholar] [CrossRef]
- Sueoka, N. Correlation between base composition of deoxyribonucleic acid and amino acid composition of protein. Proc. Natl. Acad. Sci. USA 1961, 47, 1141–1149. [Google Scholar] [CrossRef] [PubMed]
- Cherry, J.L. Highly expressed and slowly evolving proteins share compositional properties with thermophilic proteins. Mol. Biol. Evol. 2010, 27, 735–741. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, H.; Nishikawa, K. The amino acid composition is different between the cytoplasmic and extracellular sides in membrane proteins. FEBS Lett. 1992, 303, 141–146. [Google Scholar] [PubMed]
- Nakashima, H.; Nishikawa, K. Discrimination of intracellular and extracellular proteins using amino acid composition and residue-pair frequencies. J. Mol. Biol. 1994, 238, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Dosztanyi, Z.; Csizmok, V.; Tompa, P.; Simon, I. IUPred: Web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 2005, 21, 3433–3434. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Uversky, V.N.; Kurgan, L. Genes encoding intrinsic disorder in eukaryota have high GC content. Intrinsically Disord. Proteins 2016, 4, e1262225. [Google Scholar] [CrossRef] [PubMed]
- Yruela, I.; Contreras-Moreira, B. Genetic recombination is associated with intrinsic disorder in plant proteomes. BMC Genom. 2013, 14, 772. [Google Scholar] [CrossRef] [PubMed]
- Paliy, O.; Gargac, S.M.; Cheng, Y.; Uversky, V.N.; Dunker, A.K. Protein disorder is positively correlated with gene expression in Escherichia coli. J. Proteome Res. 2008, 7, 2234–2245. [Google Scholar] [CrossRef] [PubMed]
- Singh, G.P.; Dash, D. How expression level influences the disorderness of proteins. Biochem. Biophys. Res. Commun. 2008, 371, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.R.; Liao, B.Y.; Zhuang, S.M.; Zhang, J. Protein misinteraction avoidance causes highly expressed proteins to evolve slowly. Proc. Natl. Acad. Sci. USA 2012, 109, E831–840. [Google Scholar] [CrossRef] [PubMed]
- Hendsch, Z.S.; Tidor, B. Do salt bridges stabilize proteins? A continuum electrostatic analysis. Protein Sci. 1994, 3, 211–226. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.-X.; Wang, Y.-B.; Pan, Y.-J.; Li, W.-F. Differences in amino acids composition and coupling patterns between mesophilic and thermophilic proteins. Amino Acids 2008, 34, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Catanzano, F.; Barone, G.; Graziano, G.; Capasso, S. Thermodynamic analysis of the effect of selective monodeamidation at asparagine 67 in ribonuclease a. Protein Sci. 1997, 6, 1682–1693. [Google Scholar] [CrossRef] [PubMed]
- Charlesworth, B. Fundamental concepts in genetics: Effective population size and patterns of molecular evolution and variation. Nat. Rev. Genet. 2009, 10, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Bolser, D.; Staines, D.M.; Pritchard, E.; Kersey, P. Ensembl Plants: Integrating tools for visualizing, mining, and analyzing plant genomics data. Methods Mol. Biol. 2016, 1374, 115–140. [Google Scholar] [PubMed]
- Kasprzyk, A.; Keefe, D.; Smedley, D.; London, D.; Spooner, W.; Melsopp, C.; Hammond, M.; Rocca-Serra, P.; Cox, T.; Birney, E. EnsMart: A generic system for fast and flexible access to biological data. Genome Res. 2004, 14, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Hooper, C.M.; Castleden, I.R.; Tanz, S.K.; Aryamanesh, N.; Millar, A.H. SUBA4: The interactive data analysis centre for arabidopsis subcellular protein locations. Nucleic Acids Res. 2016, 45, D1064–D1074. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing. Available online: http://www.R-project.org/ (accessed on 31 October 2014).
- Kim, S. Ppcor: An R package for a fast calculation to semi-partial correlation coefficients. Commun. Stat. Appl. Methods 2015, 22, 665. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 1995, 57, 289–300. [Google Scholar]
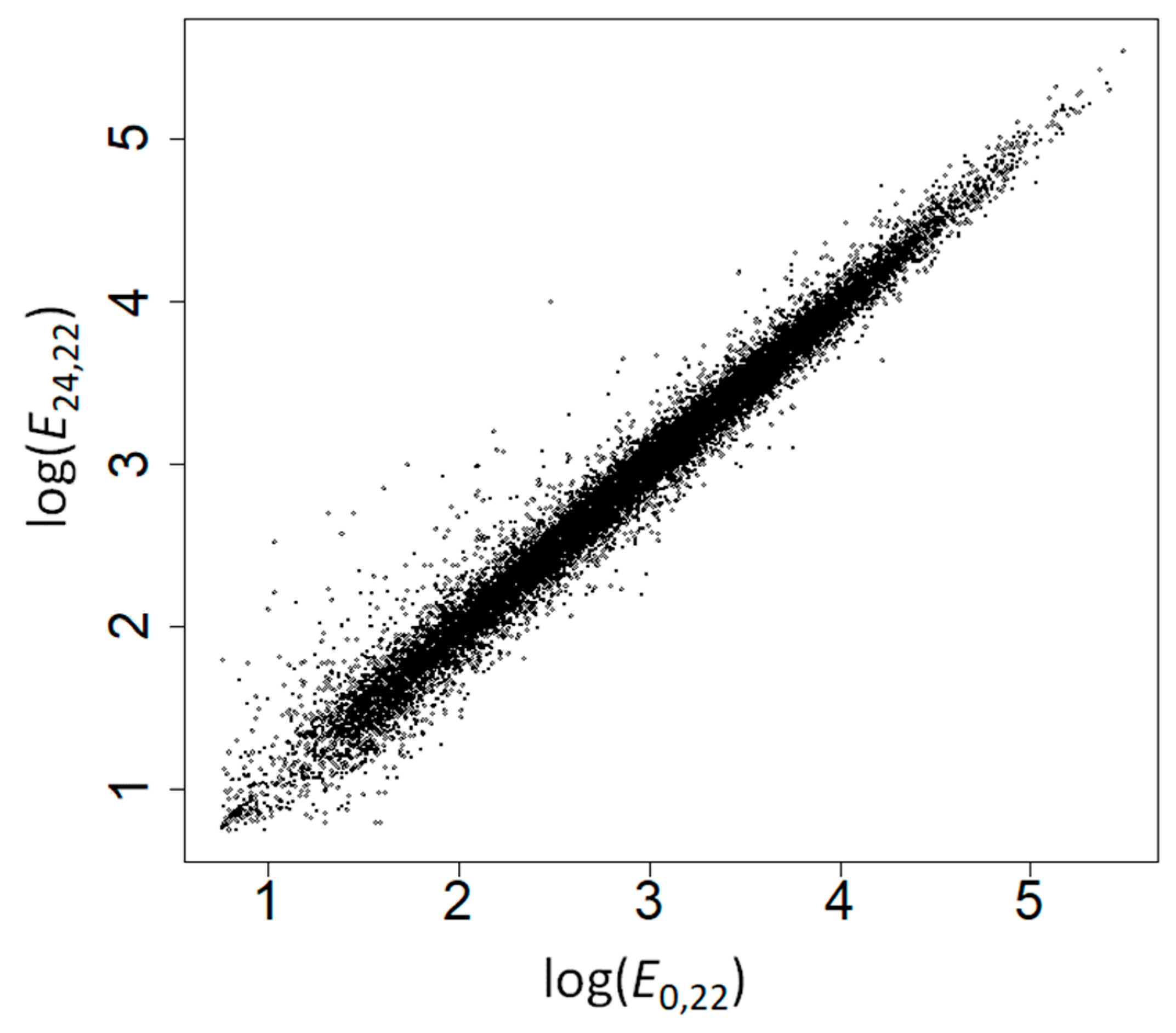
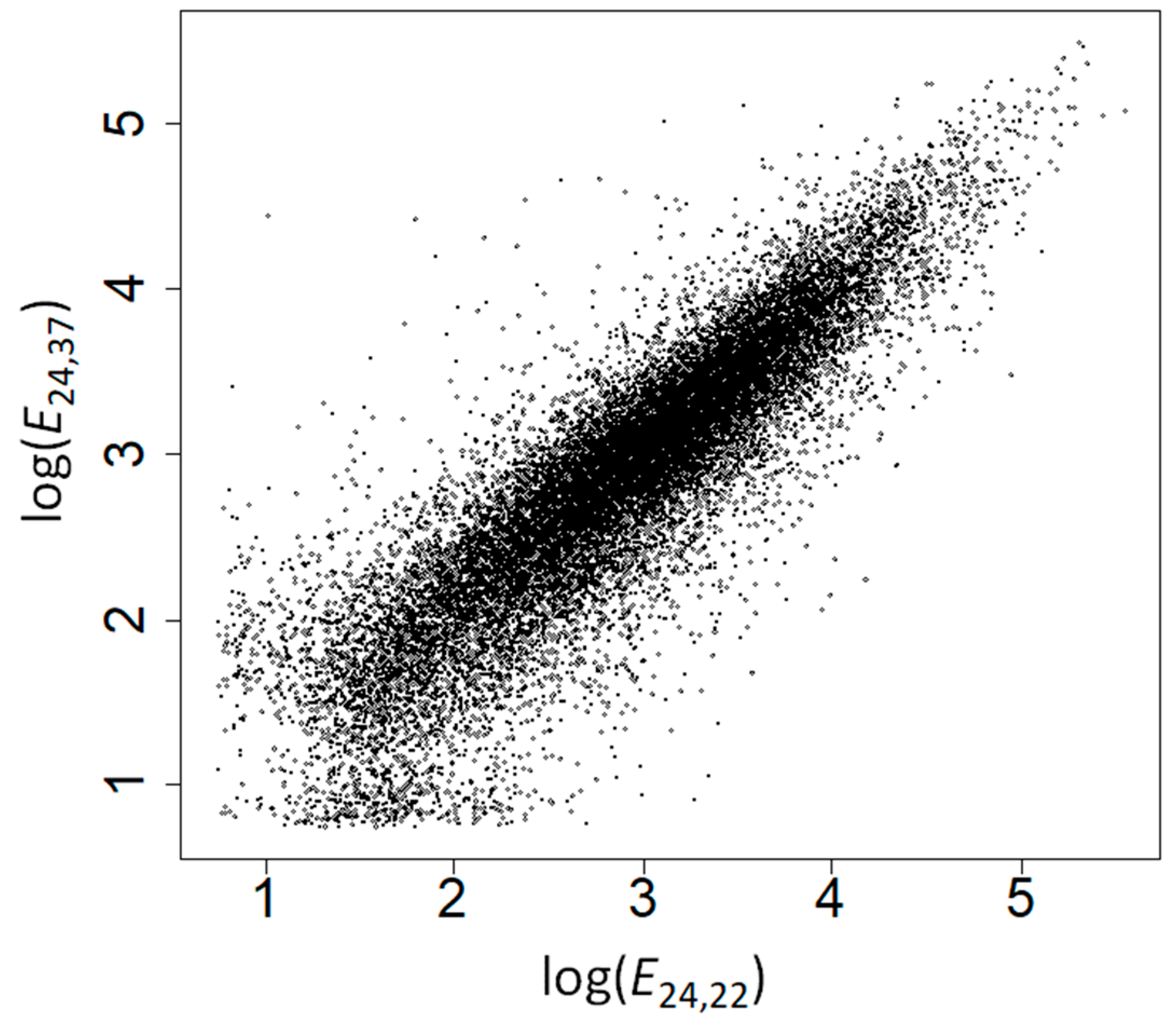
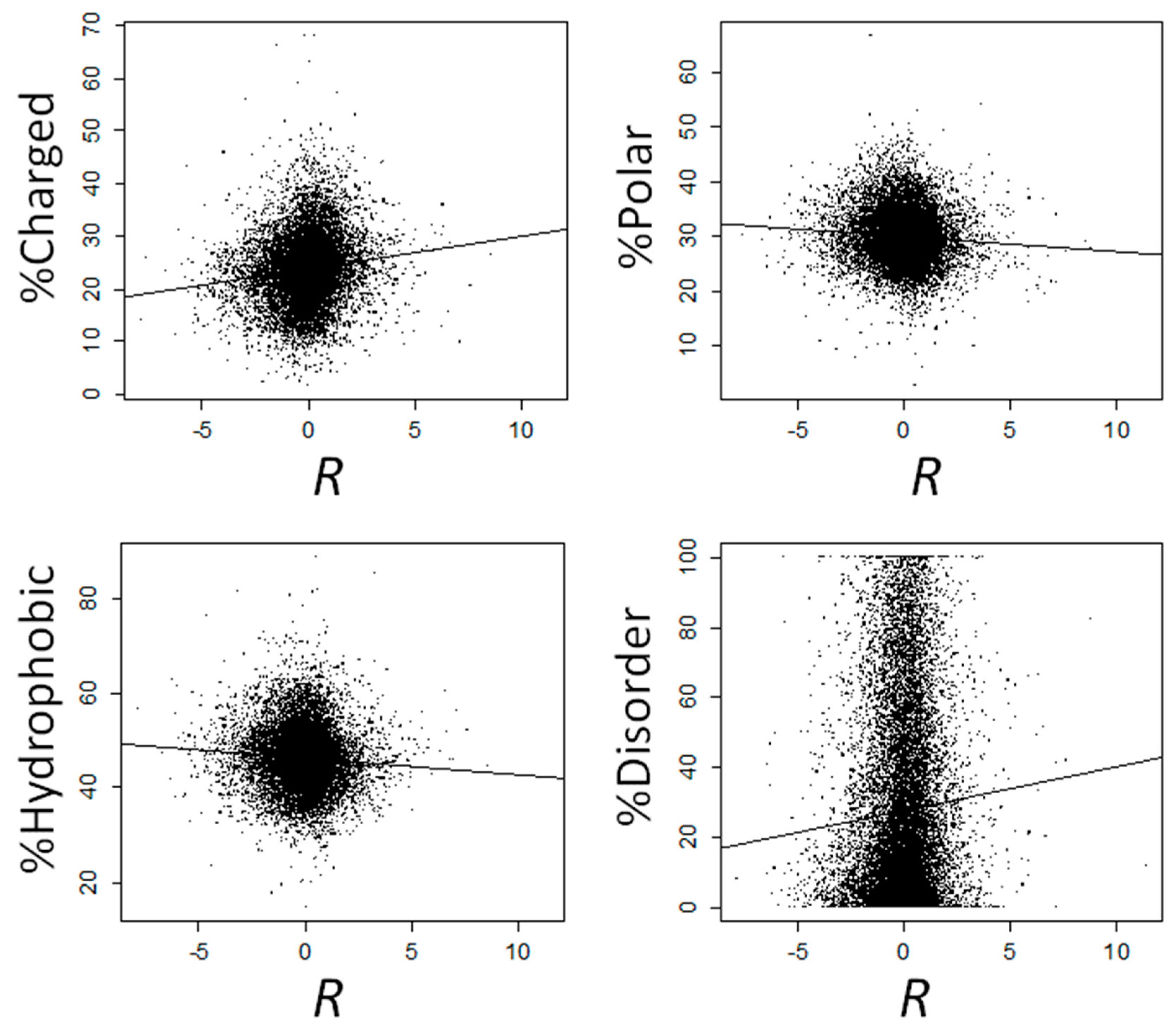
| Type | Amino Acid | No Control | Controlling for GC Content | Controlling for E24,22 | Controlling for E24,37 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ρ | p-Value | q-Value | ρ | p-Value | q-Value | ρ | p-Value | q-Value | ρ | p-Value | q-Value | ||
| Charged | Arg | 0.075 | 1.31 × 10−26 | 4.37 × 10−26 | 0.068 | 2.05 × 10−22 | 5.86 × 10−22 | 0.068 | 0.013 | 0.015 | 0.079 | 1.02 × 10−29 | 3.40 × 10−29 |
| Asp | 0.104 | 1.62 × 10−50 | 1.62 × 10−49 | 0.105 | 9.95 × 10−52 | 9.95 × 10−51 | 0.106 | 7.16 × 10−53 | 7.16 × 10−52 | 0.095 | 1.84 × 10−42 | 1.23 × 10−41 | |
| Glu | 0.118 | 5.48 × 10−64 | 1.10 × 10−62 | 0.122 | 5.60 × 10−69 | 1.12 × 10−67 | 0.115 | 2.61 × 10−61 | 5.22 × 10−60 | 0.115 | 2.60 × 10−61 | 5.20 × 10−60 | |
| Lys | 0.082 | 8.23 × 10−32 | 4.12 × 10−31 | 0.100 | 9.58 × 10−47 | 4.79 × 10−46 | 0.081 | 2.27 × 10−31 | 9.08 × 10−31 | 0.079 | 8.76 × 10−30 | 3.40 × 10−29 | |
| Total | 0.146 | 2.47 × 10−98 | 0.155 | 3.61 × 10−111 | 0.145 | 1.04 × 10−97 | 0.140 | 1.65 × 10−90 | |||||
| Polar | Asn | −0.025 | 3.86 × 10−4 | 0.001 | −0.018 | 0.011 | 0.015 | −0.044 | 4.17 × 10−10 | 8.34 × 10−10 | 0.005 | 0.433 | 0.433 |
| Cys | −0.011 | 0.127 | 0.158 | −0.009 | 0.187 | 0.208 | −0.034 | 1.54 × 10−6 | 2.57 × 10−6 | 0.026 | 2.07 × 10−4 | 3.19 × 10−4 | |
| Gln | 0.046 | 3.20 × 10−11 | 7.11 × 10−11 | 0.053 | 2.36 × 10−14 | 5.24 × 10−14 | 0.046 | 6.79 × 10−11 | 1.51 × 10−10 | 0.044 | 3.95 × 10−10 | 7.90 × 10−10 | |
| His | −0.010 | 0.134 | 0.158 | −0.009 | 0.210 | 0.221 | −0.024 | 0.001 | 0.001 | 0.010 | 0.146 | 0.154 | |
| Ser | −0.036 | 2.25 × 10−7 | 4.09 × 10−7 | −0.042 | 2.36 × 10−9 | 4.72 × 10−9 | −0.052 | 1.00 × 10−13 | 2.86 × 10−13 | −0.012 | 0.092 | 0.102 | |
| Thr | −0.099 | 1.10 × 10−45 | 7.33 × 10−45 | −0.100 | 9.24 × 10−47 | 4.79 × 10−46 | −0.098 | 1.12 × 10−44 | 7.47 × 10−44 | −0.096 | 2.75 × 10−43 | 2.75 × 10−42 | |
| Trp | −0.033 | 2.26 × 10−6 | 3.77 × 10−6 | −0.036 | 2.50 × 10−7 | 4.55 × 10−7 | −0.039 | 2.11 × 10−8 | 3.84 × 10−8 | −0.022 | 0.002 | 0.002 | |
| Tyr | −0.024 | 0.001 | 0.001 | −0.016 | 0.021 | 0.026 | −0.025 | 3.72 × 10−4 | 4.96 × 10−4 | −0.021 | 0.003 | 0.004 | |
| Total | −0.076 | 1.72 × 10−27 | −0.072 | 1.11 × 10−24 | −0.102 | 9.48 × 10−49 | −0.034 | 9.25 × 10−7 | |||||
| Hydro phobic | Ala | −0.008 | 0.280 | 0.311 | −0.020 | 0.004 | 0.006 | 0.027 | 1.32 × 10−4 | 1.89 × 10−4 | −0.060 | 1.50 × 10−17 | 3.75 × 10−17 |
| Gly | −0.054 | 1.40 × 10−14 | 3.50 × 10−14 | −0.066 | 1.99 × 10−21 | 4.98 × 10−21 | −0.028 | 5.46 × 10−5 | 8.40 × 10−5 | −0.092 | 1.17 × 10−39 | 5.85 × 10−39 | |
| Ile | −0.045 | 1.01 × 10−10 | 2.02 × 10−10 | −0.035 | 5.63 × 10−7 | 9.38 × 10−7 | −0.052 | 1.55 × 10−13 | 3.88 × 10−13 | −0.033 | 2.91 × 10−6 | 5.29 × 10−6 | |
| Leu | −0.004 | 0.547 | 0.547 | −0.004 | 0.533 | 0.533 | −0.016 | 0.021 | 0.023 | 0.015 | 0.029 | 0.034 | |
| Met | 0.006 | 0.387 | 0.407 | 0.014 | 0.042 | 0.049 | −0.001 | 0.942 | 0.942 | 0.017 | 0.017 | 0.021 | |
| Phe | −0.075 | 1.04 × 10−26 | 4.16 × 10−26 | −0.070 | 9.79 × 10−24 | 3.26 × 10−23 | −0.084 | 2.59 × 10−33 | 1.30 × 10−32 | −0.056 | 1.36 × 10−15 | 3.02 × 10−15 | |
| Pro | −0.060 | 8.03 × 10−18 | 2.29 × 10−17 | −0.074 | 1.85 × 10−26 | 7.40 × 10−26 | −0.052 | 8.41 × 10−14 | 2.80 × 10−13 | −0.070 | 7.88 × 10−24 | 2.25 × 10−23 | |
| Val | −0.017 | 0.012 | 0.017 | −0.024 | 0.001 | 0.001 | −0.006 | 0.370 | 0.390 | −0.033 | 3.30 × 10−6 | 5.50 × 10−6 | |
| Total | −0.084 | 4.08 × 10−33 | −0.096 | 1.31 × 10−43 | −0.064 | 2.88 × 10−20 | −0.109 | 2.73 × 10−55 | |||||
| Type | Amino Acid | Median Overexpressed (%) | Median Repressed (%) | p-Value | q-Value |
|---|---|---|---|---|---|
| Charged | Arg | 5.43 | 5.19 | 8.06 × 10−21 | 4.61 × 10−20 |
| Asp | 5.36 | 5.10 | 1.60 × 10−36 | 6.40 × 10−35 | |
| Glu | 6.61 | 6.15 | 8.28 × 10−44 | 6.62 × 10−42 | |
| Lys | 6.33 | 6.06 | 1.20 × 10−21 | 8.00 × 10−21 | |
| Total | 24.32 | 23.20 | 1.90 × 10−66 | ||
| Polar | Asn | 4.08 | 4.12 | 0.017 | 0.024 |
| Cys | 1.59 | 1.60 | 0.043 | 0.060 | |
| Gln | 3.27 | 3.16 | 7.77 × 10−8 | 1.88 × 10−7 | |
| His | 2.11 | 2.10 | 0.204 | 0.244 | |
| Ser | 8.79 | 8.96 | 2.27 × 10−7 | 5.19 × 10−7 | |
| Thr | 4.90 | 5.13 | 5.31 × 10−34 | 1.42 × 10−32 | |
| Trp | 1.07 | 1.11 | 4.75 × 10−4 | 0.001 | |
| Tyr | 2.65 | 2.68 | 0.132 | 0.163 | |
| Total | 29.54 | 30.04 | 2.53 × 10−20 | ||
| Hydrophobic | Ala | 6.32 | 6.30 | 0.889 | 0.889 |
| Gly | 6.18 | 6.41 | 2.77 × 10−10 | 8.21 × 10−10 | |
| Ile | 5.12 | 5.23 | 1.87 × 10−7 | 4.40 × 10−7 | |
| Leu | 9.24 | 9.27 | 0.675 | 0.720 | |
| Met | 2.38 | 2.37 | 0.399 | 0.449 | |
| Phe | 4.08 | 4.28 | 1.55 × 10−18 | 7.75 × 10−18 | |
| Pro | 4.54 | 4.71 | 2.56 × 10−12 | 8.53 × 10−12 | |
| Val | 6.67 | 6.68 | 0.178 | 0.215 | |
| Total | 45.77 | 46.43 | 6.56 × 10−21 |
| Type | Amino Acid | Median Overexpressed (%) | Median Repressed (%) | p-Value | q-Value |
|---|---|---|---|---|---|
| Charged | Arg | 5.26 | 4.80 | 7.82 × 10−9 | 1.04 × 10−7 |
| Asp | 5.51 | 4.95 | 1.62 × 10−12 | 4.32 × 10−11 | |
| Glu | 6.92 | 5.92 | 1.31 × 10−17 | 1.05 × 10−15 | |
| Lys | 6.78 | 6.17 | 1.78 × 10−7 | 1.78 × 10−6 | |
| Total | 25.30 | 22.54 | 1.50 × 10−26 | ||
| Polar | Asn | 4.04 | 4.29 | 2.81 × 10−4 | 0.001 |
| Cys | 1.66 | 1.69 | 0.031 | 0.045 | |
| Gln | 3.13 | 2.94 | 2.87 × 10−4 | 6.57 × 10−4 | |
| His | 2.03 | 2.12 | 0.023 | 0.035 | |
| Ser | 8.47 | 8.41 | 0.780 | 0.810 | |
| Thr | 4.95 | 5.26 | 3.52 × 10−6 | 1.56 × 10−5 | |
| Trp | 1.05 | 1.15 | 0.035 | 0.050 | |
| Tyr | 2.57 | 2.86 | 6.09 × 10−5 | 1.87 × 10−4 | |
| Total | 29.74 | 30.17 | 3.20 × 10−8 | ||
| Hydrophobic | Ala | 6.11 | 6.12 | 0.867 | 0.878 |
| Gly | 6.05 | 6.50 | 5.49 × 10−5 | 1.76 × 10−4 | |
| Ile | 5.25 | 5.48 | 0.001 | 0.002 | |
| Leu | 9.01 | 9.17 | 0.215 | 0.292 | |
| Met | 2.46 | 2.52 | 0.321 | 0.395 | |
| Phe | 4.12 | 4.65 | 9.55 × 10−13 | 3.82 × 10−11 | |
| Pro | 4.31 | 4.62 | 9.34 × 10−5 | 2.58 × 10−4 | |
| Val | 6.76 | 6.84 | 0.294 | 0.386 | |
| Total | 45.20 | 47.24 | 6.04 × 10−11 |
| Subcellular Location | n | Median Charged Amino Acids (%) | Median Polar Amino Acids (%) | Median Hydrophobic Amino Acids (%) | Median Intrinsic Disorder (%) | Median R |
|---|---|---|---|---|---|---|
| Cytosol | 633 | 25.46 | 26.74 | 47.31 | 15.64 | 0.131 |
| Endoplasmic reticulum | 163 | 24.12 | 27.22 | 48.68 | 10.11 | 0.147 |
| Extracellular | 197 | 18.94 | 32.87 | 48.43 | 8.61 | −0.296 |
| Golgi | 375 | 23.20 | 29.37 | 47.38 | 14.22 | 0.108 |
| Mitochondrion | 286 | 23.12 | 27.63 | 49.26 | 14.93 | 0.261 |
| Nucleus | 446 | 26.50 | 29.16 | 43.92 | 42.73 | 0.406 |
| Peroxisome | 63 | 23.16 | 26.47 | 50.00 | 10.61 | −0.207 |
| Plasma membrane | 343 | 22.21 | 28.63 | 48.73 | 14.97 | −0.195 |
| Plastid | 720 | 23.33 | 27.65 | 48.91 | 15.28 | −0.190 |
| Vacuole | 81 | 21.14 | 28.24 | 49.75 | 8.12 | −0.008 |
| Subcellular Location | Correlation R-Charged Amino Acids | Correlation R-Polar Amino Acids | Correlation R-Hydrophobic Amino Acids | Correlation R-Intrinsic Disorder | ||||
|---|---|---|---|---|---|---|---|---|
| ρ | p-Value | ρ | p-Value | ρ | p-Value | ρ | p-Value | |
| Cytosol | 0.171 | 1.54 × 10−5 | −0.142 | 3.27 × 10−4 | −0.069 | 0.082 | 0.123 | 0.002 |
| Endoplasmic reticulum | 0.061 | 0.437 | −0.015 | 0.847 | −0.112 | 0.155 | 0.226 | 0.004 |
| Extracellular | 0.054 | 0.452 | 0.021 | 0.765 | −0.073 | 0.309 | 0.068 | 0.346 |
| Golgi | −0.046 | 0.370 | 0.068 | 0.191 | −0.009 | 0.866 | 0.073 | 0.156 |
| Mitochondrion | 0.124 | 0.036 | 0.060 | 0.312 | −0.125 | 0.034 | 0.065 | 0.272 |
| Nucleus | 0.020 | 0.681 | −0.119 | 0.012 | 0.102 | 0.031 | −0.207 | 1.09 × 10−5 |
| Peroxisome | 0.016 | 0.902 | −0.104 | 0.416 | 0.080 | 0.535 | 0.237 | 0.062 |
| Plasma membrane | 0.064 | 0.234 | −0.017 | 0.750 | −0.004 | 0.947 | −0.154 | 0.004 |
| Plastid | 0.137 | 2.20 × 10−4 | 0.007 | 0.859 | −0.110 | 0.003 | 0.062 | 0.095 |
| Vacuole | 0.184 | 0.099 | 0.082 | 0.466 | −0.189 | 0.091 | 0.266 | 0.017 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alvarez-Ponce, D.; Ruiz-González, M.X.; Vera-Sirera, F.; Feyertag, F.; Perez-Amador, M.A.; Fares, M.A. Arabidopsis Heat Stress-Induced Proteins Are Enriched in Electrostatically Charged Amino Acids and Intrinsically Disordered Regions. Int. J. Mol. Sci. 2018, 19, 2276. https://doi.org/10.3390/ijms19082276
Alvarez-Ponce D, Ruiz-González MX, Vera-Sirera F, Feyertag F, Perez-Amador MA, Fares MA. Arabidopsis Heat Stress-Induced Proteins Are Enriched in Electrostatically Charged Amino Acids and Intrinsically Disordered Regions. International Journal of Molecular Sciences. 2018; 19(8):2276. https://doi.org/10.3390/ijms19082276
Chicago/Turabian StyleAlvarez-Ponce, David, Mario X. Ruiz-González, Francisco Vera-Sirera, Felix Feyertag, Miguel A. Perez-Amador, and Mario A. Fares. 2018. "Arabidopsis Heat Stress-Induced Proteins Are Enriched in Electrostatically Charged Amino Acids and Intrinsically Disordered Regions" International Journal of Molecular Sciences 19, no. 8: 2276. https://doi.org/10.3390/ijms19082276
APA StyleAlvarez-Ponce, D., Ruiz-González, M. X., Vera-Sirera, F., Feyertag, F., Perez-Amador, M. A., & Fares, M. A. (2018). Arabidopsis Heat Stress-Induced Proteins Are Enriched in Electrostatically Charged Amino Acids and Intrinsically Disordered Regions. International Journal of Molecular Sciences, 19(8), 2276. https://doi.org/10.3390/ijms19082276







