A Rice B-Box Protein, OsBBX14, Finely Regulates Anthocyanin Biosynthesis in Rice
Abstract
1. Introduction
2. Results
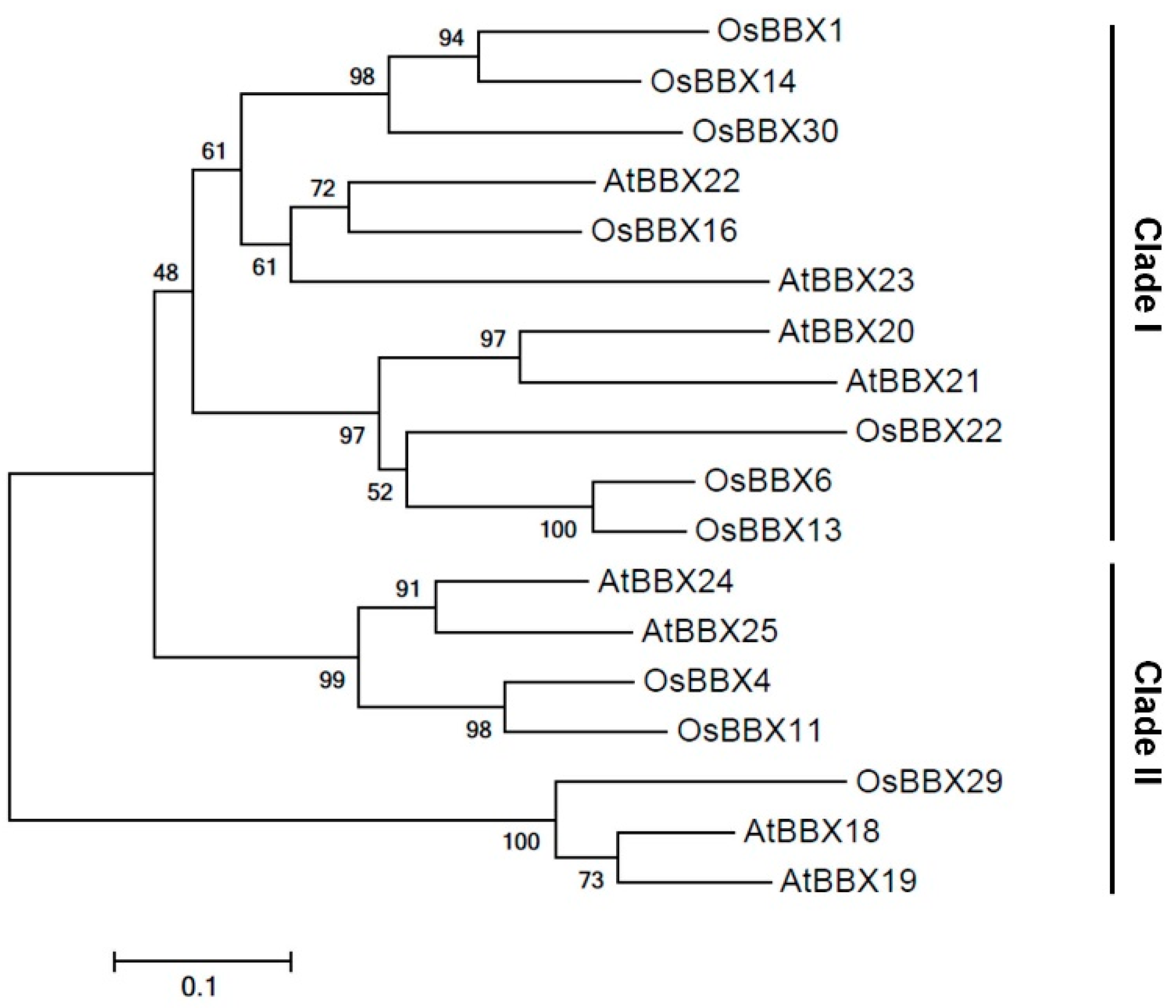
2.1. Identification of B-Box Genes from White and Black Rice Seeds
2.2. Temporal Expression of OsBBX14 and Other Regulatory Genes in Developing Rice Seeds
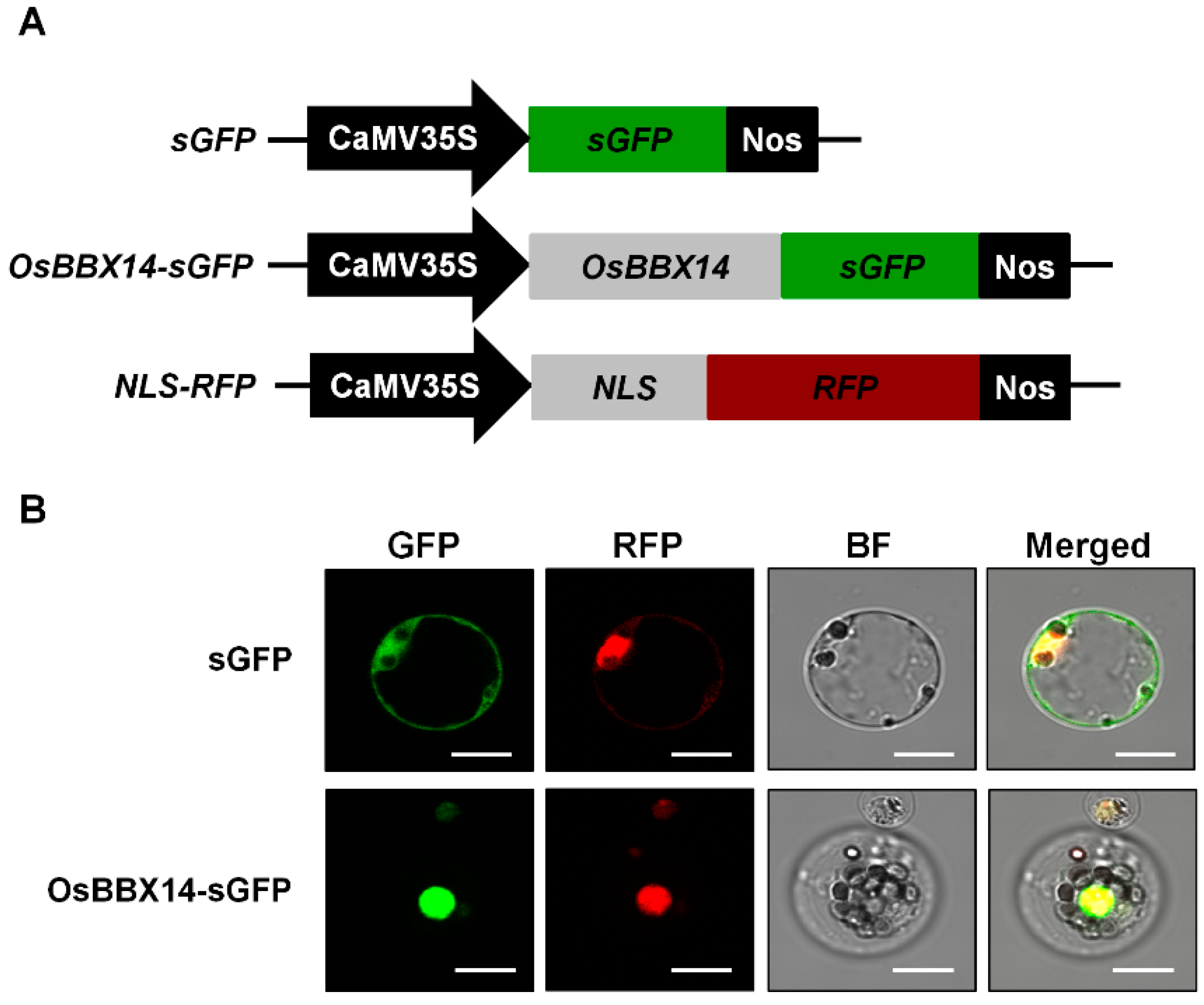
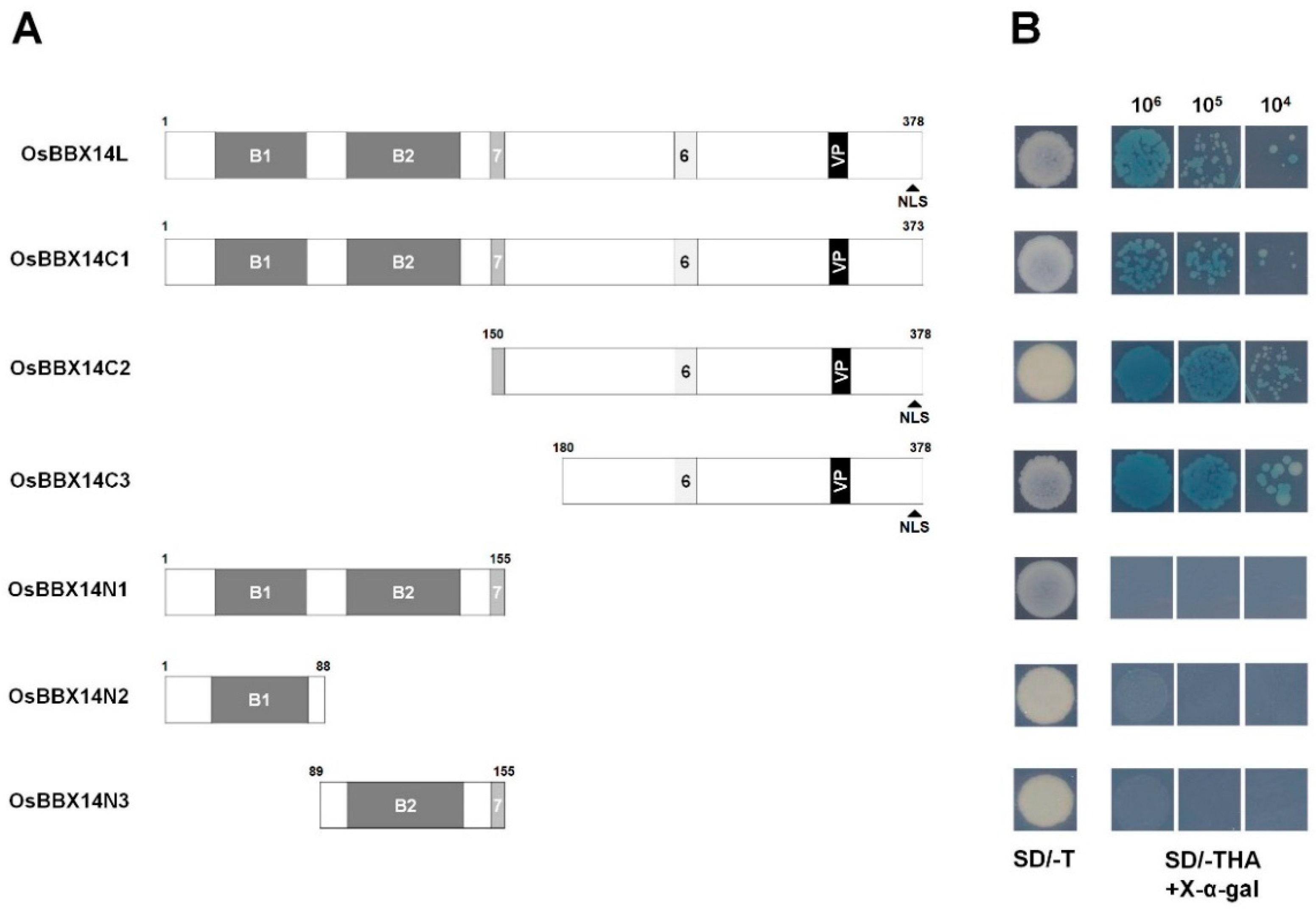
2.3. Subcellular Localization and Transcriptional Activation Activity of OsBBX14
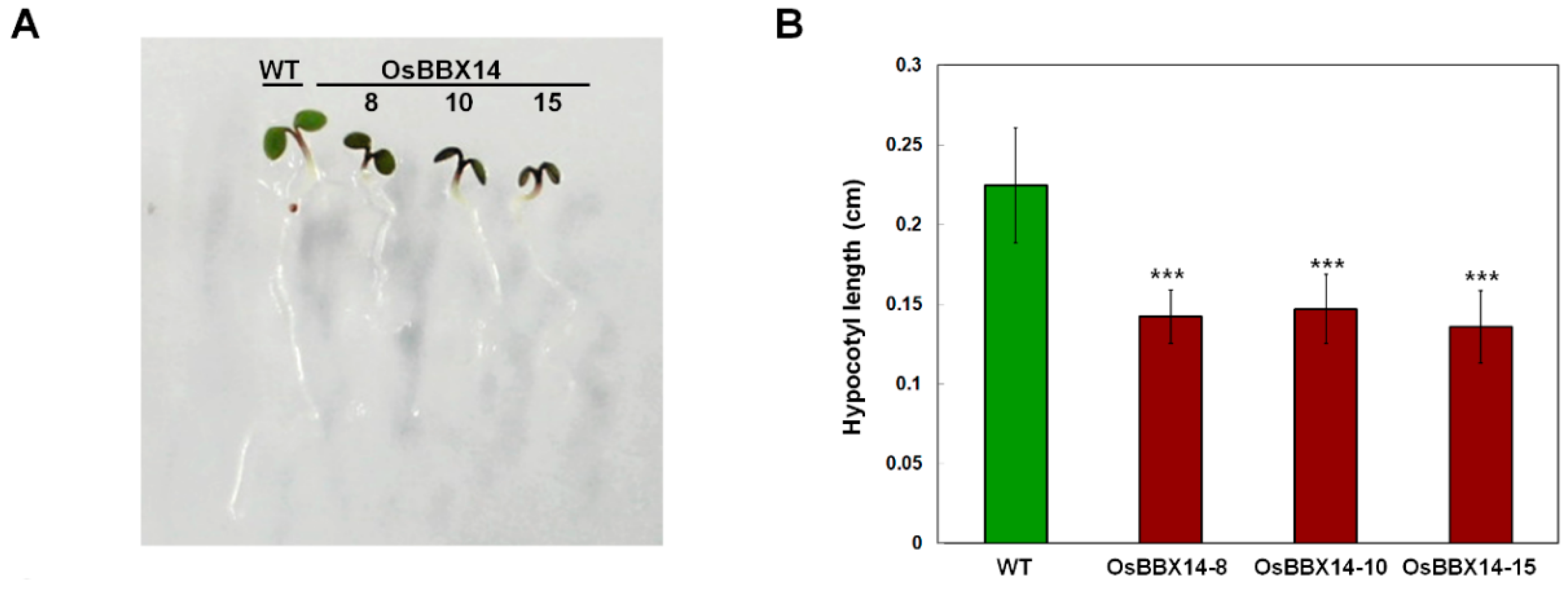
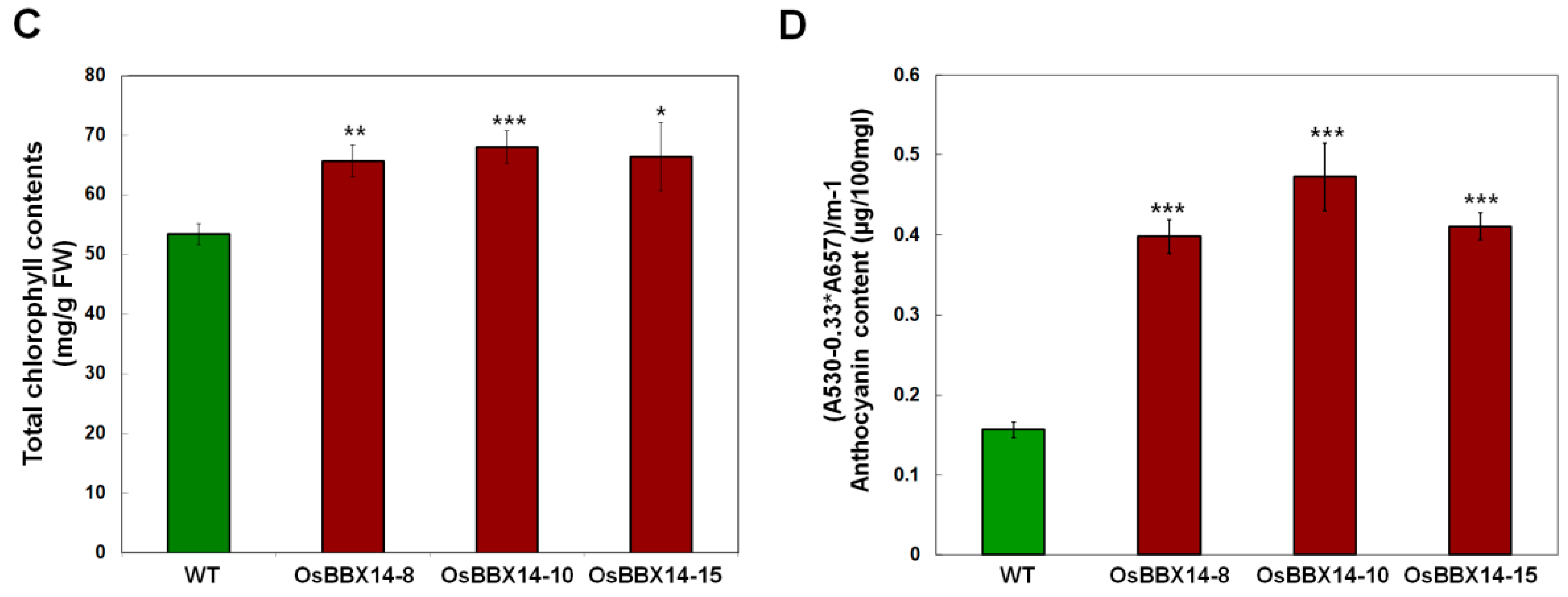
2.4. Overexpression of OsBBX14 in Arabidopsis Caused an Anthocyanin Accumulation and Short Hypocotyl Length
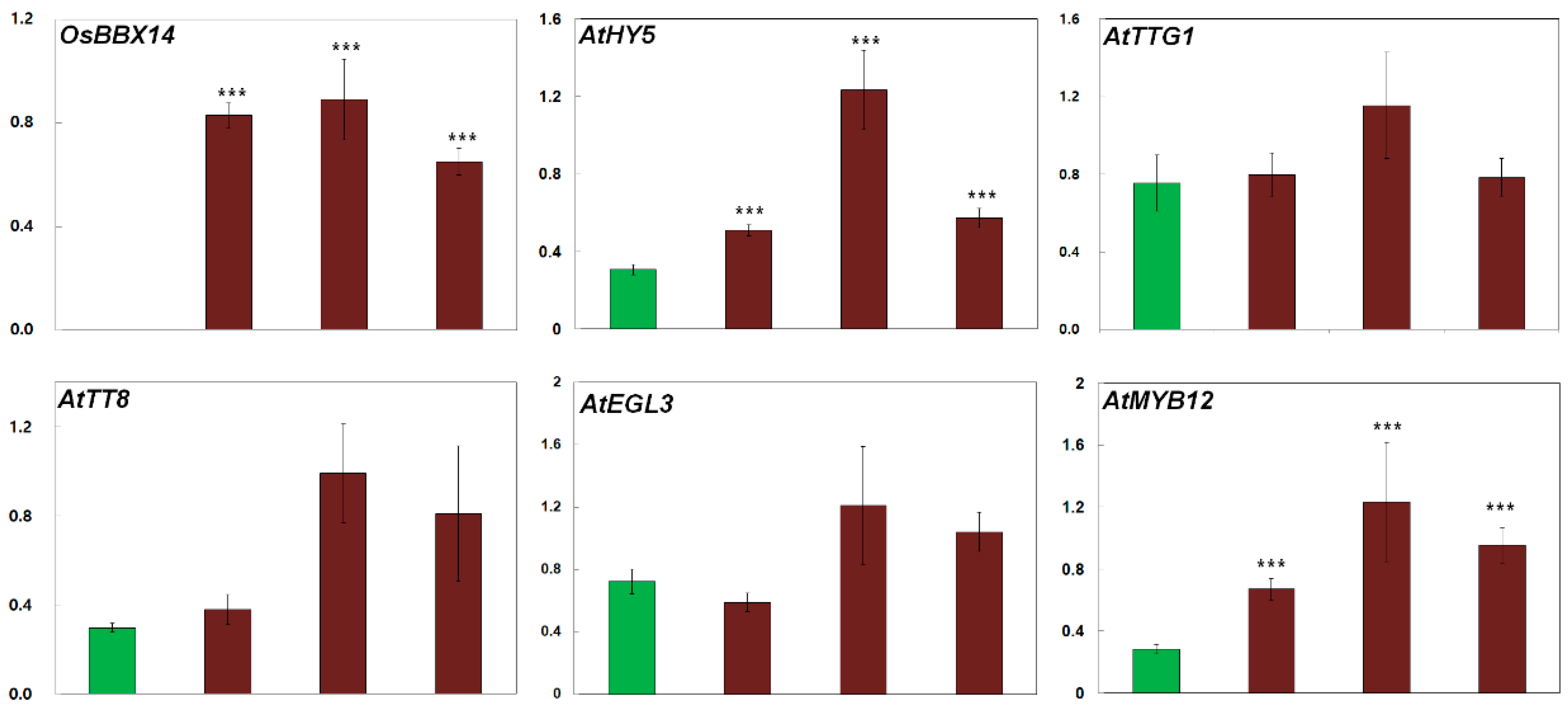
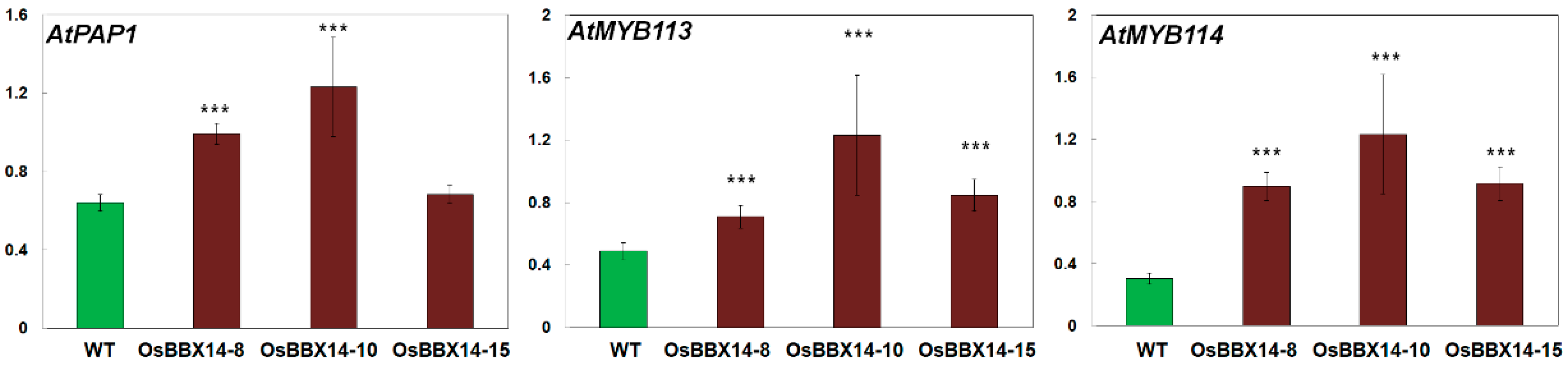
2.5. Overexpression of OsBBX14 Induced the Expression of Anthocyanin Biosynthesis Regulatory Genes in Arabidopsis
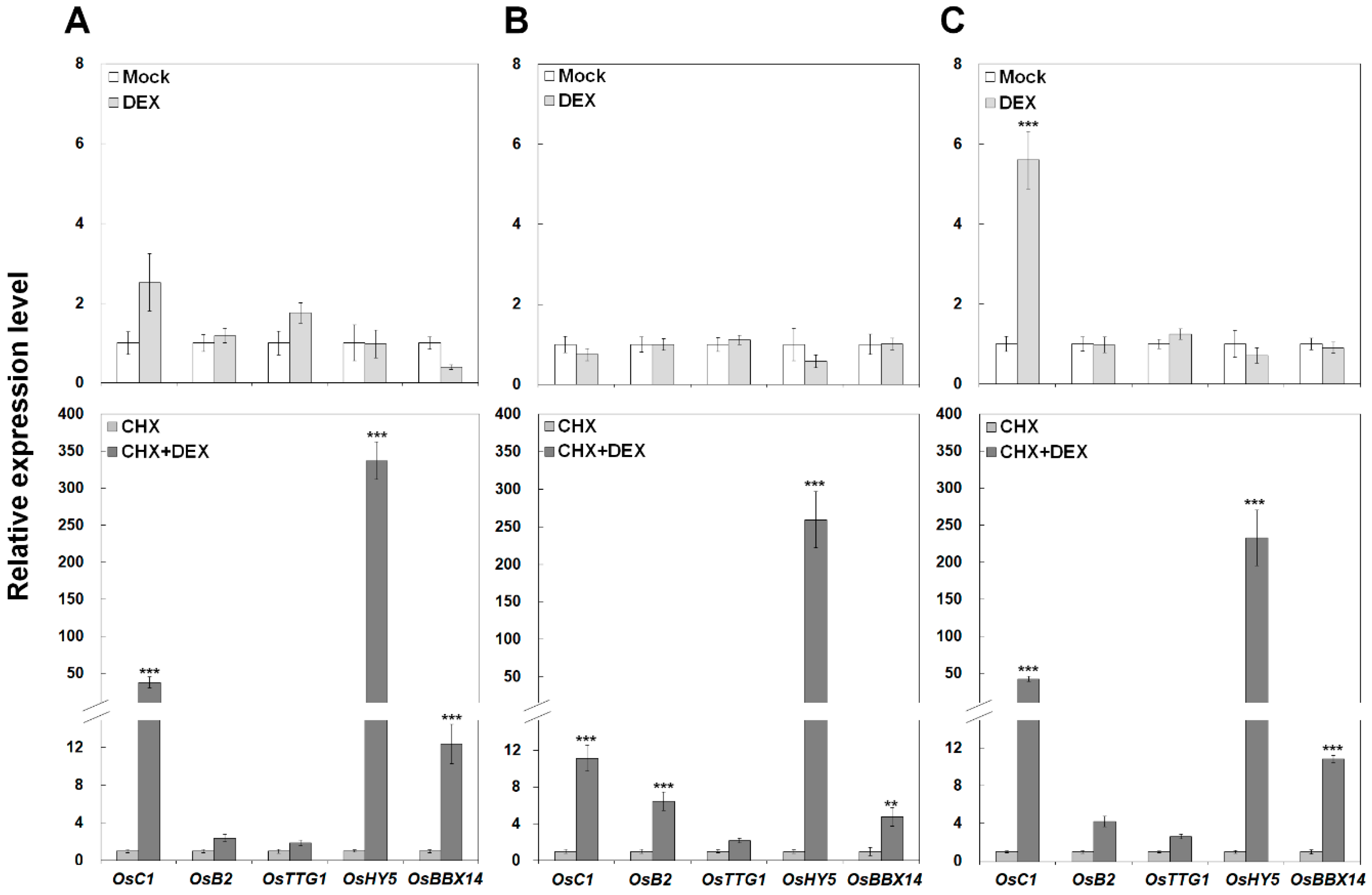
2.6. OsBBX14 Directly Activates the Expression of the Anthocyanin Regulatory Genes
2.7. Protein–Protein Interactions between OsBBX14 and OsHY5
3. Discussion
3.1. OsBBX14 Is a B-Box Protein with Transcriptional Activity
3.2. OsBBX14 Modulates the Expression of Anthocyanin Biosynthesis Regulators by Interacting with OsHY5
4. Materials and Methods
4.1. Plants Materials
4.2. Transcriptome Profiling
4.3. RNA Isolation and qPCR
4.4. Bioinformatics Analysis
4.5. Subcellular Localization Analysis and Plant Transformation
4.6. Measurement of Hypocotyl Length and Total Anthocyanin and Chlorophyll Contents
4.7. Transient Expression Assay Using a DEX-Inducible Activation System in Rice Protoplasts
4.8. Transactivation and Y2H Assays
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Oikawa, T.; Maeda, H.; Oguchi, T.; Yamaguchi, T.; Tanabe, N.; Ebana, K.; Yano, M.; Ebitani, T.; Izawa, T. The birth of a black rice gene and its local spread by introgression. Plant Cell 2015, 27, 2401–2414. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, M.T.; Thomson, M.J.; Pfeil, B.E.; McCouch, S. Caught red-handed: Rc encodes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell 2006, 18, 283–294. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Jin, L.; Xiao, P.; Lu, Y.; Bao, J. Total phenolics, flavonoids, antioxidant capacity in rice grain and their relations to grain color, size and weight. J. Cereal Sci. 2009, 49, 106–111. [Google Scholar] [CrossRef]
- Kim, J.K.; Lee, S.Y.; Chu, S.M.; Lim, S.H.; Suh, S.C.; Lee, Y.T.; Cho, H.S.; Ha, S.H. Variation and correlation analysis of flavonoids and carotenoids in Korean pigmented rice (Oryza sativa L.) cultivars. J. Agric. Food Chem. 2010, 58, 12804–12809. [Google Scholar] [CrossRef] [PubMed]
- Koes, R.; Verweij, W.; Quattrocchio, F. Flavonoids: A colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005, 10, 236–242. [Google Scholar] [CrossRef] [PubMed]
- Grotewold, E. The genetics and biochemistry of floral pigments. Annu. Rev. Plant Biol. 2006, 57, 761–780. [Google Scholar] [CrossRef] [PubMed]
- Shih, C.H.; Chu, H.; Tang, L.K.; Sakamoto, W.; Maekawa, M.; Chu, I.K.; Wang, M.; Lo, C. Functional characterization of key structural genes in rice flavonoid biosynthesis. Planta 2008, 228, 1043–1054. [Google Scholar] [CrossRef] [PubMed]
- Hichri, I.; Barrieu, F.; Bogs, J.; Kappel, C.; Delrot, S.; Lauvergeat, V. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 2011, 62, 2465–2483. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Zhang, Z.; Chen, C.; Wu, W.; Ren, N.; Jiang, C.; Yu, J.; Zhao, Y.; Zheng, X.; Yang, Q.; et al. The C-S-A gene system regulates hull pigmentation and reveals evolution of anthocyanin biosynthesis pathway in rice. J. Exp. Bot. 2018, 69, 1485–1498. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef] [PubMed]
- Jaakola, L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Albert, N.W.; Davies, K.M.; Lewis, D.H.; Zhang, H.; Montefiori, M.; Brendolise, C.; Boase, M.R.; Ngo, H.; Jameson, P.E.; Schwinn, K.E. A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 2014, 26, 962–980. [Google Scholar] [CrossRef] [PubMed]
- Shin, D.H.; Choi, M.; Kim, K.; Bang, G.; Cho, M.; Choi, S.B.; Choi, G.; Park, Y.I. HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Lett. 2013, 587, 1543–1547. [Google Scholar] [CrossRef] [PubMed]
- Maier, A.; Schrader, A.; Kokkelink, L.; Falke, C.; Welter, B.; Iniesto, E.; Rubio, V.; Uhrig, J.F.; Hülskamp, M.; Hoecker, U. Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis. Plant J. 2013, 74, 638–651. [Google Scholar] [CrossRef] [PubMed]
- Albert, N.W.; Lewis, D.H.; Zhang, H.; Irving, L.J.; Jameson, P.E.; Davies, K.M. Light-induced vegetative anthocyanin pigmentation in Petunia. J. Exp. Bot. 2009, 60, 2191–2202. [Google Scholar] [CrossRef] [PubMed]
- Datta, S.; Hettiarachchi, C.; Johansson, H.; Holm, M. SALT TOLERANCE HOMOLOG2, a B-box protein in Arabidopsis that activates transcription and positively regulates light-mediated development. Plant Cell 2007, 19, 3242–3255. [Google Scholar] [CrossRef] [PubMed]
- Gangappa, S.N.; Botto, J.F. The multifaceted roles of HY5 in plant growth and development. Mol. Plant 2016, 9, 1353–1365. [Google Scholar] [CrossRef] [PubMed]
- Gangappa, S.N.; Botto, J.F. The BBX family of plant transcription factors. Trends Plant Sci. 2014, 19, 460–470. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zhao, X.; Weng, X.; Wang, L.; Xie, W. The rice B-box zinc finger gene family: Genomic identification, characterization, expression profiling and diurnal analysis. PLoS ONE 2012, 7, e48242. [Google Scholar] [CrossRef] [PubMed]
- Khanna, R.; Kronmiller, B.; Maszle, D.R.; Coupland, G.; Holm, M.; Mizuno, T.; Wu, S.H. The Arabidopsis B-box zinc finger family. Plant Cell 2009, 21, 3416–3420. [Google Scholar] [CrossRef] [PubMed]
- Gangappa, S.N.; Crocco, C.D.; Johansson, H.; Datta, S.; Hettiarachchi, C.; Holm, M.; Botto, J.F. The Arabidopsis B-BOX protein BBX25 interacts with HY5, negatively regulating BBX22 expression to suppress seedling photomorphogenesis. Plant Cell 2013, 25, 1243–1257. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Jiang, Y.; Li, J.; Lin, F.; Holm, M.; Deng, X.W. BBX21, an Arabidopsis B-box protein, directly activates HY5 and is targeted by COP1 for 26S proteasome-mediated degradation. Proc. Natl. Acad. Sci. USA 2016, 113, 7655–7660. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.S.J.; Li, Y.H.; Chen, L.T.; Chen, W.C.; Hsieh, W.P.; Shin, J.; Neng Jane, W.; Chou, S.J.; Choi, G.; Hu, J.M.; et al. LZF1, a HY5-regulated transcriptional factor, functions in Arabidopsis de-etiolation. Plant J. 2008, 54, 205–219. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Wang, Y.; Li, Q.F.; Björn, L.O.; He, J.X.; Li, S.S. Arabidopsis STO/BBX24 negatively regulates UV-B signaling by interacting with COP1 and repressing HY5 transcriptional activity. Cell Res. 2012, 22, 1046. [Google Scholar] [CrossRef] [PubMed]
- Crocco, C.D.; Botto, J.F. BBX proteins in green plants: Insights into their evolution, structure, feature and functional diversification. Gene 2013, 531, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.C.; Kim, J.Y.; Ko, J.H.; Kang, H.; Han, K.H. Identification of direct targets of transcription factor MYB46 provides insights into the transcriptional regulation of secondary wall biosynthesis. Plant Mol. Biol. 2014, 85, 589–599. [Google Scholar] [CrossRef] [PubMed]
- Li, S. Transcriptional control of flavonoid biosynthesis: Fine-tuning of the MYB-bHLH-WD40 (MBW) complex. Plant Signal. Behav. 2014, 9, e27522. [Google Scholar] [CrossRef] [PubMed]
- Datta, S.; Johansson, H.; Hettiarachchi, C.; Irigoyen, M.; Desai, M.; Rubio, V.; Holm, M. LZF1/SALT TOLERANCE HOMOLOG3, an Arabidopsis B-Box protein involved in light-dependent development and gene expression, undergoes COP1-mediated ubiquitination. Plant Cell 2008, 20, 2324–2338. [Google Scholar] [CrossRef] [PubMed]
- Holm, M.; Hardtke, C.S.; Gaudet, R.; Deng, X.W. Identification of a structural motif that confers specific interaction with the WD40 repeat domain of Arabidopsis COP1. EMBO J. 2001, 20, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Job, N.; Yadukrishnan, P.; Bursch, K.; Datta, S.; Johansson, H. Two B-box proteins regulate photomorphogenesis by oppositely modulating HY5 through their diverse C-terminal domains. Plant Physiol. 2018, 00856. [Google Scholar] [CrossRef] [PubMed]
- Ganesan, G.; Sankararamasubramanian, H.M.; Harikrishnan, M.; Ashwin, G.; Parida, A. A MYB transcription factor from the grey mangrove is induced by stress and confers NaCl tolerance in tobacco. J. Exp. Bot. 2012, 63, 4549–4561. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Huai, J.; Shang, F.; Xu, G.; Tang, W.; Jing, Y.; Lin, R. A PIF1/PIF3-HY5-BBX23 transcription factor cascade affects photomorphogenesis. Plant Physiol. 2017, 174, 2487–2500. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Jiang, Y.; Li, J.; Holm, M.; Deng, X.W. The B-Box Domain Protein BBX21 Promotes Photomorphogenesis. Plant Physiol. 2018, 176, 2365–2375. [Google Scholar] [CrossRef] [PubMed]
- Stracke, R.; Favory, J.J.; Gruber, H.; Bartelniewoehner, L.; Bartels, S.; Binkert, M.; Funk, M.; Weisshaar, B.; Ulm, R. The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant Cell Environ. 2010, 33, 88–103. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Fujii, S.; Sasaki, D.; Baba, S.; Ohta, H.; Masuda, T.; Wada, H. Transcriptional regulation of thylakoid galactolipid biosynthesis coordinated with chlorophyll biosynthesis during the development of chloroplasts in Arabidopsis. Front. Plant Sci. 2014, 5, 272. [Google Scholar] [CrossRef] [PubMed]
- Abbas, N.; Maurya, J.P.; Senapati, D.; Gangappa, S.N.; Chattopadhyay, S. Arabidopsis CAM7 and HY5 physically interact and directly bind to the HY5 promoter to regulate its expression and thereby promote photomorphogenesis. Plant Cell 2014, 26, 1036–1052. [Google Scholar] [CrossRef] [PubMed]
- Binkert, M.; Kozma-Bognár, L.; Terecskei, K.; De Veylder, L.; Nagy, F.; Ulm, R. UV-B-Responsive Association of the Arabidopsis bZIP Transcription Factor ELONGATED HYPOCOTYL5 with Target Genes, Including Its Own Promoter. Plant Cell 2014, 26, 4200–4213. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S. Fast gapped-read alignment with Bowtie2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Choi, M.J.; Lee, J.Y.; Kim, J.K.; Ha, S.H.; Lim, S.H. Molecular and Biochemical Analysis of Two Rice Flavonoid 3′-Hydroxylase to Evaluate Their Roles in Flavonoid Biosynthesis in Rice Grain. Int. J. Mol. Sci. 2016, 17, 1549. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Tamura, K.; Jakobsen, I.B.; Nei, M. MEGA2: Molecular evolutionary genetics analysis software. Bioinformatics 2001, 17, 1244–1245. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Moon, S.J.; Min, M.K.; Choi, E.H.; Kim, J.; Koh, E.Y.; Yoon, I.; Yoo, S.D.; Kim, B.G. Functional characterization and reconstitution of ABA signaling components using transient gene expression in rice protoplasts. Front. Plant Sci. 2015, 6, 614. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.H.; Song, J.H.; Kim, D.H.; Kim, J.K.; Lee, J.Y.; Kim, Y.M.; Ha, S.H. Activation of anthocyanin biosynthesis by expression of the radish R2R3-MYB transcription factor gene RsMYB1. Plant Cell Rep. 2016, 35, 641–653. [Google Scholar] [CrossRef] [PubMed]
- Inskeep, W.P.; Bloom, P.R. Extinction coefficients of chlorophyll a and b in N, N-dimethylformamide and 80% acetone. Plant Physiol. 1985, 77, 483–485. [Google Scholar] [CrossRef] [PubMed]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, D.-H.; Park, S.; Lee, J.-Y.; Ha, S.-H.; Lee, J.-G.; Lim, S.-H. A Rice B-Box Protein, OsBBX14, Finely Regulates Anthocyanin Biosynthesis in Rice. Int. J. Mol. Sci. 2018, 19, 2190. https://doi.org/10.3390/ijms19082190
Kim D-H, Park S, Lee J-Y, Ha S-H, Lee J-G, Lim S-H. A Rice B-Box Protein, OsBBX14, Finely Regulates Anthocyanin Biosynthesis in Rice. International Journal of Molecular Sciences. 2018; 19(8):2190. https://doi.org/10.3390/ijms19082190
Chicago/Turabian StyleKim, Da-Hye, Sangkyu Park, Jong-Yeol Lee, Sun-Hwa Ha, Jun-Gu Lee, and Sun-Hyung Lim. 2018. "A Rice B-Box Protein, OsBBX14, Finely Regulates Anthocyanin Biosynthesis in Rice" International Journal of Molecular Sciences 19, no. 8: 2190. https://doi.org/10.3390/ijms19082190
APA StyleKim, D.-H., Park, S., Lee, J.-Y., Ha, S.-H., Lee, J.-G., & Lim, S.-H. (2018). A Rice B-Box Protein, OsBBX14, Finely Regulates Anthocyanin Biosynthesis in Rice. International Journal of Molecular Sciences, 19(8), 2190. https://doi.org/10.3390/ijms19082190





