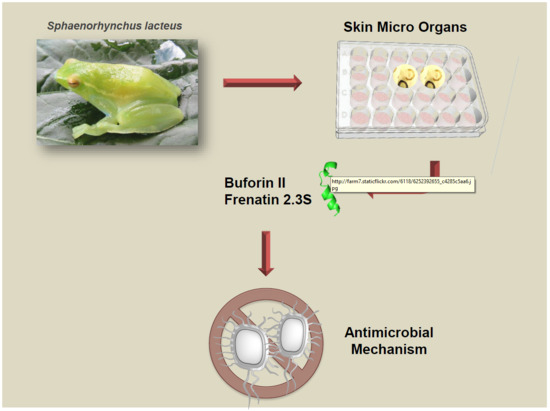
Unveiling the Multifaceted Mechanisms of Antibacterial Activity of Buforin II and Frenatin 2.3S Peptides from Skin Micro-Organs of the Orinoco Lime Treefrog (Sphaenorhynchus lacteus)
Abstract
1. Introduction
2. Results
2.1. Synthesis and Characterization of Physicochemical Properties of BF2 and F2.3S Peptides
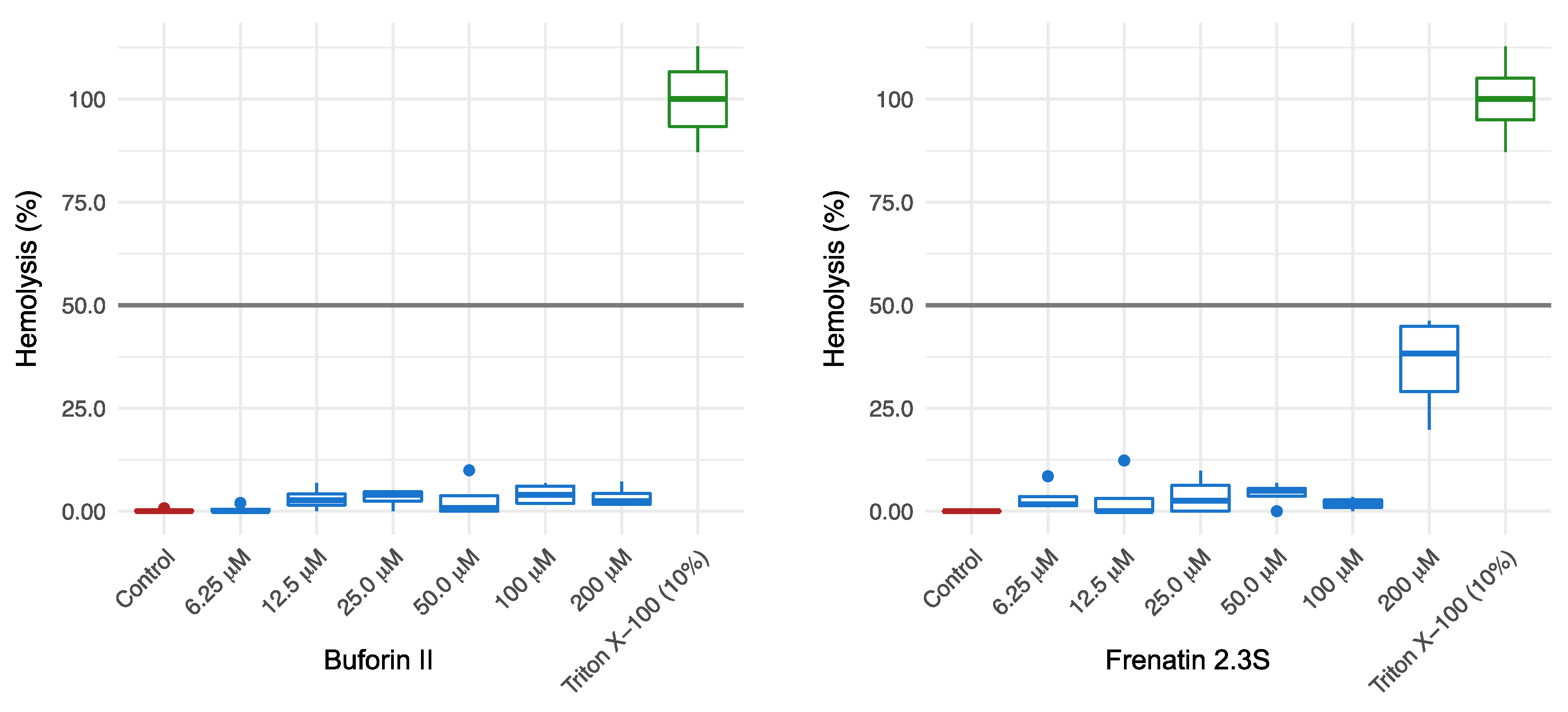
2.2. Antibacterial and Cytotoxic Activities of BF2 and F2.3S Peptides
2.3. Bacterial Membrane Depolarization, Permeabilization and Agglutination Activities of BF2 and F2.3S Peptides
2.4. Leakage and Agglutination Activities on Phospholipid Liposomes by BF2 and F2.3S Peptides
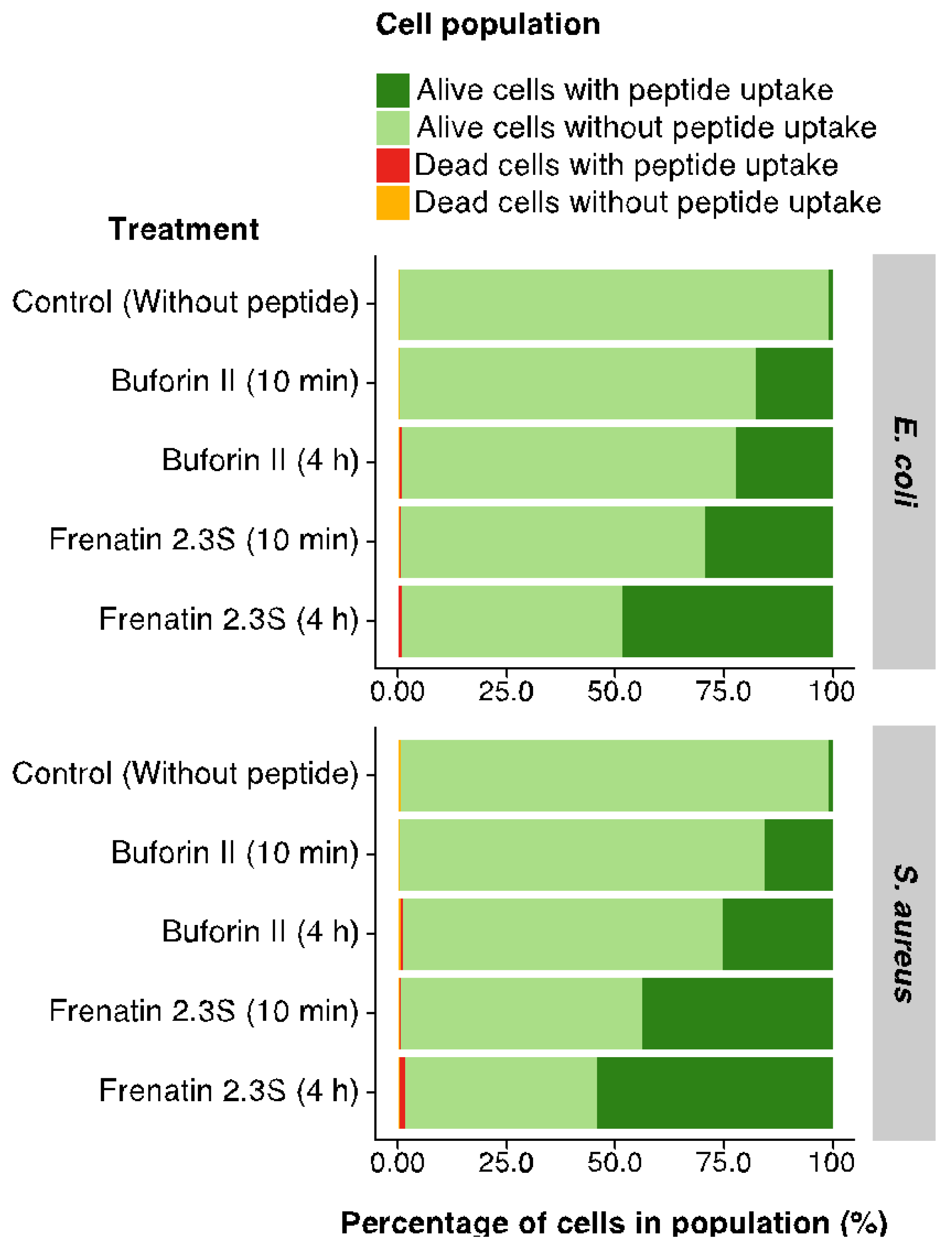
2.5. Internalization into Bacterial Cells and In Vitro DNA Interaction by BF2 and F2.3S Peptides
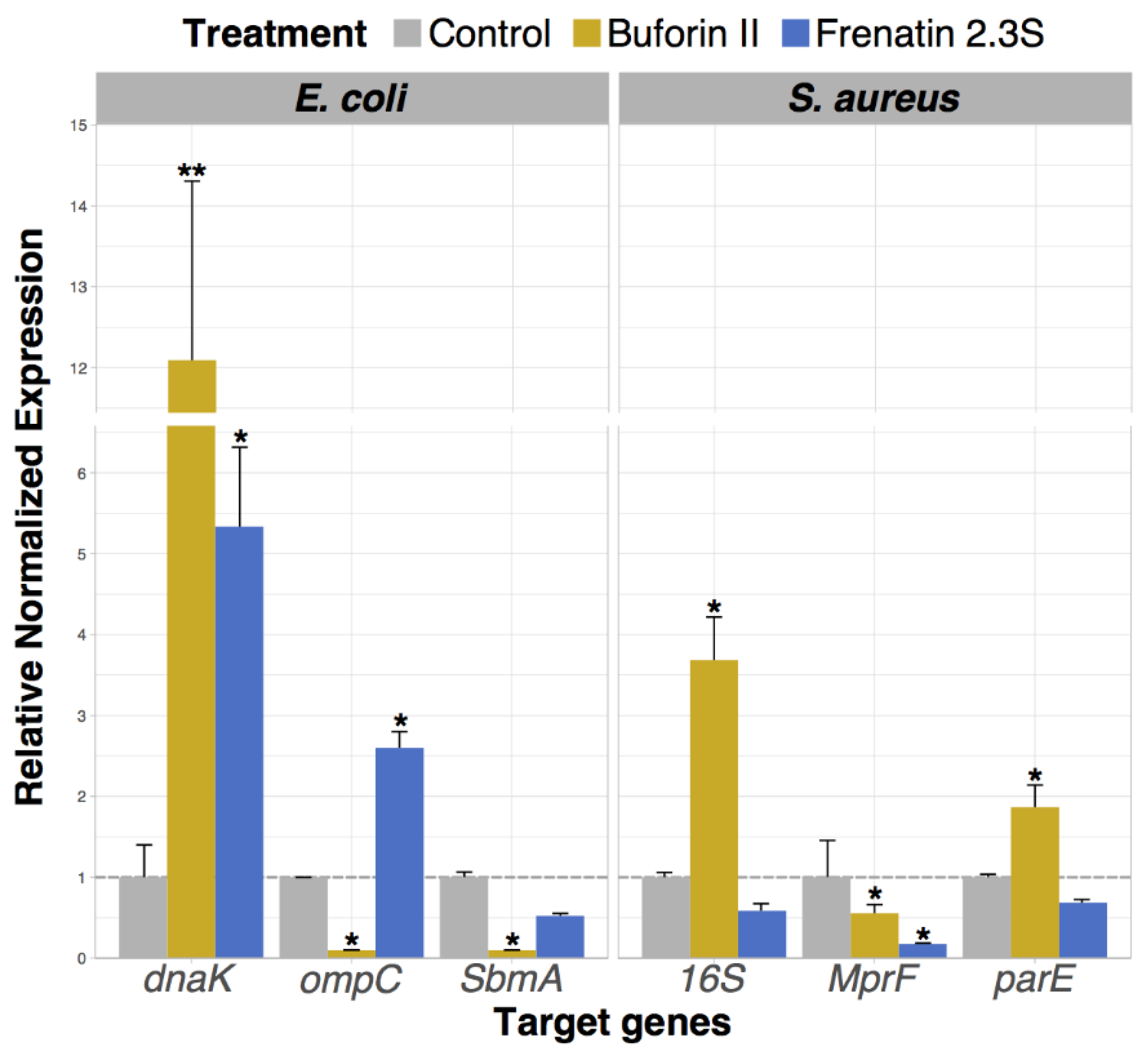
2.6. Analysis of Bacterial Gene Expression Profile Pattern Induced by Peptide Incubation
3. Discussion
4. Materials and Methods
4.1. In Silico Analysis
4.2. Peptide Synthesis
4.3. Minimum Bactericidal Concentration
4.4. Cell Viability Assay by ATP Quantification
4.5. Hemolysis Assay
4.6. Cytotoxicity Assay in Human Monocytes
4.7. Bacterial Cell Membrane Depolarization Assay
4.8. Bacterial Cell Leakage Assay
4.9. Minimum Agglutination Concentration (MAC)
4.10. Large Unilamellar Vesicle (LUV) Liposome Preparation
4.11. Liposome Membrane Leakage Activity
4.12. Dynamic Light Scattering (DLS)
4.13. Fluorescence-Activated Cell Sorter (FACS) Assay
4.14. DNA Binding Assay
4.15. Gene Expression Analysis
4.16. Real Time Quantitative PCR Polymerase Chain Reaction (RT-qPCR)
4.17. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| APD | Antimicrobial peptide databases |
| AMPs | Antimicrobial peptides |
| ANTS | 8-aminonaphthalene-1,3,6-trisulfonic acid |
| BF2 | Buforin II |
| CFU | Colony forming units |
| DLS | Dynamic light scattering |
| DOPC | 1,2-dioleoyl-sn-glycero-3-phosphocholine |
| DOPG | 1,2-Dioleoyl-sn-glycero-3-phosphoglycerol |
| DTT | Dithiothreitol |
| ED50 | 50% effective dose |
| FACS | Fluorescence-activated cell sorting |
| F2.3S | Frenatin 2.3S |
| IL | Interleukin |
| LB | Luria-Bertani media |
| LPS | Lipopolysaccharide |
| LUV | Large unilamellar vesicle |
| MAC | Minimum agglutination concentration |
| MBC | Minimum bactericidal concentrations |
| MPEx | Membrane protein explorer |
| SMOs | Skin micro-organs |
| FITC | Fluorescein |
| TNFα | Tumor necrosis factor-alpha |
References
- Kosikowska, P.; Lesner, A. Antimicrobial peptides (AMPs) as drug candidates: A patent review (2003–2015). Expert Opin. Ther. Pat. 2016, 26, 689–702. [Google Scholar] [CrossRef] [PubMed]
- Uhlig, T.; Kyprianou, T.; Martinelli, F.G.; Oppici, C.A.; Heiligers, D.; Hills, D.; Calvo, X.R.; Verhaert, P. The emergence of peptides in the pharmaceutical business: From exploration to exploitation. EuPA Open Proteom. 2014, 4, 58–69. [Google Scholar] [CrossRef]
- Rodríguez, C.; Rollins-Smith, L.; Ibáñez, R.; Durant-Archibold, A.A.; Gutiérrez, M. Toxins and pharmacologically active compounds from species of the family Bufonidae (Amphibia, Anura). J. Ethnopharmacol. 2016, 198, 235–254. [Google Scholar] [CrossRef] [PubMed]
- Le, C.-F.; Fang, C.-M.; Sekaran, S.D. Intracellular Targeting Mechanisms by Antimicrobial Peptides. Antimicrob. Agents Chemother. 2017, 61, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Hoeksema, M.; Van Eijk, M.; Haagsman, H.P.; Hartshorn, K.L. Histones as mediators of host defense, inflammation and thrombosis. Future Microbiol. 2016, 11, 441–453. [Google Scholar] [CrossRef] [PubMed]
- Park, C.B.C.; Kim, M.M.S.; Kim, S.C.S. A novel antimicrobial peptide from Bufo bufo gargarizans. Biochem. Biophys. Res. 1996, 218, 408–413. [Google Scholar] [CrossRef] [PubMed]
- Yi, G.; Bae, C.; Chang, S.; Cheong, C. Solution structure of an antimicrobial peptide buforin II. FEBS Lett. 1996, 398, 87–90. [Google Scholar] [CrossRef]
- Park, C.B.; Kim, H.S.; Kim, S.C. Mechanism of action of the antimicrobial peptide buforin II: Buforin II kills microorganisms by penetrating the cell membrane and inhibiting cellular functions. Biochem. Biophys. Res. Commun. 1998, 244, 253–257. [Google Scholar] [CrossRef] [PubMed]
- Pavia, K.E.; Spinella, S.A.; Elmore, D.E. Novel histone-derived antimicrobial peptides use different antimicrobial mechanisms. Biochim. Biophys. Acta Biomembr. 2012, 1818, 869–876. [Google Scholar] [CrossRef] [PubMed]
- Elmore, D.E. Insights into buforin II membrane translocation from molecular dynamics simulations. Peptides 2012, 38, 357–362. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.A.; Kim, H.; Lee, J.Y.; Shin, J.R.; Kim, D.J.; Cho, J.H.; Kim, S.C. Mechanism of action and specificity of antimicrobial peptides designed based on buforin IIb. Peptides 2012, 34, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Park, C.B.; Yi, K.-S.; Matsuzaki, K.; Kim, M.S.; Kim, S.C. Structure-activity analysis of buforin II, a histone H2A-derived antimicrobial peptide: The proline hinge is responsible for the cell-penetrating ability of buforin II. Proc. Natl. Acad. Sci. USA 2000, 97, 8245–8250. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, S.; Takeshima, K.; Park, C.B.; Kim, S.C.; Matsuzaki, K. Interactions of the novel anfimicrobial peptide buforin 2 with lipid bilayers: Proline as a translocation promoting factor. Biochemistry 2000, 39, 8648–8654. [Google Scholar] [CrossRef] [PubMed]
- Fleming, E.; Maharaj, N.P.; Chen, J.L.; Nelson, R.B.; Elmore, D.E. Effect of lipid composition on buforin II structure and membrane entry. Proteins Struct. Funct. Bioinform. 2008, 73, 480–491. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Peng, Y. Synergistic effect of clinically used antibiotics and peptide antibiotics against gram-positive and gram-negative bacteria. Exp. Ther. Med. 2013, 6, 1000–1004. [Google Scholar] [CrossRef] [PubMed]
- Raftery, M.J.; Waugh, R.J.; Bowie, J.H.; Wallace, J.C.; Tyler, M.J. The structures of the frenatin peptides from the skin secretion of the giant tree frog Litoria infrafrenata. J. Pept. Sci. 1996, 2, 117–124. [Google Scholar] [CrossRef]
- Pantic, J.M.; Jovanovic, I.P.; Radosavljevic, G.D.; Gajovic, N.M.; Arsenijevic, N.N.; Conlon, J.M.; Lukic, M.L. The frog skin host-defense peptide frenatin 2.1S enhances recruitment, activation and tumoricidal capacity of NK cells. Peptides 2017, 93, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Pantic, J.M.; Jovanovic, I.P.; Radosavljevic, G.D.; Arsenijevic, N.N.; Conlon, J.M.; Lukic, M.L. The potential of frog skin-derived peptides for development into therapeutically-valuable immunomodulatory agents. Molecules 2017, 22, 2071. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Camargo, C.; Méndez, M.C.; Salazar, V.; Moscoso, J.; Narváez, D.; Torres, M.M.; Florez, F.K.; Groot, H.; Mitrani, E. Frog skin cultures secrete anti-yellow fever compounds. J. Antibiot. 2016, 69, 783–790. [Google Scholar] [CrossRef] [PubMed]
- Conlon, J.M.; Mechkarska, M.; Radosavljevic, G.; Attoub, S.; King, J.D.; Lukic, M.L.; Mcclean, S. A family of antimicrobial and immunomodulatory peptides related to the frenatins from skin secretions of the Orinoco lime frog Sphaenorhynchus lacteus (Hylidae). Peptides 2014, 56, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Conlon, J.M.; Mechkarska, M.; Pantic, J.M.; Lukic, M.L.; Coquet, L.; Leprince, J.; Nielsen, P.F.; Rinaldi, A.C. An immunomodulatory peptide related to frenatin 2 from skin secretions of the Tyrrhenian painted frog Discoglossus sardus (Alytidae). Peptides 2013, 40, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Chen, T.; Walker, B.; Shaw, C. Novel frenatins from the skin of the Australasian giant white-lipped tree frog, Litoria infrafrenata: Cloning of precursor cDNAs and identification in defensive skin secretion. Peptides 2005, 26, 2445–2451. [Google Scholar] [CrossRef] [PubMed]
- Pantic, J.M.; Radosavljevic, G.D.; Jovanovic, I.P.; Arsenijevic, N.N.; Conlon, J.M.; Lukic, M.L. In vivo administration of the frog skin peptide frenatin 2.1S induces immunostimulatory phenotypes of mouse mononuclear cells. Peptides 2015, 71, 269–275. [Google Scholar] [CrossRef] [PubMed]
- König, E.; Bininda-Emonds, O.R.P.; Shaw, C. The diversity and evolution of anuran skin peptides. Peptides 2015, 63, 96–117. [Google Scholar] [CrossRef] [PubMed]
- Groot, H.; Muñoz-Camargo, C.; Moscoso, J.; Riveros, G.; Salazar, V.; Kaston Florez, F.; Mitrani, E. Skin micro-organs from several frog species secrete a repertoire of powerful antimicrobials in culture. J. Antibiot. 2012, 65, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Li, X.; Wang, Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016, 44. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999, 292, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Te Winkel, J.D.; Gray, D.A.; Seistrup, K.H.; Hamoen, L.W.; Strahl, H. Analysis of Antimicrobial-Triggered Membrane Depolarization Using Voltage Sensitive Dyes. Front. Cell Dev. Biol. 2016, 4, 29. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, S.; Chikushi, A.; Tougu, S.; Imura, Y.; Nishida, M.; Yano, Y.; Matsuzaki, K. Membrane translocation mechanism of the antimicrobial peptide buforin 2. Biochemistry 2004, 43, 15610–15616. [Google Scholar] [CrossRef] [PubMed]
- Mattiuzzo, M.; Bandiera, A.; Gennaro, R.; Benincasa, M.; Pacor, S.; Antcheva, N.; Scocchi, M. Role of the Escherichia coli SbmA in the antimicrobial activity of proline-rich peptides. Mol. Microbiol. 2007, 66, 151–163. [Google Scholar] [CrossRef] [PubMed]
- Andrä, J.; Goldmann, T.; Ernst, C.M.; Peschel, A.; Gutsmann, T. Multiple peptide resistance factor (MprF)-mediated resistance of Staphylococcus aureus against antimicrobial peptides coincides with a modulated peptide interaction with artificial membranes comprising lysyl-phosphatidylglycerol. J. Biol. Chem. 2011, 286, 18692–18700. [Google Scholar] [CrossRef] [PubMed]
- Datrie, M.; Schumann, M.; Wieprecht, T.; Winkler, A.; Beyermann, M.; Krause, E.; Matsuzaki, K.; Murase, O.; Bienert, M. Peptide helicity and membrane surface charge modulate the balance of electrostatic and hydrophobic interactions with lipid bilayers and biological membranes. Biochemistry 1996, 35, 12612–12622. [Google Scholar] [CrossRef]
- Kacprzyk, L.; Rydengård, V.; Mörgelin, M.; Davoudi, M.; Pasupuleti, M.; Malmsten, M.; Schmidtchen, A. Antimicrobial activity of histidine-rich peptides is dependent on acidic conditions. Biochim. Biophys. Acta Biomembr. 2007, 1768, 2667–2680. [Google Scholar] [CrossRef] [PubMed]
- Torrent, M.; Andreu, D.; Nogués, V.M.; Boix, E. Connecting peptide physicochemical and antimicrobial properties by a rational prediction model. PLoS ONE 2011, 6, e16968. [Google Scholar] [CrossRef] [PubMed]
- Dathe, M.; Wieprecht, T.; Nikolenko, H.; Handel, L.; Maloy, W.L.; MacDonald, D.L.; Beyermann, M.; Bienert, M. Hydrophobicity, hydrophobic moment and angle subtended by charged residues modulate antibacterial and haemolytic activity of amphipathic helical peptides. FEBS Lett. 1997, 403, 208–212. [Google Scholar] [CrossRef]
- Nicolas, P. Multifunctional host defense peptides: Intracellular-targeting antimicrobial peptides. FEBS J. 2009, 276, 6483–6496. [Google Scholar] [CrossRef] [PubMed]
- Stalmans, S.; Wynendaele, E.; Bracke, N.; Gevaert, B.; D’Hondt, M.; Peremans, K.; Burvenich, C.; De Spiegeleer, B. Chemical-functional diversity in cell-penetrating peptides. PLoS ONE 2013, 8, e71752. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Xia, Y.; Li, D.; Du, Q.; Liang, D. Relationship between peptide structure and antimicrobial activity as studied by de novo designed peptides. Biochim. Biophys. Acta Biomembr. 2014, 1838, 2985–2993. [Google Scholar] [CrossRef] [PubMed]
- Caillon, L.; Killian, J.A.; Lequin, O.; Khemtémourian, L. Biophysical Investigation of the Membrane-Disrupting Mechanism of the Antimicrobial and Amyloid-Like Peptide Dermaseptin S9. PLoS ONE 2013, 8, e75528. [Google Scholar] [CrossRef] [PubMed]
- Torrent, M.; Pulido, D.; Valle, J.; Nogués, M.V.; Andreu, D.; Boix, E. Ribonucleases as a host-defence family: Evidence of evolutionarily conserved antimicrobial activity at the N-terminus. Biochem. J. 2013, 456, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Wan, M.; van der Does, A.M.; Tang, X.; Lindbom, L.; Agerberth, B.; Haeggstrom, J.Z. Antimicrobial peptide LL-37 promotes bacterial phagocytosis by human macrophages. J. Leukoc. Biol. 2014, 95, 971–981. [Google Scholar] [CrossRef] [PubMed]
- Giacometti, A.; Cirioni, O.; Ghiselli, R.; Mocchegiani, F.; Del Prete, M.S.; Viticchi, C.; Kamysz, W.; ŁEmpicka, E.; Saba, V.; Scalise, G. Potential therapeutic role of cationic peptides in three experimental models of septic shock. Antimicrob. Agents Chemother. 2002, 46, 2132–2136. [Google Scholar] [CrossRef] [PubMed]
- Gough, M.; Hancock, R.E.W.; Kelly, N.M. Antiendotoxin activity of cationic peptide antimicrobial agents. Infect. Immun. 1996, 64, 4922–4927. [Google Scholar] [PubMed]
- Torrent, M.; Pulido, D.; Nogués, M.V.; Boix, E. Exploring new biological functions of amyloids: Bacteria cell agglutination mediated by host protein aggregation. PLoS Pathog. 2012, 8, e1003005. [Google Scholar] [CrossRef] [PubMed]
- Pulido, D.; Torrent, M.; Andreu, D.; Nogués, M.V.; Boix, E.; Nogues, M.V.; Boix, E. Two human host defense ribonucleases against mycobacteria, the eosinophil cationic protein (RNase 3) and RNase 7. Antimicrob. Agents Chemother. 2013, 57, 3797–3805. [Google Scholar] [CrossRef] [PubMed]
- Hao, G.; Shi, Y.H.; Tang, Y.L.; Le, G.W. The intracellular mechanism of action on Escherichia coli of BF2-A/C, two analogues of the antimicrobial peptide Buforin 2. J. Microbiol. 2013, 51, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Sim, S.; Wang, P.; Beyer, B.N.; Cutrona, K.J.; Radhakrishnan, M.L.; Elmore, D.E. Investigating the nucleic acid interactions of histone-derived antimicrobial peptides. FEBS Lett. 2017, 591, 706–717. [Google Scholar] [CrossRef] [PubMed]
- Calloni, G.; Chen, T.; Schermann, S.M.; Chang, H.C.; Genevaux, P.; Agostini, F.; Tartaglia, G.G.; Hayer-Hartl, M.; Hartl, F.U. DnaK Functions as a Central Hub in the E. coli Chaperone Network. Cell Rep. 2012, 1, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Wang, C.; Guo, C.; Tian, Y.; Li, H.; Peng, X. Differential regulation of OmpC and OmpF by AtpB in Escherichia coli exposed to nalidixic acid and chlortetracycline. J. Proteom. 2012, 75, 5898–5910. [Google Scholar] [CrossRef] [PubMed]
- Viveiros, M.; Dupont, M.; Rodrigues, L.; Couto, I.; Davin-Regli, A.; Martins, M.; Pagès, J.M.; Amaral, L. Antibiotic stress, genetic response and altered permeability of E. coli. PLoS ONE 2007, 2, e365. [Google Scholar] [CrossRef] [PubMed]
- Marcellini, L.; Borro, M.; Gentile, G.; Rinaldi, A.C.; Stella, L.; Aimola, P.; Barra, D.; Mangoni, M.L. Esculentin-1b(1-18)—A membrane-active antimicrobial peptide that synergizes with antibiotics and modifies the expression level of a limited number of proteins in Escherichia coli. FEBS J. 2009, 276, 5647–5664. [Google Scholar] [CrossRef] [PubMed]
- Glazebrook, J.; Ichige, A.; Walker, G.C. A Rhizobium meliloti homolog of the Escherichia coli peptide-antibiotic transport protein SbmA is essential for bacteroid development. Genes Dev. 1993, 7, 1485–1497. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, S.; Modak, J.K.; Ryan, C.S.; Garcia-Bustos, J.; Davies, J.K.; Roujeinikova, A. Mechanism of escherichia coli resistance to pyrrhocoricin. Antimicrob. Agents Chemother. 2014, 58, 2754–2762. [Google Scholar] [CrossRef] [PubMed]
- Van Boxtel, R.; Wattel, A.A.; Arenas, J.; Goessens, W.H.F.; Tommassen, J. Acquisition of carbapenem resistance by plasmid-encoded-AmpC-expressing Escherichia coli. Antimicrob. Agents Chemother. 2017, 61, e01413-16. [Google Scholar] [CrossRef] [PubMed]
- Paulsen, V.S.; Mardirossian, M.; Blencke, H.M.; Benincasa, M.; Runti, G.; Nepa, M.; Haug, T.; Stensvåg, K.; Scocchi, M. Inner membrane proteins YgdD and SbmA are required for the complete susceptibility of escherichia coli to the proline-rich antimicrobial peptide arasin 1(1-25). Microbiology 2016. [Google Scholar] [CrossRef] [PubMed]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Andersson, D.I.; Hughes, D.; Kubicek-Sutherland, J.Z. Mechanisms and consequences of bacterial resistance to antimicrobial peptides. Drug Resist. Updates 2016, 26, 43–57. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, K. Magainins as paradigm for the mode of action of pore forming polypeptides. Biochim. Biophys. Acta 1998, 1376, 391–400. [Google Scholar] [CrossRef]
- Epand, R.M.; Vogel, H.J. Diversity of antimicrobial peptides and their mechanisms of action. Biochim. Biophys. Acta Biomembr. 1999, 1462, 11–28. [Google Scholar] [CrossRef]
- Marquette, A.; Bechinger, B. Biophysical Investigations Elucidating the Mechanisms of Action of Antimicrobial Peptides and Their Synergism. Biomolecules 2018, 8, 18. [Google Scholar] [CrossRef] [PubMed]
- Katzenback, B. Antimicrobial Peptides as Mediators of Innate Immunity in Teleosts. Biology 2015, 4, 607–639. [Google Scholar] [CrossRef] [PubMed]
- Arranz-Trullén, J.; Lu, L.; Pulido, D.; Bhakta, S.; Boix, E. Host antimicrobial peptides: The promise of new treatment strategies against tuberculosis. Front. Immunol. 2017, 8, 1499. [Google Scholar] [CrossRef] [PubMed]
- Hasson, E.; Gallula, J.; Shimoni, Y.; Grad-Itach, E.; Marikovsky, M.; Mitrani, E. Skin-derived micro-organs induce angiogenesis in rabbits. J. Vasc. Res. 2006, 43, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Lipsky, B.A.; Holroyd, K.J.; Zasloff, M. Topical versus Systemic Antimicrobial Therapy for Treating Mildly Infected Diabetic Foot Ulcers: A Randomized, Controlled, Double-Blinded, Multicenter Trial of Pexiganan Cream. Clin. Infect. Dis. 2008, 47, 1537–1545. [Google Scholar] [CrossRef] [PubMed]
- Mangoni, M.L.; Mcdermott, A.M.; Zasloff, M. Antimicrobial peptides and wound healing: Biological and therapeutic considerations. Exp. Dermatol. 2016, 25, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Snider, C.; Jayasinghe, S.; Hristova, K.; White, S.H. MPEx: A tool for exploring membrane proteins. Protein Sci. 2009, 18, 2624–2628. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Sharma, A.; Singla, D.; Rashid, M.; Pal, G.; Raghava, S. Designing of peptides with desired half-life in intestine-like environment. BMC Bioinform. 2014, 15. [Google Scholar] [CrossRef] [PubMed]
- Pulido, D.; Prats-Ejarque, G.; Villalba, C.; Albacar, M.; González-López, J.J.; Torrent, M.; Moussaoui, M.; Boix, E. A novel RNase 3/ECP peptide for Pseudomonas aeruginosa biofilm eradication that combines antimicrobial, lipopolysaccharide binding, and cell-agglutinating activities. Antimicrob. Agents Chemother. 2016, 60, 6313–6325. [Google Scholar] [CrossRef] [PubMed]
- Salazar, V.A.; Rubin, J.; Moussaoui, M.; Pulido, D.; Nogués, M.V.; Venge, P.; Boix, E. Protein post-translational modification in host defense: The antimicrobial mechanism of action of human eosinophil cationic protein native forms. FEBS J. 2014, 281, 5432–5446. [Google Scholar] [CrossRef] [PubMed]
- Evans, B.C.; Nelson, C.E.; Yu, S.S.; Beavers, K.R.; Kim, A.J.; Li, H.; Nelson, H.M.; Giorgio, T.D.; Duvall, C.L. Ex vivo red blood cell hemolysis assay for the evaluation of pH-responsive endosomolytic agents for cytosolic delivery of biomacromolecular drugs. J. Vis. Exp. 2013, e50166. [Google Scholar] [CrossRef] [PubMed]
- Torrent, M.; Badia, M.; Moussaoui, M.; Sanchez, D.; Nogues, M.V.; Boix, E. Comparison of human RNase 3 and RNase 7 bactericidal action at the Gram-negative and Gram-positive bacterial cell wall. FEBS J. 2010, 277, 1713–1725. [Google Scholar] [CrossRef] [PubMed]
- Torrent, M.; Nogués, M.V.; Boix, E. Eosinophil cationic protein (ECP) can bind heparin and other glycosaminoglycans through its RNase active site. J. Mol. Recognit. 2011, 24, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Torrent, M.; Cuyás, E.; Carreras, E.; Navarro, S.; López, O.; de la Maza, A.; Nogués, M.V.; Reshetnyak, Y.K.; Boix, E. Topography studies on the membrane interaction mechanism of the eosinophil cationic protein. Biochemistry 2007, 46, 720–733. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Ream, J.A.; Lewis, L.K.; Lewis, K.A. Rapid agarose gel electrophoretic mobility shift assay for quantitating protein: RNA interactions. Anal. Biochem. 2016, 511, 36–41. [Google Scholar] [CrossRef] [PubMed]

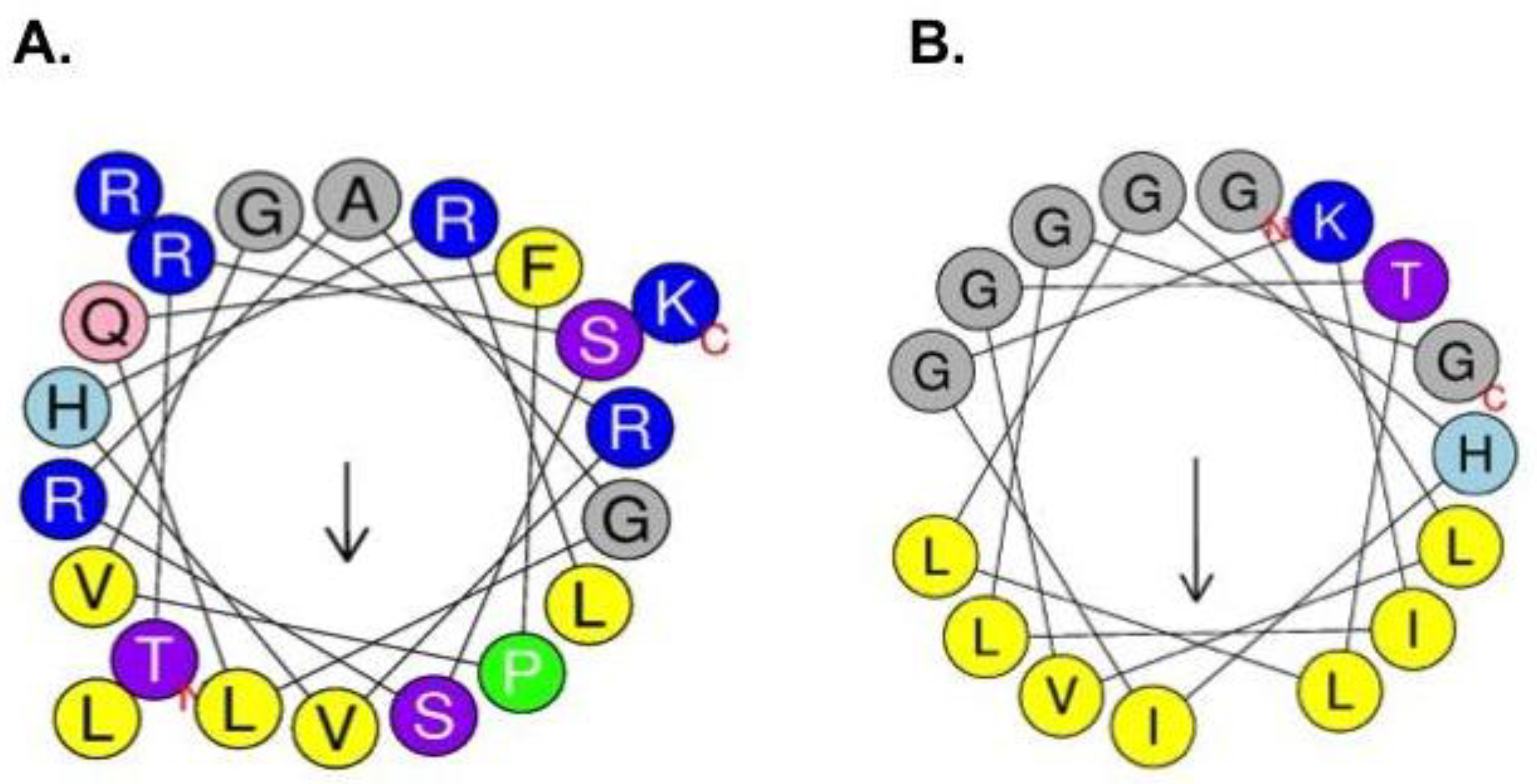

| Physicochemical Properties | Peptides | |
|---|---|---|
| BF2 | F2.3S | |
| Theoretical mass (Da) | 2434 | 1570 |
| Experimental mass (Da) 1 | 2434.88 | 1575.93 |
| Net charge | +6 | +1 |
| Isoelectric point | 12.44 | 9.07 |
| GRAVY | −0.63 | 1.17 |
| Hydrophobic ratio | 33% | 47% |
| W-W Hydrophobicity 2 (Kcal/mol) | 7.73 | 5.55 |
| Boman index (Kcal/mol) | 3.34 | −1.66 |
| Stability | Unstable | Stable |
| Half-life in vitro (Mammalian reticulocytes) 3 | 7.2 h | 30 h |
| Half-life in vivo (Yeast) 3 | >20 h | >10 h |
| Half-life in vivo (E. coli) 3 | >20 h | >10 h |
| Biological Activity | Peptide | ||
|---|---|---|---|
| BF2 | F2.3S | ||
| Gram negative | MBC100 (µM) | 0.93 | 0.62 |
| E. coli (BL21) | ED50 (µM) | 0.33 ± 0.005 | 0.23 ± 0.01 |
| ED50 (µM) | 1.3 ± 0.082 | 3.4 ± 0.062 | |
| P. aeruginosa (PA01) | ED50 (µM) | 1.8 ± 0.005 | 3.1 ± 0.003 |
| P. aeruginosa (PA14) | ED50 (µM) | >100 | >100 |
| P. aeruginosa (M8C1) * | ED50 (µM) | >100 | >100 |
| P. aeruginosa (M18C1) * | |||
| Gram positive | MBC100 (µM) | 1.87 | 1.87 |
| S. aureus (ATCC 502A) | ED50 (µM) | 0.51 ± 0.02 | 1.1 ± 0.07 |
| ED50 (µM) | 6.46 ± 0.002 | 11.99 ± 0.001 | |
| S.aureus (39413) * | ED50 (µM) | >100 | 55.82 ± 0.005 |
| S.aureus (34026) * | ED50 (µM) | >100 | >100 |
| S.aureus (36055) * | |||
| Human monocytes | CC50 (µM) | >100 | >100 |
| Biological Activity | Peptide | ||
|---|---|---|---|
| BF2 | F2.3S | ||
| Depolarization (ED50) (µM) | Bacteria | >5 | 0.1 ± 0.05 |
| Leakage (ED50) (µM) | Bacteria | 0.9 ± 0.1 | 0.1 ± 0.07 |
| Liposomes | >2 | 0.1 | |
| Agglutination (MAC) (µM) | Bacteria | 1.5 | >5 |
| Liposomes | 0.5 ± 0.002 | >2 | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muñoz-Camargo, C.; Salazar, V.A.; Barrero-Guevara, L.; Camargo, S.; Mosquera, A.; Groot, H.; Boix, E. Unveiling the Multifaceted Mechanisms of Antibacterial Activity of Buforin II and Frenatin 2.3S Peptides from Skin Micro-Organs of the Orinoco Lime Treefrog (Sphaenorhynchus lacteus). Int. J. Mol. Sci. 2018, 19, 2170. https://doi.org/10.3390/ijms19082170
Muñoz-Camargo C, Salazar VA, Barrero-Guevara L, Camargo S, Mosquera A, Groot H, Boix E. Unveiling the Multifaceted Mechanisms of Antibacterial Activity of Buforin II and Frenatin 2.3S Peptides from Skin Micro-Organs of the Orinoco Lime Treefrog (Sphaenorhynchus lacteus). International Journal of Molecular Sciences. 2018; 19(8):2170. https://doi.org/10.3390/ijms19082170
Chicago/Turabian StyleMuñoz-Camargo, Carolina, Vivian A. Salazar, Laura Barrero-Guevara, Sandra Camargo, Angela Mosquera, Helena Groot, and Ester Boix. 2018. "Unveiling the Multifaceted Mechanisms of Antibacterial Activity of Buforin II and Frenatin 2.3S Peptides from Skin Micro-Organs of the Orinoco Lime Treefrog (Sphaenorhynchus lacteus)" International Journal of Molecular Sciences 19, no. 8: 2170. https://doi.org/10.3390/ijms19082170
APA StyleMuñoz-Camargo, C., Salazar, V. A., Barrero-Guevara, L., Camargo, S., Mosquera, A., Groot, H., & Boix, E. (2018). Unveiling the Multifaceted Mechanisms of Antibacterial Activity of Buforin II and Frenatin 2.3S Peptides from Skin Micro-Organs of the Orinoco Lime Treefrog (Sphaenorhynchus lacteus). International Journal of Molecular Sciences, 19(8), 2170. https://doi.org/10.3390/ijms19082170