Constitutive Expression of Aechmea fasciata SPL14 (AfSPL14) Accelerates Flowering and Changes the Plant Architecture in Arabidopsis
Abstract
1. Introduction
2. Results
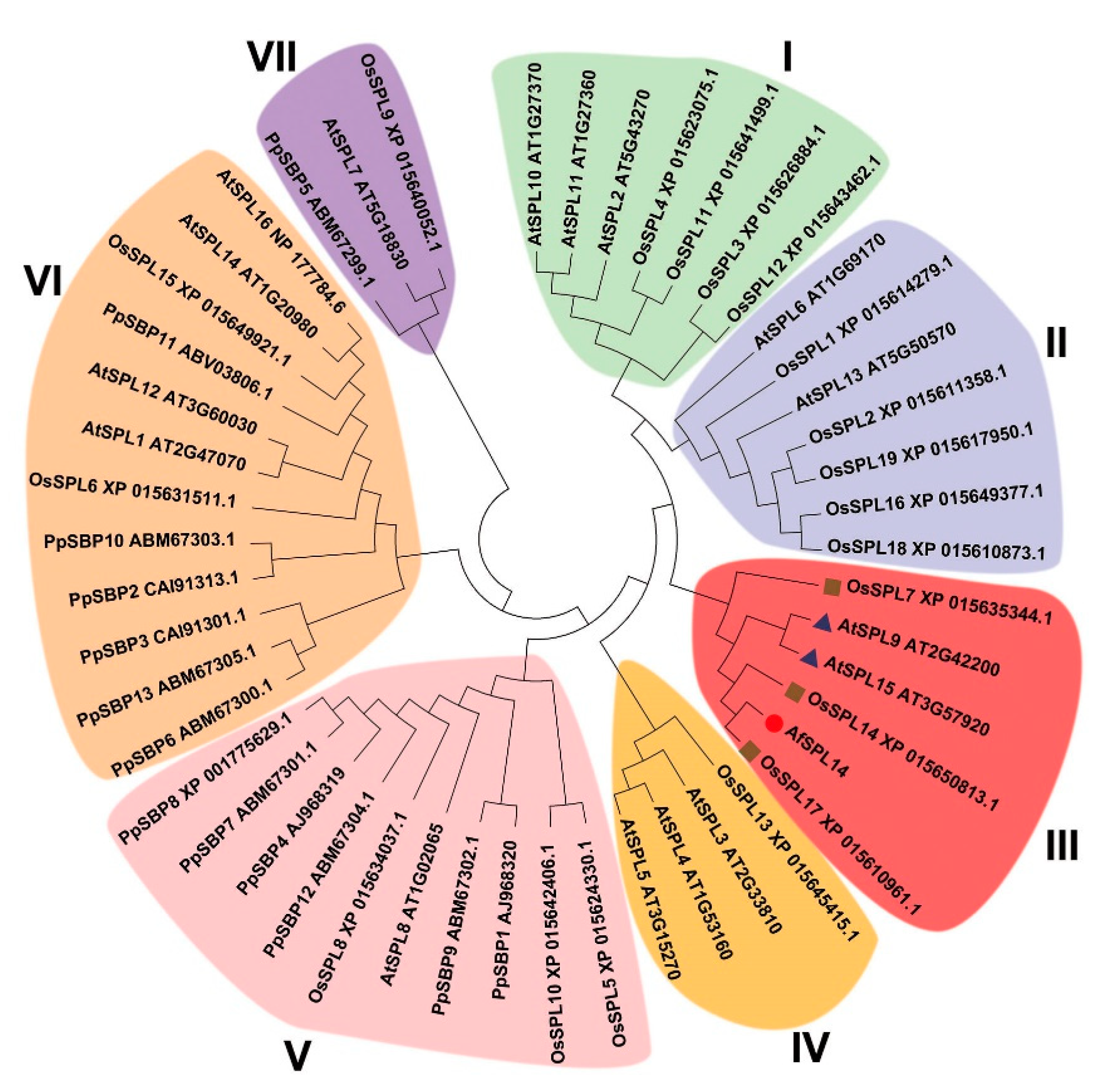
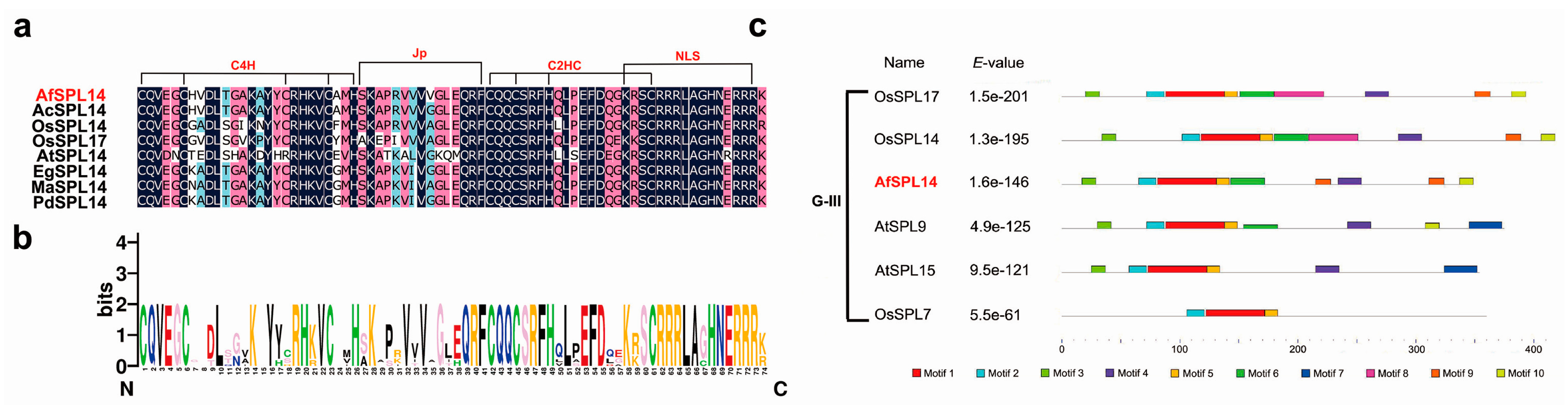
2.1. Isolation and Sequence Analysis of AfSPL14 in A. fasciata
2.2. AfSPL14 Was a Target of miR156 of A. fasciata (AfmiR156)
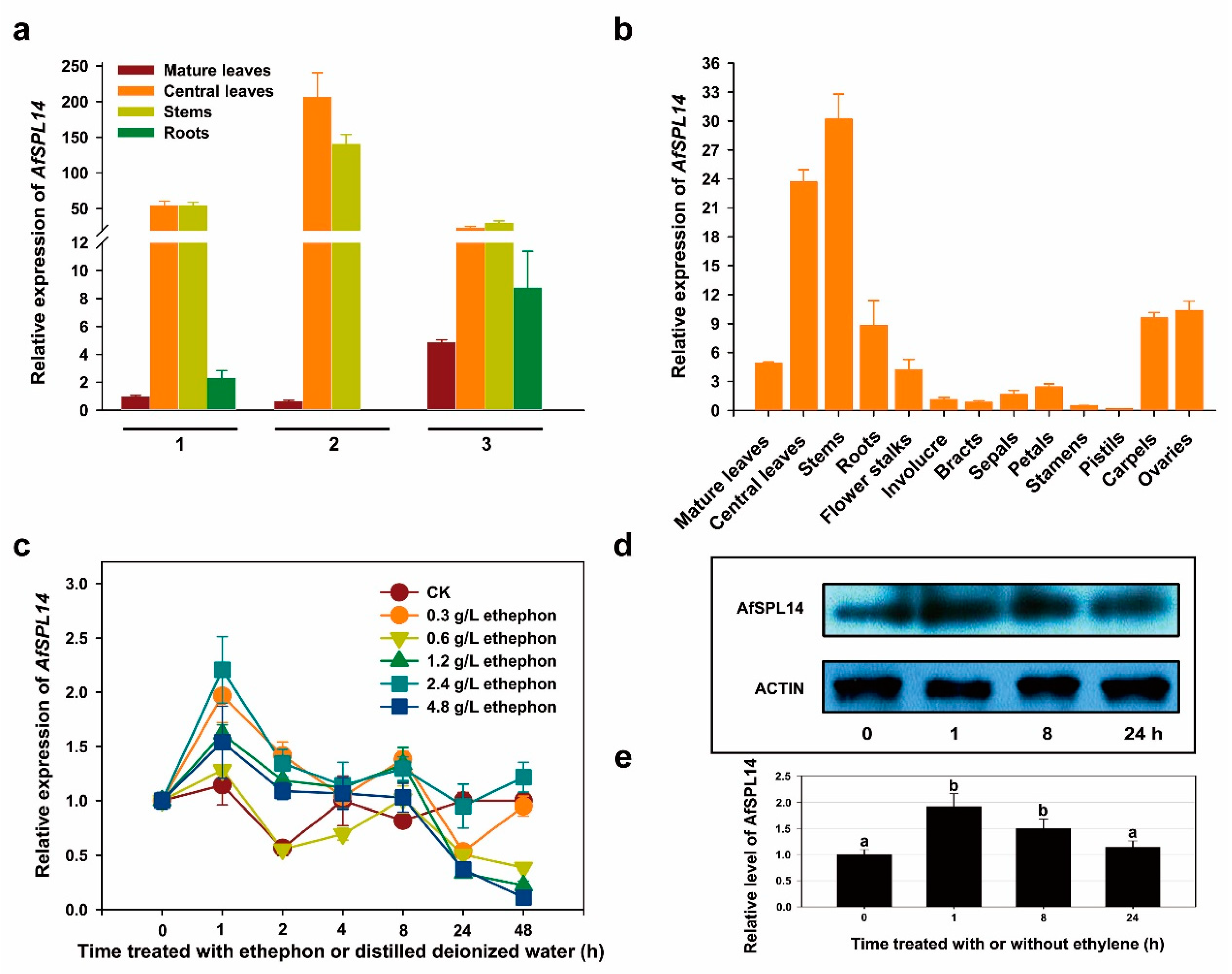
2.3. Transcript Profiling of AfSPL14 in A. fasciata
2.4. Response to Exogenous Ethephon Treatment
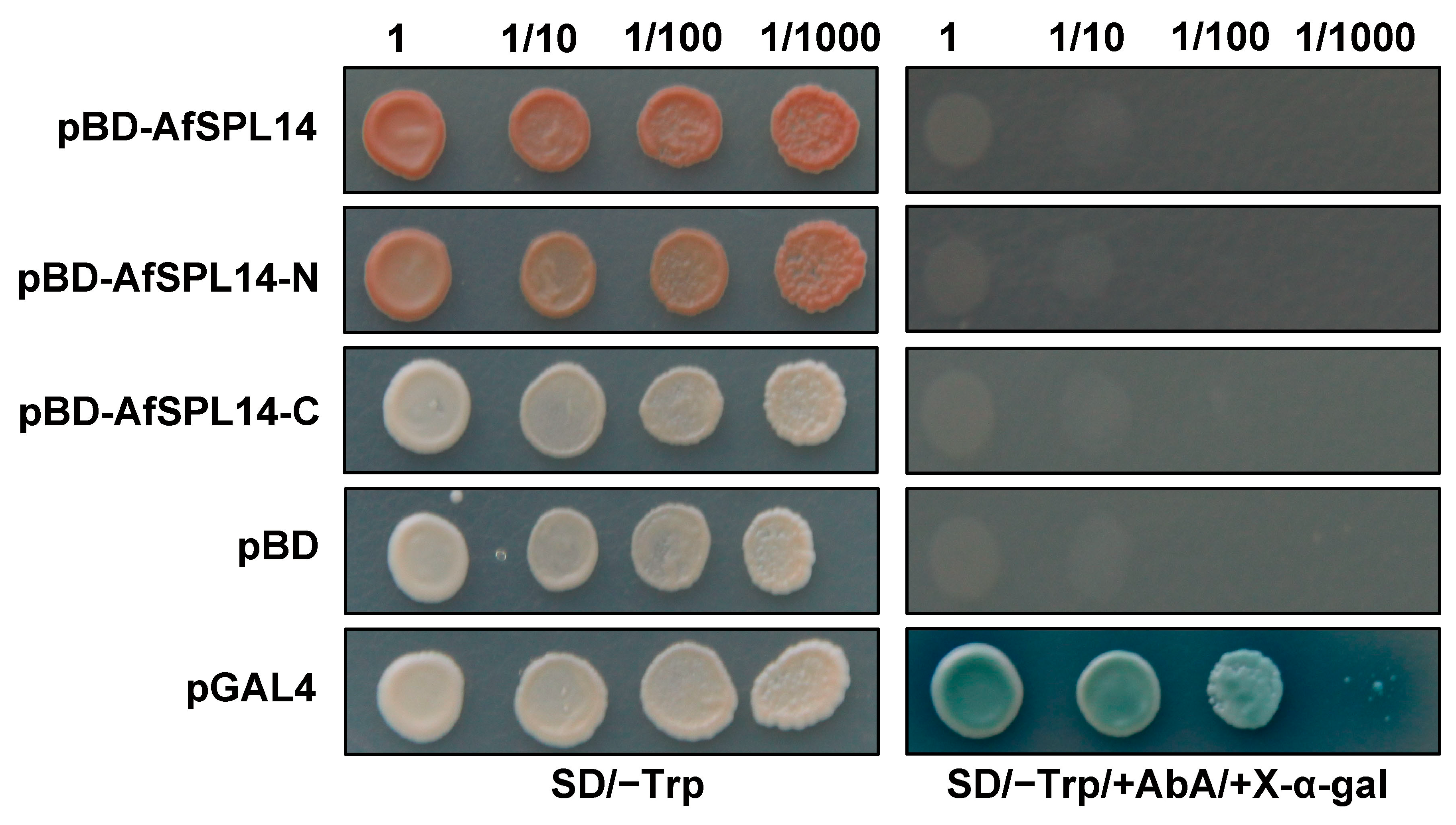
2.5. AfSPL14 Does Not Exhibit Transactivation Activity in Yeast
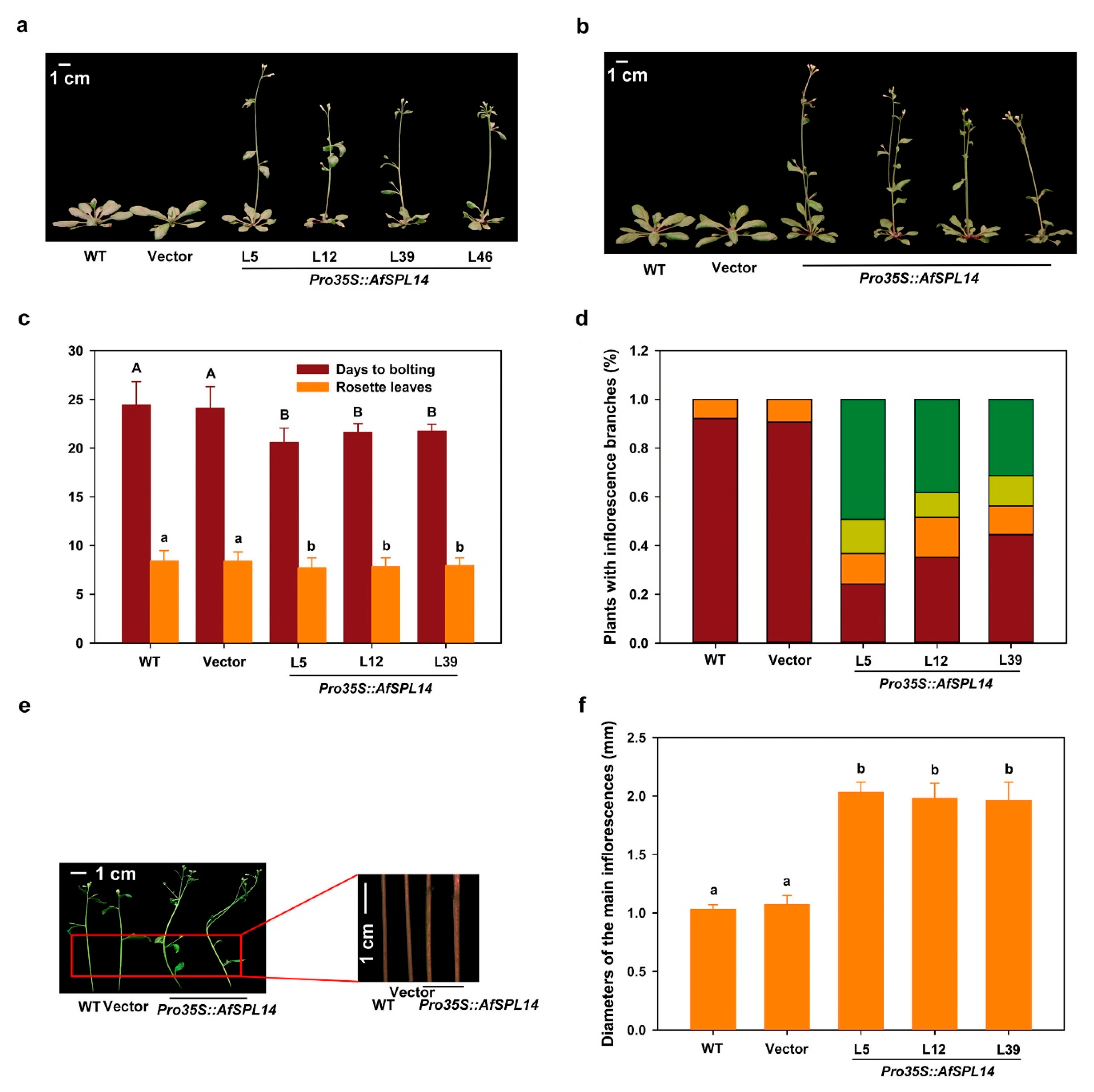
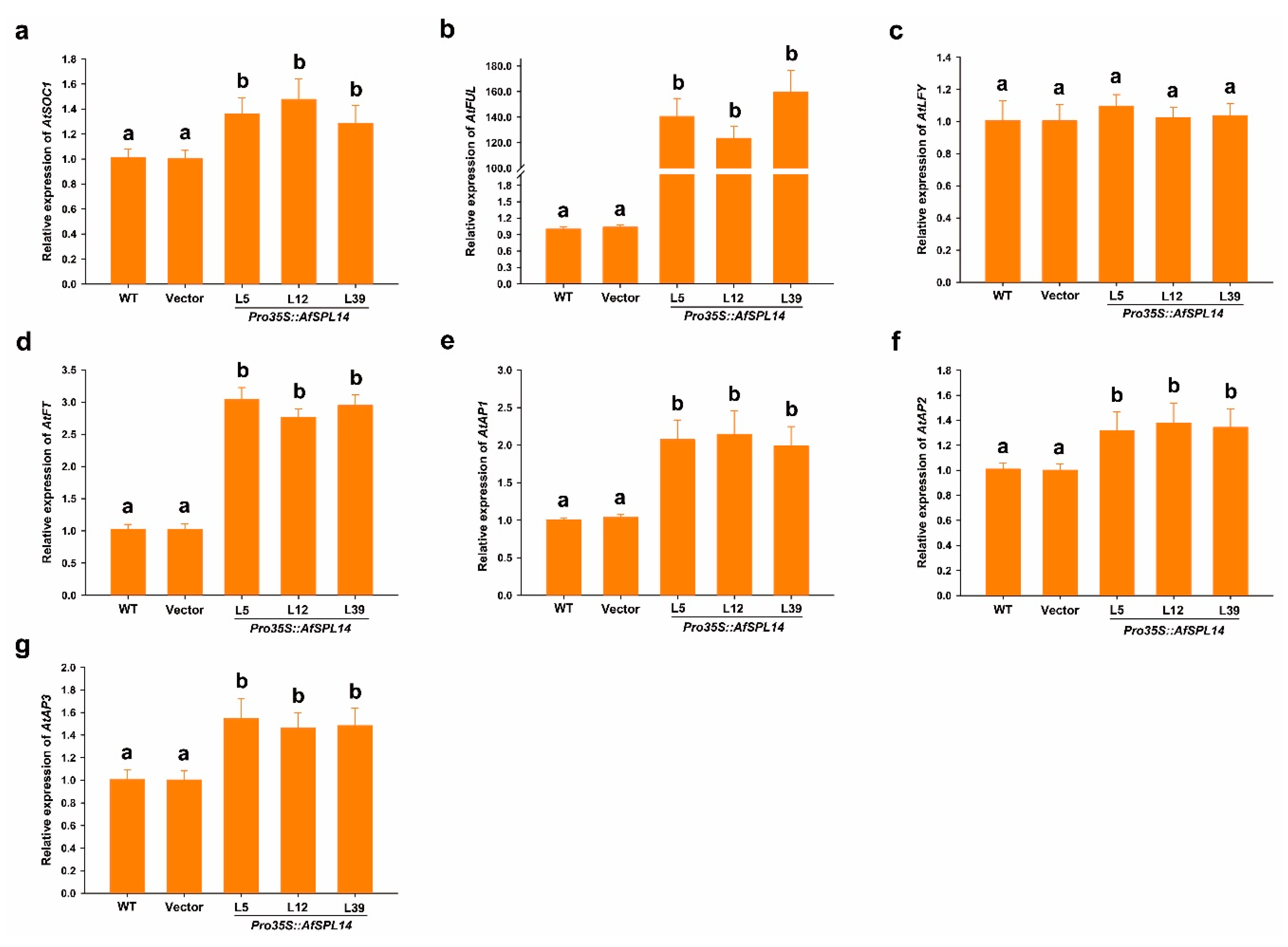
2.6. Constitutive Expression of AfSPL14 in Arabidopsis
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Sample Preparation
4.2. Isolation and Sequencing of the AfSPL14 Gene
4.3. Bioinformatic Analysis
4.4. 5′ RLM-RACE
4.5. RT-qPCR
4.6. Transgenic Plants
4.7. Transactivation Analysis of AfSPL14 in Yeast Cells
4.8. Exogenous Ethephon Treatment of A. fasciata
4.9. SDS-PAGE and Immunoblot Analysis
4.10. Data Analysis
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| SBP | SQUAMOSA PROMOTER BINDING PROTEIN |
| SPL | SBP-LIKE |
| miRNA | microRNA |
| TGA1 | TEOSINTE GLUME ARCHITECTURE |
| LD | long day |
| RACE | Rapid amplification of cDNA ends |
| UTR | Untranslated region |
| ORF | Open reading frame |
| MW | Molecular weight |
| pI | Isoelectric point |
| Jp | Joint peptide |
| NLS | Nuclear localization signal |
| RT-qPCR | Reverse transcription followed by quantitative real-time PCR |
| RLM-RACE | RNA ligase mediated rapid amplification of cDNA ends |
| DAF | Day after flowering |
| WT | Wild Type |
| SOC1 | SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 |
| FUL | FRUITFULL |
| AP1 | APETALA1 |
| LFY | LEAFY |
| FT | Flowering Locus T |
| AP2 | APETALA2 |
| AP3 | APETALA3 |
| CAM | Crassulacean acid metabolism |
| EIN3 | ETHYLENE INSENSITIVE 3 |
| GA | Gibberellic acid |
| CTAB | Hexadecyl trimethyl ammonium bromide |
| CDS | the coding sequence |
| SDS-PAGE | Sodium dodecyl sulfate polyacrylamide gel electrophoresis |
References
- Preston, J.C.; Hileman, L.C. Functional evolution in the plant SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) gene family. Front. Plant Sci. 2013, 4, 80. [Google Scholar] [CrossRef] [PubMed]
- Klein, J.; Saedler, H.; Huijser, P. A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol. Gen. Genet. 1996, 250, 7–16. [Google Scholar] [PubMed]
- Cardon, G.H.; Hohmann, S.; Nettesheim, K.; Saedler, H.; Huijser, P. Functional analysis of the Arabidopsis thaliana SBP-box gene SPL3: A novel gene involved in the floral transition. Plant J. 1997, 12, 367–377. [Google Scholar] [CrossRef] [PubMed]
- Arazi, T.; Talmor-Neiman, M.; Stav, R.; Riese, M.; Huijser, P.; Baulcombe, D.C. Cloning and characterization of micro-RNAs from moss. Plant J. 2005, 43, 837–848. [Google Scholar] [CrossRef] [PubMed]
- Riese, M.; Höhmann, S.; Saedler, H.; Münster, T.; Huijser, P. Comparative analysis of the SBP-box gene families in P. patens and seed plants. Gene 2007, 401, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, K.; Kigawa, T.; Inoue, M.; Tateno, M.; Yamasaki, T.; Yabuki, T.; Aoki, M.; Seki, E.; Matsuda, T.; Nunokawa, E.; Ishizuka, Y.; et al. A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors. J. Mol. Biol. 2004, 337, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Moreno, M.A.; Harper, L.C.; Krueger, R.W.; Dellaporta, S.L.; Freeling, M. Ligulelessl encodes a nuclear-localized protein required for induction of ligules and auricles during maize leaf organogenesis. Genes Dev. 1997, 11, 616–628. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Park, J.J.; Kim, S.L.; Yim, J.; An, G. Mutations in the rice liguleless gene result in a complete loss of the auricle, ligule, and laminar joint. Plant Mol. Biol. 2007, 65, 487–499. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.W.; Schwab, R.; Czech, B.; Mica, E.; Weigel, D. Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 2008, 20, 1231–1243. [Google Scholar] [CrossRef] [PubMed]
- Shikata, M.; Koyama, T.; Mitsuda, N.; Ohme-Takagi, M. Arabidopsis SBP-box genes SPL10, SPL11 and SPL2 control morphological change in association with shoot maturation in the reproductive phase. Plant Cell Physiol. 2009, 50, 2133–2145. [Google Scholar] [CrossRef] [PubMed]
- Nodine, M.D.; Bartel, D.P. MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Genes Dev. 2010, 24, 2678–2692. [Google Scholar] [CrossRef] [PubMed]
- Xing, S.; Salinas, M.; Hohmann, S.; Berndtgen, R.; Huijser, P. miR156-targeted and nontargeted SBP-box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell 2010, 22, 3935–3950. [Google Scholar] [CrossRef] [PubMed]
- Xing, S.; Salinas, M.; Garcia-Molina, A.; Hohmann, S.; Berndtgen, R.; Huijser, P. SPL8 and miR156-targeted SPL genes redundantly regulate Arabidopsis gynoecium differential patterning. Plant J. 2013, 75, 566–577. [Google Scholar] [CrossRef] [PubMed]
- Kropat, J.; Tottey, S.; Birkenbihl, R.P.; Depege, N.; Huijser, P.; Merchant, S. A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc. Natl. Acad. Sci. USA 2005, 102, 18730–18735. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, H.; Hayashi, M.; Fukazawa, M.; Kobayashi, Y.; Shikanai, T. SQUAMOSA promoter binding protein-like7 is a central regulator for copper homeostasis in Arabidopsis. Plant Cell 2009, 21, 347–361. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Poethig, R.S. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 2006, 133, 3539–3547. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, S.; Grande, A.V.; Bujdoso, N.; Saedler, H.; Huijser, P. The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol. Biol. 2008, 67, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.W.; Czech, B.; Weigel, D. MiR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 2009, 138, 738–749. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, A.; Wu, M.F.; Yang, L.; Wu, G.; Poethig, R.S.; Wagner, D. The microRNA-regulated SBP-box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev. Cell 2009, 17, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Stone, J.M.; Liang, X.; Nekl, E.R.; Stiers, J.J. Arabidopsis AtSPL14, a plant-specific SBP-domain transcription factor, participates in plant development and sensitivity to fumonisin B1. Plant J. 2005, 41, 744–754. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Nussbaum-Wagler, T.; Li, B.; Zhao, Q.; Vigouroux, Y.; Faller, M.; Bomblies, K.; Lukens, L.; Doebley, J.F. The origin of the naked grains of maize. Nature 2005, 436, 714–719. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Cheng, X.; Liu, P.; Sun, J. miR156-regulated TaSPLs interact with TaD53 to regulate TaTB1 and TaBA1 expression in bread wheat. Plant Physiol. 2017, 174, 1931–1948. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Xu, W.; Liu, X.; Mao, X.; Li, A.; Wang, J.; Chang, X.; Zhang, X.; Jing, R. Functional conservation and divergence among homoeologs of TaSPL20 and TaSPL21, two SBP-box genes governing yield-related traits in hexaploid wheat. Plant Physiol. 2017, 174, 1177–1191. [Google Scholar] [CrossRef] [PubMed]
- Gou, J.; Fu, C.; Liu, S.; Tang, C.; Debnath, S.; Flanagan, A.; Ge, Y.; Tang, Y.; Jiang, Q.; Larson, P.R.; Wen, J.; Wang, Z.Y. The miR156-SPL4 module predominantly regulates aerial axillary bud formation and controls shoot architecture. New Phytol. 2017, 216, 829–840. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Wang, Y.; Xue, D.; Wang, J.; Yan, M.; Liu, G.; Dong, G.; Zeng, D.; Lu, Z.; Zhu, X.; et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 2010, 42, 541–544. [Google Scholar] [CrossRef] [PubMed]
- Miura, K.; Ikeda, M.; Matsubara, A.; Song, X.J.; Ito, M.; Asano, K.; Matsuoka, M.; Kitano, H.; Ashikari, M. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 2010, 42, 545–549. [Google Scholar] [CrossRef] [PubMed]
- Givnish, T.J.; Barfuss, M.H.; Van Ee, B.; Riina, R.; Schulte, K.; Horres, R.; Gonsiska, P.A.; Jabaily, R.S.; Crayn, D.M.; Smith, J.A.; et al. Phylogeny, adaptive radiation, and historical biogeography in Bromeliaceae: Insights from an eight-locus plastid phylogeny. Am. J. Bot. 2011, 98, 872–895. [Google Scholar] [CrossRef] [PubMed]
- Zanella, C.M.; Janke, A.; Palma-Silva, C.; Kaltchuk-Santos, E.; Pinheiro, F.G.; Paggi, G.M.; Soares, L.E.; Goetze, M.; Buttow, M.V.; Bered, F. Genetics, evolution and conservation of Bromeliaceae. Genet. Mol. Biol. 2012, 35, 1020–1026. [Google Scholar] [CrossRef] [PubMed]
- Bartholomew, D.P.; Paull, R.E.; Rohrbach, K.G. The Pineapple-Botany, Production and Uses; CABI Publishing: Wallingford, UK, 2003; p. 176. [Google Scholar]
- Trusov, Y.; Botella, J.R. Silencing of the ACC synthase gene ACACS2 causes delayed flowering in pineapple [Ananas comosus (L.) Merr.]. J. Exp. Bot. 2006, 57, 3953–3960. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.; Duan, J.; Xie, J.; Wei, C.; Liu, Y.; Liu, S.; Sun, G. Isolation and characterization of a FLOWERING LOCUS T homolog from pineapple (Ananas comosus (L.) Merr). Gene 2012, 505, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.L.; Duan, J.; Xie, J.H.; Liu, Y.G.; Wei, C.B.; Liu, S.H.; Zhang, J.X.; Sun, G.M. Cloning and expression analysis of a PISTILLATA homologous gene from pineapple (Ananas comosus L. Merr). Int. J. Mol. Sci. 2012, 13, 1039–1053. [Google Scholar] [CrossRef] [PubMed]
- Lei, M.; Li, Z.Y.; Wang, J.B.; Fu, Y.L.; Ao, M.F.; Xu, L. AfAP2-1, An age-dependent gene of Aechmea fasciata, responds to exogenous ethylene treatment. Int. J. Mol. Sci. 2016, 17, 303. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Hou, H.; Li, X.; Xiang, J.; Yin, X.; Gao, H.; Zheng, Y.; Bassett, C.L.; Wang, X. Genome-wide identification and analysis of the SBP-box family genes in apple (Malus x domestica Borkh.). Plant Physiol. Biochem. 2013, 70, 100–114. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Int. Conf. Intell. Syst. Mol. Biol. 1994, 2, 28–36. [Google Scholar]
- Xie, K.; Wu, C.; Xiong, L. Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol. 2006, 142, 280–293. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, J.; Zhang, X.; Lei, M.; Fu, Y.; Zhang, J.; Wang, Z.; Xu, L. Transcriptome sequencing determined flowering pathway genes in Aechmea fasciata treated with ethylene. J. Plant Growth Regul. 2016, 35, 316–329. [Google Scholar] [CrossRef]
- Salinas, M.; Xing, S.; Hohmann, S.; Berndtgen, R.; Huijser, P. Genomic organization, phylogenetic comparison and differential expression of the SBP-box family of transcription factors in tomato. Planta 2012, 235, 1171–1184. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Liu, X.; Zhao, G.; Mao, X.; Li, A.; Jing, R. Molecular characterization and expression analysis of Triticum aestivum squamosa-promoter binding protein-box genes involved in ear development. J. Integr. Plant Biol. 2013, 56, 571–581. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Ling, L. Genome-wide identification and evolutionary analysis of the SBP-box gene family in castor bean. PLoS ONE 2014, 9, e86688. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Sun, L.; Zhou, Y.; Yang, W.; Cheng, T.; Wang, J.; Zhang, Q. Identifiation and expression analysis of the SQUAMOSA promoter-binding protein (SBP)-box gene family in Prunus mume. Mol. Genet. Genom. 2015, 290, 1701–1715. [Google Scholar] [CrossRef] [PubMed]
- Shalom, L.; Shlizerman, L.; Zur, N.; Doron-Faigenboim, A.; Blumwald, E.; Sadka, A. Molecular characterization of SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) gene family from Citrus and the effect of fruit load on their expression. Front. Plant Sci. 2015, 6, 389. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.X.; Jin, J.H.; He, Y.M.; Lu, B.Y.; Li, D.W.; Chai, W.G.; Khan, A.; Gong, Z.H. Genome-wide identification and analysis of the SBP-box family genes under Phytophthora capsici stress in pepper (Capsicum annuum L.). Front. Plant Sci. 2016, 7, 504. [Google Scholar] [CrossRef] [PubMed]
- Preston, J.C.; Jorgensen, S.A.; Orozco, R.; Hileman, L.C. Paralogous SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes differentially regulate leaf initiation and reproductive phase change in petunia. Planta 2016, 243, 429–440. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Hao, M.; Wang, W.; Mei, D.; Tong, C.; Wang, H.; Liu, J.; Fu, L.; Hu, Q. Genomic identification, characterization and differential expression analysis of SBP-box gene family in Brassica napus. BMC Plant Biol. 2016, 16, 196. [Google Scholar] [CrossRef] [PubMed]
- Song, A.; Gao, T.; Wu, D.; Xin, J.; Chen, S.; Guan, Z.; Wang, H.; Jin, L.; Chen, F. Transcriptome-wide identification and expression analysis of chrysanthemum SBP-like transcription factors. Plant Physiol. Biochem. 2016, 102, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.H.; Wu, Q.S.; Huang, X.; Liu, S.H.; Zhang, H.N.; Zhang, Z.; Sun, G.M. Molecular cloning and characterization of four genes encoding ethylene receptors associated with pineapple (Ananas comosus L.) flowering. Front. Plant Sci. 2016, 7, 710. [Google Scholar] [CrossRef] [PubMed]
- Bi, F.; Meng, X.; Ma, C.; Yi, G. Identification of miRNAs involved in fruit ripening in Cavendish bananas by deep sequencing. BMC Genom. 2015, 16, 776. [Google Scholar] [CrossRef] [PubMed]
- Srikanth, A.; Schmid, M. Regulation of flowering time: All roads lead to Rome. Cell. Mol. Life Sci. 2011, 68, 2013–2037. [Google Scholar] [CrossRef] [PubMed]
- Fornara, F.; de Montaigu, A.; Coupland, G. SnapShot: Control of flowering in Arabidopsis. Cell 2010, 141, 550. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.H.; Seo, Y.H.; Seo, P.J.; Reyes, J.L.; Yun, J.; Chua, N.H.; Park, C.M. The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 2007, 19, 2736–2748. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Park, M.Y.; Conway, S.R.; Wang, J.W.; Weigel, D.; Poethig, R.S. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 2009, 138, 750–759. [Google Scholar] [CrossRef] [PubMed]
- Hyun, Y.; Richter, R.; Vincent, C.; Martinez-Gallegos, R.; Porri, A.; Coupland, G. Multi-layered regulation of SPL15 and cooperation with SOC1 integrate endogenous flowering pathways at the Arabidopsis shoot meristem. Dev. Cell 2016, 37, 254–266. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.H.; Lee, H.J.; Ryu, J.Y.; Park, C.M. SPL3/4/5 integrate developmental aging and photoperiodic signals into the FT-FD module in Arabidopsis flowering. Mol. Plant 2016, 9, 1647–1659. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Li, W.; Miura, K.; Ashikari, M.; Kyozuka, J. Control of tiller growth of rice by OsSPL14 and Strigolactones, which work in two independent pathways. Plant Cell Physiol. 2012, 53, 1793–1801. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Ma, C.; Xu, Y.; Wang, T.; Chen, Y.; Lu, J.; Zhang, L.; Jiang, C.Z.; Hong, B.; Gao, J. Control of chrysanthemum flowering through integration with an aging pathway. Nat. Commun. 2017, 8, 829. [Google Scholar] [CrossRef] [PubMed]
- Ling, L.Z.; Zhang, S.D. Exploring the evolutionary differences of SBP-box genes targeted by miR156 and miR529 in plants. Genetica 2012, 140, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wu, K.; Yuan, Q.; Liu, X.; Liu, Z.; Lin, X.; Zeng, R.; Zhu, H.; Dong, G.; Qian, Q.; et al. Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 2012, 44, 950–954. [Google Scholar] [CrossRef] [PubMed]
- Chuck, G.S.; Brown, P.J.; Meeley, R.; Hake, S. Maize SBP-box transcription factors unbranched2 and unbranched3 affect yield traits by regulating the rate of lateral primordia initiation. Proc. Natl. Acad. Sci. USA 2014, 111, 18775–18780. [Google Scholar] [CrossRef] [PubMed]
- Si, L.; Chen, J.; Huang, X.; Gong, H.; Luo, J.; Hou, Q.; Zhou, T.; Lu, T.; Zhu, J.; Shangguan, Y.; et al. OsSPL13 controls grain size in cultivated rice. Nat. Genet. 2016, 48, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Holsters, M.; de Waele, D.; Depicker, A.; Messens, E.; van Montagu, M.; Schell, J. Transfection and transformation of Agrobacterium tumefaciens. Mol. Genet. Genom. 1978, 163, 181–187. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lei, M.; Li, Z.-y.; Wang, J.-b.; Fu, Y.-l.; Ao, M.-f.; Xu, L. Constitutive Expression of Aechmea fasciata SPL14 (AfSPL14) Accelerates Flowering and Changes the Plant Architecture in Arabidopsis. Int. J. Mol. Sci. 2018, 19, 2085. https://doi.org/10.3390/ijms19072085
Lei M, Li Z-y, Wang J-b, Fu Y-l, Ao M-f, Xu L. Constitutive Expression of Aechmea fasciata SPL14 (AfSPL14) Accelerates Flowering and Changes the Plant Architecture in Arabidopsis. International Journal of Molecular Sciences. 2018; 19(7):2085. https://doi.org/10.3390/ijms19072085
Chicago/Turabian StyleLei, Ming, Zhi-ying Li, Jia-bin Wang, Yun-liu Fu, Meng-fei Ao, and Li Xu. 2018. "Constitutive Expression of Aechmea fasciata SPL14 (AfSPL14) Accelerates Flowering and Changes the Plant Architecture in Arabidopsis" International Journal of Molecular Sciences 19, no. 7: 2085. https://doi.org/10.3390/ijms19072085
APA StyleLei, M., Li, Z.-y., Wang, J.-b., Fu, Y.-l., Ao, M.-f., & Xu, L. (2018). Constitutive Expression of Aechmea fasciata SPL14 (AfSPL14) Accelerates Flowering and Changes the Plant Architecture in Arabidopsis. International Journal of Molecular Sciences, 19(7), 2085. https://doi.org/10.3390/ijms19072085



